Abstract
Members of the gammaproteobacterial clade NOR5/OM60 regularly form an abundant part, up to 11%, of the bacterioplankton community in coastal systems during the summer months. Here, we report the nearly complete genome sequence of one cultured representative, Congregibacter litoralis strain KT71, isolated from North Sea surface water. Unexpectedly, a complete photosynthesis superoperon, including genes for accessory pigments, was discovered. It has a high sequence similarity to BAC clones from Monterey Bay [Beja O, Suzuki MT, Heidelberg JF, Nelson WC, Preston CM, et al. (2002) Nature 415:630–633], which also share a nearly identical gene arrangement. Although cultures of KT71 show no obvious pigmentation, bacteriochlorophyll a and spirilloxanthin-like carotenoids could be detected by HPLC analysis in cell extracts. The presence of two potential BLUF (blue light using flavin adenine dinucleotide sensors), one of which was found adjacent to the photosynthesis operon in the genome, indicates a light- and redox-dependent regulation of gene expression. Like other aerobic anoxygenic phototrophs (AAnPs), KT71 is able to grow neither anaerobically nor photoautotrophically. Cultivation experiments and genomic evidence show that KT71 needs organic substrates like carboxylic acids, oligopeptides, or fatty acids for growth. The strain grows optimally under microaerobic conditions and actively places itself in a zone of ≈10% oxygen saturation. The genome analysis of C. litoralis strain KT71 identifies the gammaproteobacterial marine AAnPs, postulated based on BAC sequences, as members of the NOR5/OM60 clade. KT71 enables future experiments investigating the importance of this group of gammaproteobacterial AAnPs in coastal environments.
Keywords: marine bacteria, NOR5/OM60 clade, whole genome analysis, Congregibacter litoralis, bacteriochlorophyll a
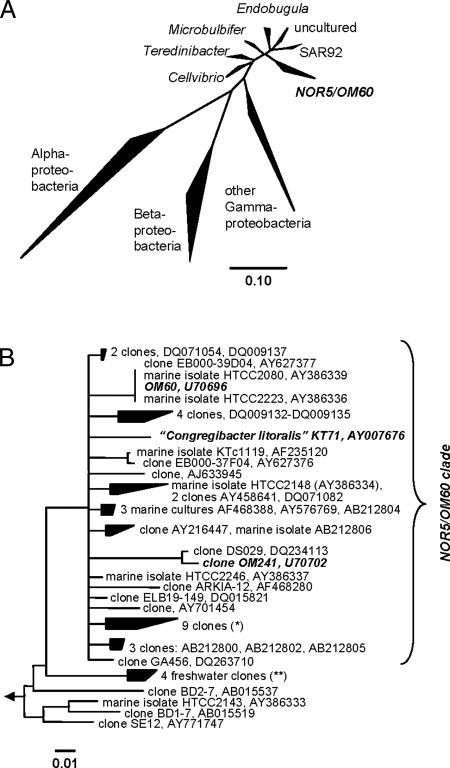
In 1999, Eilers et al. (1) isolated a bacterial strain designated KT71 from a surface water sample taken near the North Sea island Helgoland, by direct plating on complex low-nutrient media. Phylogenetic analysis showed that KT71 was the first cultured representative of a cosmopolitan gammaproteobacterial lineage, which we in the following refer to as the NOR5/OM60 clade (Fig. 1). The first indication for this clade dates back to 1997, when Rappe et al. retrieved two 16S rRNA clones, OM60 and OM241, from the continental shelf off Cape Hatteras, NC (2). In the following years, many sequences have been retrieved that were related to the clone OM60 (e.g., refs. 3–8). By the end of 2005, >180 partial and full length 16S rRNA sequences available within the public databases were related to KT71 and OM60.
Fig. 1.
Phylogenetic affiliation of KT71. (A) Parsimony tree of the NOR5/OM60 clade including representative neighboring clades within the Gammaproteobacteria. (B) Consensus tree of the NOR5/OM60 clade reconstructed with 86 almost-full-length sequences (>1,350 nt). All treeing methods and filters resolved a stable branching order for the NOR5/OM60 clade within the Gammaproteobacteria. Within the NOR5/OM60 clade, the branching order could not be unambiguously resolved based on the currently available dataset, which is indicated by a multifurcation (73). (∗), AY212565, DQ015838, DQ015860, DQ015829, DQ015807, DQ015840, AY135664, AY135666, and AY135673; (∗∗), AY212617, AY212664, AY693815, and AY212676.
KT71 is a Gram-negative, pleomorphic, strictly aerobic, and motile bacterium. It is of an average size of 2 × 0.5 μm, has a generation time of 4.5 h, and often grows in flocs. Based on this conspicuous trait and the site of isolation, the name Congregibacter litoralis has been proposed. A full taxonomic description of strain KT71 is currently ongoing (B.M.F., S.S., and R.A., unpublished work). Several more strains belonging to the NOR5/OM60 clade were isolated off the coast of Oregon, in sterilized seawater, using a high-throughput dilution-to-extinction technique (9, 10). Meanwhile, representatives of the NOR5/OM60 clade were also isolated from Arctic sea ice (11) and coastal sediments (12, 13).
FISH with rRNA-targeted oligonucleotide probes for NOR5/OM60 confirmed this clade as an abundant component of the bacterioplankton community in the North Sea around the island Helgoland (1). By the end of July 1998, up to 8% of the total bacterioplankton community comprised members of the NOR5/OM60 clade (1). A second peak of NOR5/OM60 cells was visible in mid-June (6%). However, NOR5/OM60 was not detected by FISH during the winter months, October to March, suggesting a marked seasonality. The fraction of DNA-synthesizing NOR5/OM60 cells seems to be quite variable. Active DNA synthesis could be detected in August but not in May 2002, even though NOR5/OM60 was present in high numbers in both samples (6% and 11% of total bacterioplankton cell, respectively) (14).
