Abstract
The microbial ecology of human skin is complex, but little is known about its species composition. We examined the diversity of the skin biota from the superficial volar left and right forearms in six healthy subjects using broad-range small subunit rRNA genes (16S rDNA) PCR-based sequencing of randomly selected clones. For the initial 1,221 clones analyzed, 182 species-level operational taxonomic units (SLOTUs) belonging to eight phyla were identified, estimated as 74.0% [95% confidence interval (C.I.), ≈64.8–77.9%] of the SLOTUs in this ecosystem; 48.0 ± 12.2 SLOTUs were found in each subject. Three phyla (Actinobacteria, Firmicutes, and Proteobacteria) accounted for 94.6% of the clones. Most (85.3%) of the bacterial sequences corresponded to known and cultivated species, but 98 (8.0%) clones, comprising 30 phylotypes, had <97% similarity to prior database sequences. Only 6 (6.6%) of the 91 genera and 4 (2.2%) of the 182 SLOTUs, respectively, were found in all six subjects. Analysis of 817 clones obtained 8–10 months later from four subjects showed additional phyla (numbering 2), genera (numbering 28), and SLOTUs (numbering 65). Only four (3.4%) of the 119 genera (Propionibacteria, Corynebacteria, Staphylococcus, and Streptococcus) were observed in each subject tested twice, but these genera represented 54.4% of all clones. These results show that the bacterial biota in normal superficial skin is highly diverse, with few well conserved and well represented genera, but otherwise low-level interpersonal consensus.
Keywords: human microbial ecology, small subunit rRNA genes, clone library, microbial biota diversity, normal human skin
The human skin has been considered to harbor a complex microbial ecosystem (1), with transient, short-term resident and long-term resident biota, based on the consistency with which they are isolated (2). Staphylococcus, Micrococcus, Corynebacterium, Brevibacteria, Propionibacteria, and Acinetobacter species, among others, are regularly cultivated from normal skin (3, 4). Staphylococcus aureus, Streptococcus pyogenes, and Pseudomonas aeruginosa may be transient colonizers, especially in pathological conditions (5–7). Environmental factors, such as temperature, humidity, and light exposure, and host factors, including gender, genotype, immune status, and cosmetic use (4), all may affect microbial composition, population size, and community structure. Disease may result from microecologic shifts. As a complex microbial ecosystem with potential roles in inflammatory diseases, little is known about the species composition in cutaneous samples.
Our knowledge of the human skin biota, chiefly through cultivation-based studies (1, 8), is considerably limited in assessing compositions of complex microbial communities (9, 10). In contrast, broad-range PCR primers targeted to highly conserved regions makes possible the amplification of small subunit rRNA genes (16S rDNA) sequences from all bacterial species (11–14), and the extensive and rapidly growing 16S rDNA database facilitates identification of sequences to the species or genus level (15). Such techniques are increasingly used for identifying bacterial species in complex environmental niches (16–18), including the human mouth (19, 20), esophagus (21), stomach (22), intestine (23), feces (24), and vagina (25), and for clinical diagnosis (26, 27). Because there only have been limited applications to human skin (28), we now report use of these techniques to characterize the composition of its biota. Because human skin is extensive and variable in its characteristics, we sampled a single site, the volar forearm, to maximize homogeneity and allow analysis of bilateral conservation.
Results
Clone Libraries.
From the six healthy subjects, we initially analyzed 1,345 clones from their superficial forearm skin samples. Ten species (found in 121 clones) present in both control and skin samples, and three chimeric 16S rDNA sequences were excluded [supporting information (SI) Table 2], leaving 1,221 sequences for further phylogenetic analysis (mean ± SD of 203.5 ± 2.7 clones per subject) (SI Table 3).
Classification of the Clones.