In 2004, KT71 was selected for whole-genome sequencing as part of the Gordon and Betty Moore Foundation (GBMF) Marine Microbiology Initiative. Here, we present data derived from the analysis of the genome of strain KT71 and from ecophysiological experiments addressing some of the predictions derived from genome annotation.
Results and Discussion
Structure and Phylogenetic Analysis of the Photosynthesis (PS) Operon.
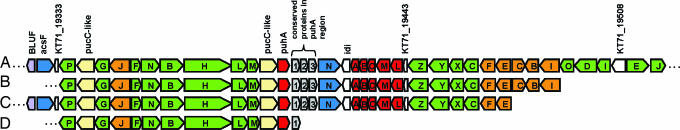
The genome annotation of KT71 revealed the presence of a full PS superoperon (15) (KT71_19323–19518) on the smaller of the two large scaffolds [supporting information (SI) Fig. 4]. Both a smooth tetranucleotide signature (SI Fig. 5) (16) and the absence of transposons in the vicinity make it highly unlikely that the PS operon has been obtained by a recent lateral gene transfer. The operon consists of the typical subclusters crtEF-bchCXYZ-puf and bchFNBHLM (Fig. 2) but differs in the global and local arrangement from cultured alpha- and betaproteobacterial anoxygenic phototrophs (17). In these, the puf gene cluster coding for the light-harvesting complex I (LHC I) and the photosynthetic reaction center are usually arranged in the order of pufBA-LMC (18). In KT71, it is switched to pufLMC-BA. Interestingly, this gene arrangement is identical to that in two BAC clones, EBAC65D09 (AE008919) and EBAC29C02 (AE008920), retrieved from coastal bacterioplankton sampled at Moss Landing, CA (Fig. 2) (17). Further analysis also revealed identical arrangement for the crt and bch genes on both the BAC clones and KT71. A third BAC clone (EBAC69B03, GenBank accession no. AY458648) shares an identical arrangement of the bchP-pucC-bchG-crtJ-bchFNBHLM-pucC-puhH region with KT71. Based on comparative sequence analyses, these BAC clones were postulated to originate from Gammaproteobacteria (17). A gene-by-gene comparison of the BAC clones EBAC65D09, EBAC29C02, and EBAC69B03 with KT71 showed a high average sequence identity on the amino acid level of 56%, 55%, and 62%, respectively (Fig. 2; SI Table 1). We conclude that the three BAC clones 65D09, 69B03, and 29C02 indeed originate from Gammaproteobacteria, more precisely from members of the NOR5/OM60 clade.
Fig. 2.
Comparison of PS operons. (A) KT71. (B–D) BAC clones EBAC65D09 (B), EBAC29C02 (C), and EBAC69B03 (D). Green, bch genes; red, puf gene; orange, crt genes; blue, hem genes; 1, similar to 23.7-kDa protein (KT71_19398); 2, similar to 17.4-kDa protein (KT71_19403); 3, similar to 30.4-kDa protein (KT71_19408). Dots indicate the presence of genes not belonging to the PS operon.
Genes coding for a LHC II and a pucC-like transcriptional regulator were found clustered together on scaffold 1 (pucBAC: KT71_03072, KT71_03077, and KT71_03082). The LHC II complex proteins were most closely related to those of Rhodopseudomonas palustris (65% and 69% sequence identity in amino acids; best blast hit).
Pigment Analysis.
Cell extracts of KT71 were subjected to spectrochromatographic analysis. Pigments found after consecutive acetone and methanol extractions followed by HPLC analysis showed the typical profiles for bacteriochlorophyll a (Bchla) with the main peaks at 360, 580, and 776 nm (Fig. 3A) and a carotenoid-like compound with absorption maxima at 470, 494, and 532 nm, respectively (Fig. 3B). The latter absorption maxima are almost identical to those described for spirilloxanthin detected in Roseateles depolymerans (19). Significant amounts of Bchla were detected only in cultures of KT71 growing with light on the oligotrophic MPM-m (1) medium for an extended time (bacteriochlorophyll concentration of 680 μg·liter−1 after 4 mo). In contrast, Bchla was never found in cultures grown to stationary phase in the nutrient-rich SYPG medium or in MPM-m medium without illumination. Genomic analysis showed that indeed all genes for the synthesis of spirilloxanthin are present. They are clustered together in the PS operon (crtJ and cluster crtFECBI), except for the crtD gene coding for a methoxyneurosporene dehydrogenase, which is found 300 kb separated on the first scaffold (KT71_07854).
Fig. 3.
Pigment analysis. (A) Absorption spectrum of Bchla extracted from KT71. Retention time, 15.16 min. (B) Absorption spectrum of spirilloxanthin-like carotenoid from KT71. Retention time, 17.33 min. Note: This curve was fitted (thick line) to better visualize the characteristic peaks.
Photoautotrophy vs. Photoheterotrophy.
KT71 has all the components necessary for a fully functional photosystem typical of anoxygenic phototrophs: the LHC I and II, a reaction center, and carotenoid pigments. Physiological tests indicated that KT71 is not able to grow autotrophically. None of the key genes for autotropic carbon fixation, like ribulose-1,5-bisphosphate-carboxylase/oxygenase (Calvin-cycle), ATP-citrate-lyase (reductive citrate cycle), or CO-dehydrogenase/acetyl-CoA-synthase (reductive acetyl-CoA pathway), were found in the genome, which is typical for aerobic anoxygenic phototrophs (AAnPs) (20). Most likely, KT71 is able to gain energy from light by a light-dependent cyclic electron transport through the photosystem and the generation of a proton gradient (20). A proton-driven ATP synthase complex was annotated (KT71_04845–04885). Alternatively, the proton gradient might be converted into a sodium gradient by proton/sodium antiporters, five of which have been found in the KT71 genome (e.g., KT71_06212 and KT71_09322). The sodium gradient in turn may drive a sodium-dependent ATPase (KT71_09367) or may be directly used by the flagella motor (KT71_00645).
First-growth experiments with KT71 suggest an enhanced cell yield with light. Two flasks containing 960 ml of minimal MPM-m medium were inoculated with 4 ml of a stationary-phase culture and incubated for 4 wk at room temperature. From the culture grown with light from a 60-W light bulb, 32.4 mg of cell mass (dry weight) could be harvested, whereas from the parallel culture grown in the dark, only 17.6 mg of cell mass (dry weight) could be obtained. These experiments have to be regarded as preliminary, because no parallel experiments were done. Future experiments should also address starvation survival, because Breznak et al. (21) could show that the survival half-time of the facultative anaerobic anoxygenic phototroph Rhodospirillum rubrum (Alphaproteobacteria) was ≈29-fold longer if grown with ambient-light intensities than without light.
Putative Regulation of PS.
Annotation identified two genes containing a member of the sensor family BLUF (blue light using flavin adenine dinucleotide). One of the BLUF sensors was detected directly upstream of the PS superoperon. It contains the BLUF domain at the N terminus of the ORF KT71_19323. In Rhodobacter sphaeroides BLUF forms part of the AppA protein, which regulates the expression of the PS cluster by sensing and integrating both the light and redox regimes (22). BAC clone EBAC29C02 also contains a BLUF sensor with a similar structure directly upstream of the PS operon suggesting an involvement of the BLUF sensor in the light regulation of the PS operon. Interestingly, in direct vicinity to the second BLUF sensor (KT71_09447), a two component response regulator (KT71_09452) could be found, suggesting, that this BLUF sensor forms part of a two component system (SI Table 2).
Microaerophily.
KT71 is a strictly aerobic organism with a clear preference for low-oxygen niches. Typical enzymes necessary for the detoxification of oxygen, a bifunctional catalase-peroxidase (KT71_02962) and a superoxide dismutase (KT71_19732), could be annotated in the genome. It did not grow with nitrate as sole electron acceptor, nor was it able to ferment. No gene encoding a dissimilatory nitrate reductase was found in the genome. A putative sulfite/nitrite reductase-like enzyme (KT71_15541) was annotated but is most likely involved in the assimilatory nitrate or sulfate reduction.
In deep-agar cultures, KT71 forms distinct bands several millimeters below the surface. The position of the visible cell layer depended on the substrate concentration in the medium and was closer to the surface at higher substrate concentrations. To determine the exact oxygen concentration for optimal growth of KT71, oxygen profiles were measured in cultures grown in SYPG medium with 0.15% (wt/vol) agar (soft agar). An oxygen profile measured with microsensors from the surface of the soft agar down to a depth of 8 mm is shown in SI Fig. 6. The highest cell density was visible at an oxygen saturation of ≈10% (30 μM O2). Experiments with varying substrate concentrations showed that KT71 exhibits an excellent chemotaxis for suboxic oxygen conditions. KT71 is motile and possesses a complete flagellum operon (gene loci KT71_00565–00780). Next to the aa3-type terminal cytochrome c oxidase (KT71_04625–04640), KT71 harbors a cbb3-type cytochrome c oxidase (fixNOQP, KT71_16991–17006). Such terminal cytochrome c oxidases with high oxygen affinity are expressed only under reduced oxygen conditions in Bradyrhizobium japonicum (23). In R. sphaeroides, the same enzyme complex was shown to be involved in the signal transduction and functions as a redox sensor (24) (SI Table 2), which might be also the case in KT71.
Substrate Spectrum.
Substrate utilization tests indicate that KT71 prefers complex substrate mixture for growth (e.g., yeast extract or Trypticase peptone), whereas many monomeric substrates given as sole source of carbon and energy are used a little or not at all. As an exception, KT71 can grow well with carbon sources such as glutamate, pyruvate, and fatty acids, most likely due to the fact they play central roles in the metabolism of this organism. Glutamate is a central metabolite and is presumably taken up by a proton/sodium-glutamate symport protein (KT71_01885). It is further fed by two glutamate dehydrogenases into the trichloroacetic acid cycle (KT71_16246 and KT71_18661) or into the proline synthesis pathway (glutamate-5-kinase, KT71_02697). Pyruvate is presumably being metabolized by a pyruvate-dehydrogenase (KT71_00115) and further metabolized by the citric acid cycle.
Annotation identified all genes necessary to perform the complete pentose phosphate pathway. This pathway plays a central role in the anabolism of nucleotides and amino acids as well as the generation of reducing power by NADPH synthesis. Laboratory experiments have shown that KT71 is not able to use glucose as sole source of carbon and energy. Alonso and Pernthaler (25) could not detect any glucose uptake of NOR5/OM60 in situ under both oxic and anoxic conditions in the North Sea for the entire NOR5/OM60 clade. In the genome, all genes for glycolysis are present, except for the initial activating enzymes. Neither a glucose phosphorylating glucokinase nor an intact phosphotransferase system (PTS) was found. For the latter, only the specific phosphorcarrier HPr (KT71_10197) and a single-chain EIIA of the PTS could be annotated (KT71_10207).
The genome contains several genes coding for putative lipase/esterases and proteases/peptidases that might be involved in the breakdown of lipids and peptides. In the laboratory, no hydrolysis of the polysaccharides starch, cellulose, or chitin by KT71 could be detected, in line with the annotation of the genome. A lipase/esterase activity could be confirmed by the hydrolysis of the artificial substrates Tween 80 (Polyoxyethylenesorbitan monooleate) and Tween 20 (Polyoxyethylenesorbitan monolaurate). Gelatin and casein were tested negatively as potential substrates for proteases and peptidases. Although proteases can have a high specificity for distinct substrates, this finding points to a preferred utilization of oligopeptides or partly degraded proteins by KT71. Two transporters for oligopeptides with up to five amino acids were found, oppABC (KT71_06839–06854) and oppF (KT71_00435). Culture experiments show that KT71 is able to synthesize all essential amino acids and most of the vitamins, except for biotin, thiamin, and vitamin B12. Two TonB-dependent vitamin B12 sensors (KT71_17391 and KT71_18621) and an ABC vitamin B12 transporter system btuCDF (KT71_17396–17411) were found in the genome. The annotation is consistent with this auxotrophy and the inability to use many substrates (e.g., glucose) as single sources of carbon and energy.