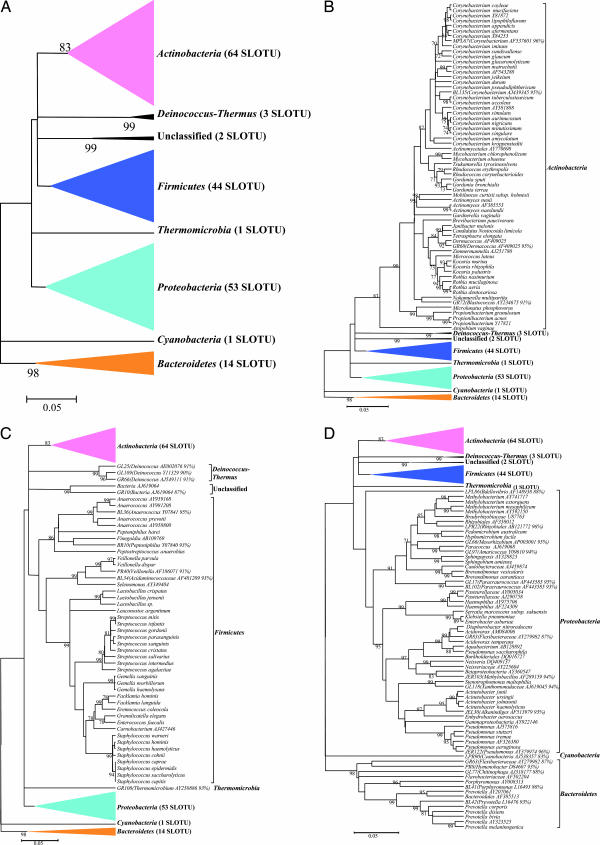
The programs SEQUENCE MATCH at RDP II (Version 9.39) and BLAST in GenBank were used to compare the clone sequences to known 16S rDNA sequences for assignment to the closest taxon. In total, 182 species-level operational taxonomic units (SLOTUs) were detected (SI Table 4); 1,123 clones (92.0%) represented 152 known species [Ribosomal Database Project (RDP) similarity scores ≥0.873 and homology ≥97%] (21). Of the 1,221 clones, 983 (80.5%) had similarity scores ≥0.873 with cultivated type strains. Fifty-eight (4.8%) of the remaining 238 clones had similarity scores ≥0.873 with 16S rDNA from a fully defined cultured, but non-type bacterial strain. These 1,041 clones were classified as belonging to culture-defined bacterial species, representing 131 SLOTUs. Of the remaining 180 clones, 82 (6.7%) had similarity scores ≥0.873 only to PCR-generated 16S rDNA clones in the database; these sequences were classified as 16S rDNA clones, representing 21 SLOTUs. The remaining 98 sequences (8.0%), not sufficiently homologous (<0.873 similarity score) with culture-defined species or with existing 16S rDNA clones, were classified as unknowns, representing 30 phylotypes, based on the putative genus or higher taxon they most closely resembled (GenBank accession nos. DQ130020–DQ130049). The taxonomic assignments were confirmed by phylogenetic analysis (Fig. 1).
Fig. 1.
Phylogenetic analysis of bacterial 16S rDNA detected in normal skin from six subjects. (A) From 1,221 clones, sequences representing eight bacterial phyla and 182 SLOTUs were observed. Alignments were done with Greengenes, and misalignments were manually curated in ARB (48), evolutionary distances were calculated with the Jukes–Cantor algorithm, and phylogenetic trees were determined by the Neighbor-Joining method; with 1,000 trees generated, bootstrap confidence levels are shown at tree nodes for values ≥70%. (B) Phylogenetic tree of the 628 clones within the phylum Actinobacteria. SLOTU designations are located at the termination of each branch. The 16S rDNA clones and cultivated non-type strains represent potential bacterial species; these clones are represented by the nearest species, followed by the GenBank accession number of the best-matched sequence. Unknowns are represented by the serial number of the clone used in this study, followed by the closest match, and percent sequence identity. (C) Phylogenetic tree of the 345 clones representing the phyla Firmicutes, Deinococcus-Thermus, Thermomicrobia, and unclassified organisms. Designations are as described for B. The clones representing Deinococcus-Thermus, Thermomicrobia, and unclassified were from the same subject. (D) Phylogenetic tree of the 248 clones representing the phyla Proteobacteria, Bacteroidetes, and Cyanobacteria. Designations are as described for B.
Estimation of SLOTU Richness.