Storage Compounds.
Two highly similar genes coding for cyanophycin synthetases were found in tandem (KT71_18591 and KT71_18596; 38% identical amino acids). Cyanophycin synthetase is described as a homodimer but was also considered to form heterodimers of the type CphA and CphA′ (26). Both genes have high similarity to the cphA genes in the Gammaproteobacteria Colwellia psychoerythraea 34H and Francisella tularensis (59% and 56% identity for the long CphA and 33% and 30% amino acid identity for the short CphA′, respectively). Cyanophycin is a polymer of aspartic acid and arginine. It was first found as a storage compound in cyanobacteria and subsequently detected in many heterotrophic bacteria. The polymer forms insoluble granula inside the cell that can be extracted with diluted acids (27). Cells containing highly refractile granulas could be mainly observed in stationary cultures of KT71 grown under conditions of a high ratio of nitrogen to carbon. Cyanophycin was identified in these granula by a negative reaction with the lipophilic stain, Nile blue A, and dissolution in diluted HCl (see SI Fig. 7). A cyanophycinase was not annotated, but most likely the polypeptide is degraded by an unknown peptidase. The formation of polyphosphate is not yet confirmed by physiological tests but two enzymes, an inorganic polyphosphate/ATP-NAD kinase (KT71_14354) and a polyphosphate kinase (KT71_16696), were found in the genome.
Formation of Aggregates and Polysaccharide Production.
In pure cultures of KT71, the formation of large flocs was observed (SI Fig. 8A). There is microscopic evidence that members of the NOR5/OM60 clade attaches also in nature to macroscopic particles (SI Fig. 8B). Genome analyses revealed several features consistent with aggregation. Several loci in the KT71 genome code for the synthesis of type IV pili or fimbriae (28). The formation of pili seems to be regulated by a sensory mechanism encoded by the genes pilS (KT71_19657) and pilR (KT71_19662; SI Table 2). In addition, two operons were found containing exopolymer producing proteins (KT71_09752–09807 and KT71_06404–06469). These operons comprised genes for polysaccharide length-determinant proteins [KT71_09767 and KT71_06439), (exo)-polysaccharide biosynthesis protein (KT71_09772 and KT_066454), polysaccharide polymerases (KT71_009807), polysaccharide export proteins (KT71_09762 and KT71_06444)], and some glycosyltransferases (e.g., KT71_06459, KT71_06434, KT71_09787, and KT71_06429). Interestingly, each of the operons contains a two-component sensor kinases/response regulator (SI Table 2). Based on the current annotation, it is not clear to which stimuli they respond.
Sulfur Metabolism.
KT71 most likely uses the APS/PAPS pathway to obtain reduced sulfur compounds. Three genes coding for the key enzymes of that pathway were annotated in KT71, a sulfate adenylyltransferase (KT71_10572), an adenylylsulfate kinase (KT71_10567), and a phosphoadenosine phosphosulfate reductase (KT71_06329). Genome annotation revealed that the gene cluster soxH-RCDXYZA-B is potentially involved in the oxidation of reduced sulfur compounds (KT71_03447–03482 and KT71_03497). This cluster contains the core gene set soxXYZAB, which is found in many species capable of oxidizing reduced sulfur compounds (29). A comparison with other sulfur-oxidizing organisms shows that the gene arrangement soxH-RCDXYZA-B is unique to KT71 and has not been found in any of the species described to date. Unlike in Silicibacter pomeryoi (30), the supplementation of media with the inorganic sulfur compounds thiosulfate or sulfur did not significantly promote growth of KT71 in cultivation experiments using different carbon sources. The inability to gain additional energy by the oxidation of reduced inorganic sulfur compounds may be due to the lack of several sox genes compared with the exemplary cluster of sox genes found in the genome of Paracoccus pantotrophus or S. pomeroyi (30). Of special interest is the lack of the gene soxV that codes in P. pantotrophus GB17 for a membrane protein that is predicted to transfer electrons from the cytoplasma to the periplasmic thioredoxin soxW (31). It was shown that inactivation of SoxV in P. pantotrophus and the phototrophic bacterium Rhodovulum sulfidophilum leads to a phenotype that is unable to use thiosulfate for energy conservation (32, 33). Despite this finding, the possibility exists that KT71 can use alternative sulfur compounds like dimethylsulfoniopropionate or dimethylsulfide that were not tested yet. These compounds are present in high amounts after algal blooms and are metabolized by Roseobacter species (34, 35).
C. litoralis: A Typical Shelf Bacterium?
KT71 was isolated from coastal surface water in the rather shallow German Bight. There, the water column is close to oxygen saturation during most of the year. It came as a surprise that KT71 avoids sites with oxygen saturation and grows optimally under microaerobic conditions. In coastal areas, suboxic conditions are found, temporarily, in large macroscopic aggregates (36), and permanently a few millimeters below the sediment surface (37). Therefore, we hypothesize that the habitat range of KT71 includes particles and sediment surfaces.
Shallow shelf areas are characterized by extensive mixing of the water, sediment interface by tides, or wind stress. Resuspension of sediment particles into the water column is followed by periods of sedimentation. Thereby, in temperate coastal systems like the German Bight, marine microorganisms are faced with pronounced fluctuations of multiple parameters such as substrate, nutrient, and oxygen concentrations, as well as light levels on a daily and seasonal scale. Based on our genomic and ecophysiological data, KT71 seems well adapted to such a dynamic shallow shelf environment.