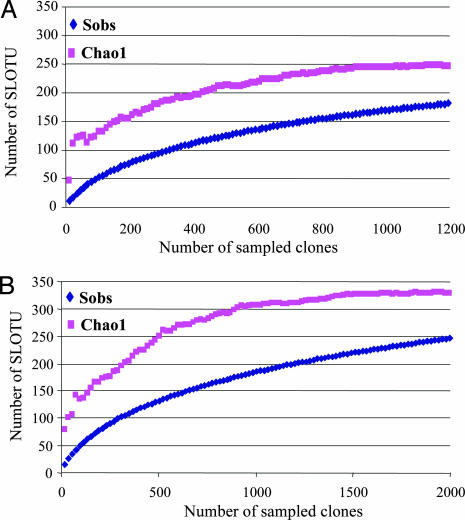
The total number of SLOTUs present in the superficial forearm skin samples from the six subjects was calculated by using the Chao1 estimator (29), based on the distribution of singletons. We estimated that the bacterial biota from those specimens contains ≈246 SLOTUs [95% confidence interval (C.I.), range 217–301]; the 182 SLOTUs observed represent 74.0% (95% C.I., 64.8–77.9%) of the estimated species or phylotypes (Fig. 2A). In host-specific analyses, the study identified ≈58.2–97.5% of the individual SLOTUs, indicating extensive intrahost diversity. Including results from resampling of four of the subjects with 817 clones (SI Table 3), the estimated total species richness was 328 (range 295–385) with 247 (range 232–262) SLOTUs observed, indicating 75.3% coverage (68.1–78.6%) (Fig. 2B).
Fig. 2.
Collector's curves of observed and estimated SLOTU richness of pooled forearm skin samples from six healthy subjects. (A) Each curve reflects the series of observed (Sobs) or estimated (Chao1) richness values obtained as the 1,221 16S rDNA clones are added to the data set in an arbitrary order. As reported for the human colon (23), as an increasing proportion of SLOTUs have been encountered, the Chao1 curve rises less steeply; however, additional SLOTUs continue to be identified to the end of the sampling. Although 182 (95% C.I., range 169–195) SLOTUs were observed, the Chao1 score of total species richness, estimates that the skin bacterial biota in the six subjects contains ≈246 SLOTUs (95% C.I., range 217–301). Based on this prediction, the present study identified 74.0% (95% C.I., 64.8–77.9%) of the SLOTUs in this bacterial ecosystem. (B) When results from resampling of four of the subjects with 817 new clones were included, the number of SLOTUs was 247 (95% C.I., range 232–262), and the Chao1 was 328 (95% C.I., range 295–385), indicating coverage of 75.3% (95% C.I., range 68.1–78.6).
Distribution of the Clones at the Phylum Level.
The 182 SLOTUs belong to seven bacterial phyla: Actinobacteria, Proteobacteria, Firmicutes, Bacteroidetes, Deinococcus-Thermus, Thermomicrobia, Cyanobacteria, and nine clones were unclassified (Fig. 1). Actinobacteria, Firmicutes, and Proteobacteria were observed in all six subjects, representing 94.6% of the 1,221 clones, accounting for 64 (51.4% of the clones), 44 (23.8%), and 53 SLOTUs (19.4%), respectively (Table 1). In comparison, these three phyla also predominate in the human distal esophagus (21) and stomach (22), but represent only about half of the clones sampled in the oral cavity (19) or from feces and colon (23) (SI Table 5).
Table 1.
Phylum diversity represented in 1,221 16S rDNA clones from normal forearm skin of six subjects
| Phylum | Known species |
Novel phylotypes |
No. of subjects with phylum | ||
|---|---|---|---|---|---|
| No. of clones (% of total) | No. of SLOTU | No. of clones (% of total) | No. of SLOTU | ||
| Actinobacteria | 614 (50.3) | 60 | 14 (1.1) | 4 | 6 |
| Firmicutes | 285 (23.3) | 40 | 5 (0.4) | 4 | 6 |
| Proteobacteria | 203 (16.6) | 42 | 34 (2.8) | 11 | 6 |
| Bacteroidales | 13 (1.1) | 9 | 6 (0.5) | 5 | 4 |
| Cyanobacteria | 0 | 0 | 2 (0.2) | 1 | 1 |
| Deinococcus-Thermus | 0 | 0 | 35 (2.9) | 3 | 1 |
| Thermomicrobia | 0 | 0 | 1 (0.1) | 1 | 1 |
| Unclassified | 8 (0.7) | 1 | 1 (0.1) | 1 | 1 |
| Total | 1,123 (92.0) | 152 | 98 (8.0) | 30 | |
Distribution of the Clones at the Genus Level.