Organic particles are nutrient-rich hotspots in the otherwise oligotrophic water column (38). By attaching to their surfaces, KT71 may directly use mono- and oligomeric substrates or may benefit, as a commensal, from the exoenzymatic activities of polymer-degrading bacteria such as Rhodopirellula baltica (39) and Gramella forsetii (40). The possibility that KT71 is actively shaping its environment by facilitating the formation of “marine snow” by polysaccharide production needs to be addressed in future studies. Particle association also serves as a transport mechanism to the sediment surface. There, KT71 may thrive on low-molecular-weight substrates like peptides or lipids that accumulate on the sea floor (41).
The presence of a complete mercury-resistance operon (KT71_16196–16226) in the genome of KT71 is consistent with a prevalence of this strain in the suboxic zone of sediments. It is known that low-redox potentials and the degradation of complex organic matter in upper sediment layers lead to the mobilization of active mercury species in the form of inorganic ions (Hg2+) or weak inorganic complexes (see, e.g., ref. 42). In a recent study, depth profiles of reactive mercury species were determined in North Sea sediment, and it was found that peak values are reached at the sediment water interface (43). Hence, genes that confer resistance to toxic mercury ions may be much more important for bacteria dwelling in the surface sediment than for bacteria indigenous to the water column.
A specific highlight of KT71 is the presence of a PS superoperon. It is becoming more clear that photoheterotrophy is widespread among marine microorganisms (17, 44). By the light-driven generation of a proton gradient, KT71 might be able to survive extended periods of starvation, e.g., during the winter period. The storage compounds cyanophycin and polyphosphate are yet another adaptation to famine situations. Interestingly, in contrast to all other AAnPs known to date, KT71 produces only trace amounts of carotenoids and shows no obvious pigmentation (SI Fig. 8c). Because a major function of carotenoids is the protection of cells from damage by UV radiation, this may reflect an adaptation to low-light zones, i.e., depths of several meters in the water column or subsurface sediment layers in shallow water. Recently, strongly pigmented strains closely related to KT71 were isolated from surface sediments in the Wadden Sea, suggesting the ability of members of the NOR5/OM60 clade to adapt also to high light conditions (J. Harder, personal communication). These strains will also allow us to determine whether PS is a general feature of the NOR5/OM60 clade.
Significance of the NOR5/OM60 Clade Represented by KT71.
It has been estimated that AAnPs account for ≥10% of the bacterioplankton community in the oligotrophic open ocean (45–47). More recent studies have shown that AAnPs may be less important in the open ocean (≈1%) (48) but can reach up to 15% abundance in eutrophic and mesotrophic coastal areas (49, 50). Currently, the alphaproteobacterial Roseobacter clade is considered to be the dominant group of marine AAnPs (35, 51, 52). The discovery of BAC clones with PS operons showing best BLAST hits to Gammaproteobacteria (17) clearly suggested the existence of a second group of AAnPs. The genome analysis of C. litoralis strain KT71 identifies this microorganism as a cultured representative of the gammaproteobacterial marine AAnPs, enabling future experiments investigating the importance of gammaproteobacterial AAnPs in coastal environments by using KT71 as a model organism.
Materials and Methods
Sequencing and Assembly.
Sequencing of KT71 was done in a conventional whole-genome shotgun sequencing approach. Two genomic libraries with insert sizes of 4 and 40 kb were made as described in Goldberg et al. (53). The prepared plasmid and fosmid clones were sequenced from both ends to provide paired-end reads at the J. Craig Venter Science Foundation Joint Technology Center on Applied Biosystems 3730XL DNA sequencers (Applied Biosystems, Foster City, CA). Whole-genome random shotgun sequencing produced 38,544 good reads averaging 892 bp in length, for a total of ≈34.38 Mbp of microbial DNA sequence (=7.53 × coverage).
The genome of KT71 contains ≈4.36 Mb with an average GC content of 57.7% and is deposited under GenBank accession no. AAOA00000000. Successful reads for each organism were used as input for the Celera Assembler (54). Data are released to the GBMF Marine Microbial Genome Sequencing Project web site (https://research.venterinstitute.org/moore) and GenBank. A genome report compliant with the “Minimum Information about a Genome Sequence specification” is available from the Genome Catalog at www.genomics.ceh.ac.uk/genomecatalogue.
Analysis of the Genome Architecture.
The cumulative GC-skew ([G−C]/[G+C]) was computed with a custom PERL script (SI Fig. 9). Genome-wide fluctuations of the GC-skew, AT-skew, GC-content, DNA curvature and DNA bending, (SI Fig. 4) were computed with custom PERL scripts and the programs banana and btwisted from the EMBOSS suite (55), respectively, and visualized with GeneWiz (56). The positions of all genes and subsets of related genes (SI Fig. 4) were visualized with the program GenomePlot (57). Genome-wide fluctuations in tetranucleotide composition (SI Fig. 5) were calculated and plotted with the program TETRA (16).
Gene Prediction and Annotation of the Genome Sequence.
Potential protein-coding genes were identified by the Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP) of the National Center for Biotechnology Information. PGAAP combines three distinct gene finders, GenMark (58), GenMark.hmm (59), and Glimmer2 (60), and parses the individual results using a conflict-resolving strategy. BLAST analysis of the intergenic regions resulted in finding one additional short ORF (pufB, KT71_19424). Transfer RNA genes were identified by using tRNAScan-SE (61), and ribosomal RNA genes were detected by standard BLAST similarity searches (62) against public nucleotide databases.
Annotation of the genome sequence was performed with the GenDB v2.2 annotation system (63). For each ORF, similarity searches against various sequence databases (NCBI nr, NCBI nt, and SwissProt) and protein family databases (Pfam, Prosite, InterPro, and COG) were performed. In addition, potential signal peptides were predicted with SignalP Ver. 2.0 (64), and potential transmembrane helices were predicted with TMHMM Ver. 2.0 (65). Based on this information, all ORFs were automatically annotated in a fuzzy logic-based approach (66). This automatic annotation was extensively manually checked and refined for each ORF. Predicted protein-coding genes were functionally classified according to COG Ver. 3 (67) (see SI Text).
Comparative Sequence Analyses.
16S rRNA gene sequences related to KT71 were downloaded from the National Center for Biotechnology Information and imported into a 16S rRNA database. Phylogenetic reconstructions were done with the ARB package (www.arb-home.de) (68) by using maximum parsimony, maximum likelihood, and neighbor-joining methods with different filters and matrices (see SI Text).
Physiological Tests.
KT71 was routinely grown and maintained either in the oligotrophic MPM-m medium described by ref. 1 (see SI Tables 3–7 for details) or in the complex medium SYPG containing the following compounds per liter of distilled water: 35.0 g of sea salts, 0.5 g of yeast extract, 0.25 g of Trypticase peptone, and 0.1 g of sodium l-glutamate. Utilization of substrates was tested in a mineral medium containing (per liter) 35.0 g of sea salts, 0.1 g of NH4Cl, 0.05 g of K2HPO4, and 10 ml of a vitamin solution (see DSMZ medium 141, www.dsmz.de). Standard tests for the detection of enzymes like catalase, oxidase, lipase/esterase, and proteases were done according to the protocols given in ref. 69. Oxygen measurements in soft-agar medium were done with a Clark-type oxygen microelectrode, as described (70).
HPLC Analysis of Photosynthetic Pigments.
Pigments from cell pellets of KT71 were obtained by freeze drying and consecutive acetone and methanol extraction with sonication. HPLC analyses of cell extracts were done on a Waters 2690 Separation Modul (Waters, Eschborn, Germany) with a 250 × 4.6-mm vortex column packed with Eurospher-100 C 18 (particle size, 5 μm; Knauer, Berlin, Germany) and a Waters 996 Photodiode Array Detector. The mobile phase was chosen after Wright and Jeffrey (71). Bchla was identified by retention-time coinjection of a Bchla standard from Rhodopseudomonas sphaeroides (Sigma–Aldrich, Taufkirchen, Germany) and spectrography at 384 nm. The quantity of Bchla was estimated from the areas under the peaks, which were calibrated with the Bchla standard. No standard was available for the spirilloxanthin-like compound, but the carotenoid was identified by comparison with similar spectra at similar retention times from, e.g., ref. 72.
Supplementary Material
Acknowledgments
Jakob Pernthaler initially suggested sequencing KT71 in the framework of the Marine Microbiology Sequencing Initiative of the GBMF. Sequencing, assembly, and annotation efforts were supported by the GBMF as part of its Marine Microbial Sequencing Project (www.moore.org/marinemicro). We thank Granger Sutton and his team for the ongoing development and maintenance of the Celera Assembler and related tools and Aaron Sutton for help with specific analysis issues. The JCVI software team, under the leadership of Saul A. Kravitz, manages assembly, annotation, and Web delivery of data for the GBMF-funded project (http://research.venterinstitute.org/moore). We thank Robert Friedman for his leadership of the GBMF-funded project. Many thanks for help with pigment extraction and HPLC analysis to Gabriele Klockgether and Raphaela Shoon. Armin Gieseke is acknowledged for help with the microsensor measurements. Jens Harder shared unpublished results on strongly pigmented strains closely related to KT71. We are indebted to Gunnar Gerdts and Antje Wichels from the Alfred Wegener Institute for Polar and Marine Research for constant support for field work on Helgoland. We thank Oded Beja, Margarete Schüler, Marcel Kuypers, and Anke Meyerdierks for helpful suggestions and discussions. We acknowledge the J. Craig Venter Institute (JCVI) Joint Technology Center, under the leadership of Yu-Hui Rogers, for producing the genomic libraries and the sequence data. This project was funded by the GBMF, the Max Planck Society, and the FP6-EU Network of Excellence Marine Genomics Europe (Grant GOCE-CT-2004-505403).
Abbreviations
- PS
photosynthesis
- LHC
light-harvesting complex
- Bchla
bacteriochlorophyll a
- AAnP
aerobic anoxygenic phototroph
- GBMF
Gordon and Betty Moore Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AAOA00000000).
See Commentary on page 2561.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608046104/DC1.