The skin samples from the six subjects yielded sequences representing 91 genera (SI Table 6). Six genera were observed in all six subjects, comprising 768 (62.9%) of the 1,221 clones analyzed, including Propionibacterium (22.0% of all clones), Corynebacterium (19.0%), Staphylococcus (11.1%), Streptococcus (5.8%), Acinetobacter (3.7%), and Finegoldia (1.3%). However, of the 91 genera, 62 (68.1%) were identified in only one subject, 8 (8.8%) in two subjects, and 6 (6.6%) in three subjects; only 15 genera (16.5%) were observed in ≥4 subjects, indicating substantial interpersonal variation at the genus level. When the results from resampling of four of the subjects were included (see below), 119 genera were observed with only 40 (33.6%) shared at the two time points. Only four (3.4%) of the 119 genera were detected in all subjects tested at both time points, but represented 54.4% of all of the clones [Propionibacterium (21.1%), Corynebacterium (14.3%), Staphylococcus (10.9%), and Streptococcus (8.1%)].
Distribution of the Clones at the Species Level.
The number of SLOTUs in each subject ranged from 32 to 67 (Mean: 48.0 ± 12.2) (SI Table 4), with 13.5 ± 5.8 present on both forearms. Sixteen SLOTUs were observed in ≥4 of the 6 subjects, comprising 29.8–79.5% of clones in each subject (mean 54.1 ± 17.4%) (SI Table 7). Four species (Propionibacterium acnes, Corynebacterium tuberculostearicum, Streptococcus mitis, and Finegoldia AB109769) detected in all six subjects, accounted for 31.0% of the clones analyzed. In contrast, 71.4% of the individual SLOTUs were isolated from single subjects only. For example, a phylotype (closest species Deinococcus AJ549111) was detected in only one subject, but represented ≈14% of her clones. Propionibacterium granulosum, Corynebacterium singulare, and Corynebacterium appendixes were found only in the three male subjects.
Distribution of the Clones in Left and Right Arms.
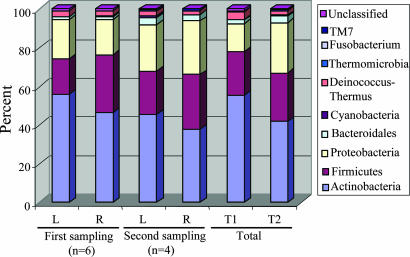
We studied symmetry of the left and right arms; at the phylum level, differences between a subject's two arms were generally small (SI Table 8), and in composite varied little over time (Fig. 3). The 10 most common SLOTUs accounted for 46.7% and 49.6% of all clones in the left and right arms, respectively (SI Table 9). However, in individual hosts, the SLOTUs that were presented in both arms represented 50.0%–77.0% (67.9 ± 9.7%) of all clones from that subject.
Fig. 3.
Distribution of 2,038 16S rDNA clones from left and right forearm, by phylum. At the first sampling, 1,221 clones were obtained from the six subjects, and 817 clones were later obtained from four of these six. Thus, in total, 2,038 clones were studied, with percents amongst the 10 phyla indicated by the color designations. For the four individuals sampled twice (subjects T1 and T2), there was little difference in the overall phylum distribution between the two time points.
Detection of Previously Uncharacterized Phylotypes.
About 8% (range: 1.0–29.3% in different subjects) of the 16S rDNA sequences generated from the normal skin samples did not match any known bacterial sequences present in public databases. In total, 30 previously uncharacterized phylotypes (98 clones) were detected, corresponding to seven bacterial phyla and one unclassified phylum (Fig. 1). Thermomicrobia, Deinococcus-Thermus, Cyanobacteria, and the unclassified phylum each were detected in only one subject. Among these phyla, 35 clones had 100% similarity to three phylotypes within the genus Deinococcus. One phylotype (2 clones), with 94% sequence similarity to unclassified Cyanobacteria AJ538357, was assigned to phylum Cyanobacteria. One clone, with 93% sequence similarity to Thermomicrobium AY250886 belonged in phylum Thermomicrobia, and one clone, with only 87% sequence similarity to bacteria AJ619064, could not be classified.