References
- 1.Eilers H, Pernthaler J, Peplies J, Glöckner FO, Gerdts G, Amann R. Appl Environ Microbiol. 2001;67:5134–5142. doi: 10.1128/AEM.67.11.5134-5142.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rappe MS, Kemp PF, Giovannoni SJ. Limnol Oceanogr. 1997;42/1:811–826. [Google Scholar]
- 3.Eilers H, Pernthaler J, Glöckner FO, Amann R. Appl Environ Microbiol. 2000;66:3044–3051. doi: 10.1128/aem.66.7.3044-3051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly KM, Chistoserdov AY. FEMS Microbiol Ecol. 2001;35:85–95. doi: 10.1111/j.1574-6941.2001.tb00791.x. [DOI] [PubMed] [Google Scholar]
- 5.Crump B, Armbrust E, Baross J. Appl Environ Microbiol. 1999;65:3192–3204. doi: 10.1128/aem.65.7.3192-3204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beja O, Suzuki MT, Koonin EV, Aravind L, Hadd A, Nguyen LP, Villacorta R, Amjadi M, Garrigues C, Jovanovich SB, et al. Environ Microbiol. 2000;2:516–529. doi: 10.1046/j.1462-2920.2000.00133.x. [DOI] [PubMed] [Google Scholar]
- 7.Acinas SG, Antón J, Rodríguez-Valera F. Appl Environ Microbiol. 1999;65:514–522. doi: 10.1128/aem.65.2.514-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schäfer H, Bernard L, Courties C, Lebaron P, Servais P, Pukall R, Stackebrandt E, Troussellier M, Guindulain T, Vives-Rego J, Muyzer G. FEMS Microbiol Ecol. 2001;34:243–253. doi: 10.1111/j.1574-6941.2001.tb00775.x. [DOI] [PubMed] [Google Scholar]
- 9.Connon SA, Giovannoni SJ. Appl Environ Microbiol. 2002;68:3878–3885. doi: 10.1128/AEM.68.8.3878-3885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho J-C, Giovannoni SJ. Appl Environ Microbiol. 2004;70:432–440. doi: 10.1128/AEM.70.1.432-440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brinkmeyer R, Knittel K, Jürgens J, Weyland H, Amann R, Helmke E. Appl Environ Microbiol. 2003;69:6610–6619. doi: 10.1128/AEM.69.11.6610-6619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agogue H, Casamayor EO, Bourrain M, Obernosterer I, Joux F, Herndl GJ, Lebaron P. FEMS Microbiol Ecol. 2005;54:269–280. doi: 10.1016/j.femsec.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Maeda T, Hayakawa K, You M, Sasaki M, Yamaji Y, Furushita M, Shiba T. Microb Environ. 2005;20:253–257. [Google Scholar]
- 14.Pernthaler A, Pernthaler J. Appl Environ Microbiol. 2005;71:4638–4644. doi: 10.1128/AEM.71.8.4638-4644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauer CE, Buggy JJ, Yang Z, Marrs BL. Mol Gen Genet. 1991;228:433–444. doi: 10.1007/BF00260637. [DOI] [PubMed] [Google Scholar]
- 16.Teeling H, Waldmann J, Lombardot T, Bauer M, Glöckner FO. BMC Bioinformatics. 2004;5:163. doi: 10.1186/1471-2105-5-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beja O, Suzuki MT, Heidelberg JF, Nelson WC, Preston CM, Hamada T, Eisen JA, Fraser CM, DeLong EF. Nature. 2002;415:630–633. doi: 10.1038/415630a. [DOI] [PubMed] [Google Scholar]
- 18.Yutin N, Beja O. Environ Microbiol. 2005;7:2027–2033. doi: 10.1111/j.1462-2920.2005.00843.x. [DOI] [PubMed] [Google Scholar]
- 19.Suyama T, Shigematsu T, Takaichi S, Nodasaka Y, Fujikawa S, Hosoya H, Tokiwa Y, Kanagawa T, Hanada S. Int J Syst Bacteriol. 1999;49:449–457. doi: 10.1099/00207713-49-2-449. [DOI] [PubMed] [Google Scholar]
- 20.Yurkov VV, Beatty JT. Microbiol Mol Biol Rev. 1998;62:695–724. doi: 10.1128/mmbr.62.3.695-724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breznak JA, Potrikus CJ, Pfennig N, Ensign JC. J Bacteriol. 1978;134:381–388. doi: 10.1128/jb.134.2.381-388.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braatsch S, Klug G. Photosynth Res. 2004;79:45–57. doi: 10.1023/B:PRES.0000011924.89742.f9. [DOI] [PubMed] [Google Scholar]
- 23.Preisig O, Anthamatten D, Hennecke H. Proc Natl Acad Sci USA. 1993;90:3309–3313. doi: 10.1073/pnas.90.8.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh J-I, Kaplan S. J Biol Chem. 2002;277:16220–16228. doi: 10.1074/jbc.M200198200. [DOI] [PubMed] [Google Scholar]
- 25.Alonso C, Pernthaler J. Appl Environ Microbiol. 2005;71:1709–1716. doi: 10.1128/AEM.71.4.1709-1716.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krehenbrink M, Oppermann-Sanio F-B, Steinbüchel A. Arch Microbiol. 2002;177:371–380. doi: 10.1007/s00203-001-0396-9. [DOI] [PubMed] [Google Scholar]
- 27.Elbahloul Y, Krehenbrink M, Reichelt R, Steinbüchel A. Appl Environ Microbiol. 2005;71:858–866. doi: 10.1128/AEM.71.2.858-866.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattick JS. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 29.Friedrich CG, Bardischewsky F, Rother D, Quentmeier A, Fischer J. Curr Opin Microbiol. 2005;8:253–259. doi: 10.1016/j.mib.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 30.Moran MA, Buchan A, Gonzalez JM, Heidelberg JF, Whitman WB, Kiene RP, Henriksen JR, King GM, Belas R, Fuqua C, et al. Nature. 2004;432:910–913. doi: 10.1038/nature03170. [DOI] [PubMed] [Google Scholar]
- 31.Bardischewsky F, Fischer J, Bettina H, Friedrich CG. Microbiology. 2006;152:465–472. doi: 10.1099/mic.0.28523-0. [DOI] [PubMed] [Google Scholar]
- 32.Appia-Ayme C, Berks BC. Biochem Biophys Res Commun. 2002;296:737–741. doi: 10.1016/s0006-291x(02)00936-1. [DOI] [PubMed] [Google Scholar]