Resampling of Subjects.
With resampling of four of the subjects 8–10 months later, the additional 817 clones studied yielded an additional two phyla (TM7 and Fusobacteria), 28 genera, and 65 SLOTUs (SI Table 10). Of the total 2,038 clones, 1,893 (92.9%) represented 203 known species; the remaining 145 sequences (7.1%) represented 44 previously uncharacterized phylotypes (<97% homologies with GenBank sequences) (GenBank accession nos. DQ130020–DQ130049 and DQ847437–DQ847450). Culture-defined bacterial species, representing 169 SLOTUs, were identified from 1,764 (86.6%) of 2,038 clones, most commonly, Propionibacterium acnes (20.2%), Enhydrobacter aerosaccus (6.7%), and Corynebacterium tuberculostearicum (5.2%).
Intrahost and Interhost Variation.
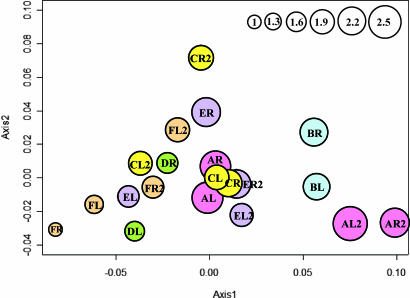
Similarities in SLOTU distributions between skin samples were evaluated by using double principal coordinate analysis (DPCoA). In four of the six subjects at time 1 (subjects A, B, C, and F), samples from the left and right forearms were closely related to each other (Fig. 4). Because samples from four of the six subjects (subjects A, C, E, and F) were obtained 8–10 months later, we could ascertain the temporal stability of populations. For example, for subject A, the two samples from time 2 clustered closely, as did those from time 1 (Fig. 4), but the two time points were not clustered; for subject C, the two samples from time 1 were closely related, whereas those from time 2 were more distant. Three hypotheses on the grouping of samples were tested. First, analysis using all 12 samples from the first time point showed that those from the same subject (left and right arms) were more similar to each other than to samples from other subjects (P < 0.001). The same result was confirmed for the 8 samples collected at the second time point (P = 0.016). Second, in analysis of 16 samples from four subjects (with both arms at both time points), those obtained at the same time from a subject were more similar to each other than the samples obtained at different times, even from the same arm (P = 0.007). Third, analyzing these same 16 samples, those from the same subject at the separate time points were not significantly more similar to each other than to samples from other subjects (P = 0.29). The P test, which allows pair-wise comparison of samples, also was used to evaluate the similarities amongst the skin samples (SI Table 11). Samples from the same subject at the same time point were not significantly different from each other, whereas the samples from the same subject at different time points could be significantly different, consistent with the results from the DPCoA.
Fig. 4.
DPCoA of SLOTU relatedness in 20 forearm skin samples obtained from six subjects. Subjects were designated A–F, and at each sampling both left (L) and right (R) forearm skin was examined. In four subjects, new specimens were obtained 8–10 months later (e.g., designated FL2). In a representation of the first two orthogonal principal axes, based on a sample dissimilarity matrix, samples from the same subject at the same time point are plotted by using the same color, with circle size proportional to the sample's Rao diversity index. The scale (top right) indicates the relative diversity of circles, with the test sample of smallest diversity (FR) indexed as 1.0.
Discussion
Sequence-based environmental microbial surveys indicate that cultivation methods substantially under-represent the extent of bacterial diversity. From environments including seawater, lakes, sediments, and soil (11), < 1% of bacteria can be cultivated; in contrast, cultivability of the human microbiota, such as in the gut, is estimated to be substantially higher (10–50%) (11, 20, 23, 30). Analysis of the 2,038 16S rDNA clones from the superficial forearm skin of six healthy subjects revealed a bacterial species-rich microbiota: the 10 phyla, 119 genus-level taxonomic units, and 247 SLOTUs represented in the microbiota correspond to the first approximation of our understanding of the composition of this ecological niche in humans, with ≈75% species coverage estimated.
Most (86.5%) clones from normal superficial skin could be classified as culture-defined bacterial species. In a pilot study of two samples from one subject, we examined the microbial diversity determined by broad-range 16S rDNA PCR-based sequencing and by standard aerobic and anaerobic cultivation methods (data not shown). Only eight (16.0%) of the 50 species determined by rDNA amplification, including Propionibacterium, Staphylococcus, and Bacillus species, could be cultivated from the same specimens. Although we considered several other putative species identified at the 16S level as likely to be cultivable, the bacteria actually present may be more fastidious than expected, even when related to cultivable species (31, 32). Not surprisingly, the bacterial diversity revealed by amplification of 16S rDNA directly from normal skin in the six subjects far exceeded that shown by cultivation methods (28).
Nevertheless, about half (54.4%) of the clones were identified as representing the genera Propionibacteria, Corynebacteria, Staphylococcus and Streptococcus, long recognized as forming part of the normal human skin biota (1, 33). Eight bacterial genera have been cultivated from skin, and are considered residential (33); our study detected seven of these genera (except Dermabacter), as well as ≈100 others not previously cultured (4, 28, 33–35). Similarly, of the ≈40 cultivatable species that are believed to be residential (4, 28, 33), we found evidence for 21 amongst the 247 observed SLOTUs. Sequences corresponding to species, including Prevotella, that are common in the oropharynx and gastrointestinal tract (21, 22, 36), also were found in human skin.
However, the overall microbial biota observed differed substantially amongst the six subjects. Although four genera (Propionibacteria, Corynebacteria, Staphylococcus, and Acinetobacter) were common, only 2.2% of the SLOTUs and 6.6% of the genera were found in all six subjects. In contrast, 71.4% of all of the individual SLOTUs and 68.1% of the genera were identified only from a single individual. Although 200–400 clones were studied from each subject, future sampling more intensively should reduce the proportion of individual-specific taxons. The data indicate that the superficial skin biota is highly diversified, with a low level of interpersonal consensus, similar to that observed for the gastrointestinal tract (23). If the finding that several gender-associated species in this small sample is confirmed, the differences could reflect variation in skin surface pH between men and women (37), among other factors.
Analysis of the variation indicates a high level of conservation between the two analogous sites (left and right) at any moment (Fig. 4). Over time, as indicated by the resampling, the host-specific conservation persists, but without a strong relationship to the prior sample from that site. These observations indicate that the bacterial biota is dynamic over the time period sampled, affecting both sides similarly, and fluctuating to an extent similar to that occurring in other individuals. In total, these data and the phylum analyses (Fig. 3), provide a picture of a largely stable predominant scaffold of a relatively small number of bacterial genera and species, supporting a relatively high frequency of transients, affecting both arms similarly; these findings parallel our observations of the fungal biota at the same sites (38).
Previously uncharacterized phylotypes were common in this study, some displaying >10% sequence dissimilarity from published sequences (SI Table 4). Each previously uncharacterized phylotype was identified only in a single subject, consistent with the observed low level of interpersonal consensus. That 7.1% of clones and 17.8% of phylotypes were previously uncharacterized, based on a criterion of <97% similarity, is much lower than in other human niches, including the intestine (23), feces (39), tongue dorsum (40), and human subgingival crevice (20). The presence of a few dominant species in the skin, use of varying definitions in different studies (19, 23, 41), as well as the progressive recognition of human-associated species, may partially explain this difference. As the field of human microbial ecology advances, it will be critical to set universal standards for comparisons of bacterial communities.
Topologically, the human body can be viewed as a tubular structure. The inner surface is lined by the mucosa of the orodigestive tract, whereas the outer surface is lined by keratinized skin. Despite such marked differences in tissue structure and environmental contact, the relationship between the bacterial biota on the inner and outer surfaces is apparent at high phylogenetic hierarchy (SI Table 5). Of the five phyla-Actinobacteria, Firmicutes, Proteobacteria, Bacteroidetes, and Fusobacteria-shared by four comprehensively studied mucosal sites [oral cavity (19), distal esophagus (21) stomach (22), and colon (23)], all except Fusobacteria also are found on skin. The preponderance (96.15%) of 16S rDNA clones sampled from the skin, similar to those from the inner mucosal surfaces, belong to the shared phyla. However, the relative abundance of the common phyla differs. The normal skin bacterial biota is the only one in which Actinobacteria predominate. Our study shows that at lower hierarchy, there is a core set of organisms that comprise most of the skin biota, but uncommon taxa comprising the balance of the population differ significantly amongst individuals and between sample times. Similarly, analyses of esophageal and colonic bacterial biota indicate that the most abundant taxa are generally well conserved but low-abundance members might not be shared between bacterial communities (42).