- 33.Bardischewsky F, Friedrich CG. FEMS Microbiol Lett. 2001;202:215–220. doi: 10.1111/j.1574-6968.2001.tb10806.x. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez JM, Simo R, Massana R, Covert JS, Casamayor EO, Pedros-Alio C, Moran MA. Appl Environ Microbiol. 2000;66:4237–4246. doi: 10.1128/aem.66.10.4237-4246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zubkov MV, Fuchs BM, Archer SD, Kiene RP, Amann R, Burkill PH. Environ Microbiol. 2001;3:304–311. doi: 10.1046/j.1462-2920.2001.00196.x. [DOI] [PubMed] [Google Scholar]
- 36.Ploug H. Limnol Oceanogr. 2001;46:1624–1631. [Google Scholar]
- 37.de Beer D, Wenzhoefer F, Ferdelman TG, Boehme SE, Huettel M, van Beusekom JEE, Boettcher ME, Musat N, Dubilier N. Limnol Oceanogr. 2005;50:113–127. [Google Scholar]
- 38.Azam F, Long RA. Nature. 2001;414:495–498. doi: 10.1038/35107174. [DOI] [PubMed] [Google Scholar]
- 39.Glöckner FO, Kube M, Bauer M, Teeling H, Lombardot T, Ludwig W, Gade D, Beck A, Borzym K, Heitmann K, et al. Proc Natl Acad Sci USA. 2003;100:8298–8303. doi: 10.1073/pnas.1431443100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bauer M, Kube M, Teeling H, Richter M, Lombardot T, Allers E, Würdemann CA, Quast C, Kuhl H, Knaust F, et al. Environ Microbiol. 2006;8:2201–2213. doi: 10.1111/j.1462-2920.2006.01152.x. [DOI] [PubMed] [Google Scholar]
- 41.Grossart H-P, Brinkhoff T, Martens T, Duerselen C, Liebezeit G, Simon M. Limnol Oceanogr. 2004;49:2212–2222. [Google Scholar]
- 42.Ramalhosa E, Segade SR, Pereira E, Vale C, Duarte A. Water Res. 2006;40:2893–2900. doi: 10.1016/j.watres.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 43.Divis P, Leermakers M, Docekalová H, Gao Y. Anal Bioanal Chem. 2005;382:1715–1719. doi: 10.1007/s00216-005-3360-8. [DOI] [PubMed] [Google Scholar]
- 44.Beja O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich S, Gates CM, Feldman RA, Spudich JL, et al. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 45.Kolber ZS, Van Dover CL, Niederman RA, Falkowski PG. Nature. 2000;407:177–179. doi: 10.1038/35025044. [DOI] [PubMed] [Google Scholar]
- 46.Kolber ZS, Plumley FG, Lang AS, Beatty JT, Blankenship RE, VanDover CL, Vetriani C, Koblizek M, Rathgeber C, Falkowski PG. Science. 2001;292:2492–2495. doi: 10.1126/science.1059707. [DOI] [PubMed] [Google Scholar]
- 47.Cottrell MT, Mannino A, Kirchman DL. Appl Environ Microbiol. 2006;72:557–564. doi: 10.1128/AEM.72.1.557-564.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goericke R. Limnol Oceanogr. 2002;47:290–295. [Google Scholar]
- 49.Schwalbach MS, Fuhrman JA. Limnol Oceanogr. 2005;50:620–628. [Google Scholar]
- 50.Sieracki ME, Gilg IC, Thier EC, Poulton NJ, Goericke R. Limnol Oceanogr. 2006;51:38–46. [Google Scholar]
- 51.Shiba T. Syst Appl Microbiol. 1991;14:140–145. [Google Scholar]
- 52.Selje N, Simon M, Brinkhoff T. Nature. 2004;427:445–448. doi: 10.1038/nature02272. [DOI] [PubMed] [Google Scholar]
- 53.Goldberg SMD, Johnson J, Busam D, Feldblyum T, Ferriera S, Friedman R, Halpern A, Khouri H, Kravitz SA, Lauro FM, et al. Proc Natl Acad Sci USA. 2006;103:11240–11245. doi: 10.1073/pnas.0604351103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KHJ, Remington KA, et al. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 55.Rice P, Longden I, Bleasby A. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 56.Hallin PF, Binnewies TT, Ussery DW. Microbiology. 2004;150:3091–3093. doi: 10.1099/mic.0.27582-0. [DOI] [PubMed] [Google Scholar]
- 57.Gibson R, Smith DR. Bioinformatics. 2003;19:1449–1450. doi: 10.1093/bioinformatics/btg152. [DOI] [PubMed] [Google Scholar]
- 58.Borodovsky M, McIninch J. Comput Chem. 1993;17:123–133. [Google Scholar]
- 59.Lukashin A, Borodovsky M. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delcher A, Harmon D, Kasif S, White O, Salzberg S. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lowe T, Eddy S. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Altschul S, Madden T, Schaffer A, Zhang JH, Zhang Z, Miller W, Lipman D. FASEB J. 1998;12:A1326–A1326. [Google Scholar]
- 63.Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, Clausen J, Kalinowski J, Linke B, Rupp O, Giegerich R, Pühler A. Nucleic Acids Res. 2003;31:2187–2195. doi: 10.1093/nar/gkg312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nielsen H, Brunak S, von Heijne G. Protein Eng. 1999;12:3–9. doi: 10.1093/protein/12.1.3. [DOI] [PubMed] [Google Scholar]
- 65.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 66.Quast C. Bremen, Germany: University of Bremen; 2006. Master's thesis. [Google Scholar]
- 67.Tatusov R, Fedorova N, Jackson J, Jacobs A, Kiryutin B, Koonin E, Krylov D, Mazumder R, Mekhedov S, Nikolskaya A, et al. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, et al. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gerhardt P. Methods for General and Molecular Bacteriology. Washington, DC: Am Soc Microbiol; 1994. [Google Scholar]
- 70.Revsbech NP. Limnol Oceanogr. 1989;34:474–478. [Google Scholar]
- 71.Wright SW, Jeffrey SW. In: Phytoplankton Pigments in Oceanography: Guidelines to Modern Methods. Jeffrey SW, Mantoura RFC, Wright SW, editors. Paris: UNESCO; 1997. pp. 327–342. [Google Scholar]
- 72.Hiraishi A, Kuraishi H, Kawahara K. Int J Syst Evol Microbiol. 2000;50:1113–1118. doi: 10.1099/00207713-50-3-1113. [DOI] [PubMed] [Google Scholar]
- 73.Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumeier J, Bachleitner M, Schleifer K-H. Electrophoresis. 1998;19:554–568. doi: 10.1002/elps.1150190416. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.