Our molecular genetic analysis of bacterial rDNA amplicons generated directly from superficial human skin samples raises the possibility of better understanding the microbial ecology of the skin, and for studying the role of novel microbes or microbial communities in the pathogenesis of dermatoses. Further studies are warranted, including assessment of the transience, stability, host- and site-specificities of this complex bacterial biota, and whether or not there exist particular associations with skin diseases.
Materials and Methods
Subjects.
Specimens from superficial skin were obtained from the left and right forearms of six healthy subjects (three males and three females); second samples were obtained 8–10 months later from four of these subjects (SI Table 3). The mean age of the subjects was 38 years of age (range, 21–54 years of age); all were in good health and had not received any antibiotics for at least one month. The study was approved by the New York University Institutional Review Board, and all subjects provided written informed consent (38).
Specimen Processing and 16S rDNA PCR Amplification.
Methods for skin sampling have been described (38). Universal bacterial primers 8F and 1510R were used to amplify PCR products ≈1.5 kb, based on positions 8 to 1513 of the Escherichia coli 16S rRNA gene, as described (21, 43, 44). More details are provided in SI Materials and Methods.
16S rDNA Clone Libraries.
The PCR products were separated from free PCR primers by using a PCR purification kit (Qiagen, Valencia, CA), ligated with the pGEM-T-Easy vector (Promega, Madison, WI), used to transform E. coli DH5α competent cells, and clones analyzed as in SI Materials and Methods. For identification of closest relatives, the newly determined sequences were compared with those available in the Ribosomal Database Project (RDP) II (release 9.39) (45) and GenBank (www.ncbi.nlm.gov) databases, by using the standard nucleotide-nucleotide BLAST program to ascertain their closest relatives.
Phylogenetic Analysis.
All sequences were examined for chimerism by using Chimera Detection at Ribosomal database Project (RDP) II (release 8.1) and Bellerophon (46). In total, only three clones were removed from the phylogenetic analysis. The remaining sequences were compared with those of RDP II (release 9.39) (45) and in GenBank to identify SLOTUs, as reported (21). The sequences were aligned with NAST at Greengenes (http://greengenes.lbl.gov/cgi-bin/nph-index.cgi) (47). Misalignments were manually curated in ARB (48), and then hypervariable regions were masked by using MASK COLUMNS at Greengenes (47). The phylogenetic trees were generated by using MEGA 3.1 (49). Evolutionary distances were calculated with the Jukes–Cantor algorithm (50). The statistical strength of the Neighbor-Joining method was assessed by bootstrap resampling (1,000 replicates) (51).
Statistical Analyses.
The total number of SLOTUs that may be present in the sampled human skin and its associated confidence interval were calculated by using a nonparametric richness estimator, Chao1, as described (29). DPCoA (52) and the P test (53, 54) were used to evaluate sample diversity and the relationships among samples, as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported in part by a Senior Scholar Award from the Ellison Medical Foundation; by Diane Belfer Program in Human Microbial Ecology in Health and Disease Grants R01 GM63270, CA97946, and AI063477; and by General Clinical Research Center Core Grant NIH/NCRR M01 RR00096 from the National Institutes of Health.
Abbreviations
- SLOTU
species-level operational taxonomic unit
- RDP
Ribosomal Database Project
- C.I.
confidence interval
- DPCoA
double principal coordinate analysis
- 16S rDNA
small subunit rRNA genes.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The 16S rDNA sequences of clones representing previously uncharacterized phylotypes as defined in this study have been deposited in the GenBank database (accession nos. DQ130020–DQ130049 and DQ847437–DQ847450).
This article contains supporting information online at www.pnas.org/cgi/content/full/0607077104/DC1.
References
- 1.Fredricks DN. J Investig Dermatol Symp Proc. 2001;6:167–169. doi: 10.1046/j.0022-202x.2001.00039.x. [DOI] [PubMed] [Google Scholar]
- 2.Holland KT, Bojar RA. Am J Clin Dermatol. 2002;3:445–449. doi: 10.2165/00128071-200203070-00001. [DOI] [PubMed] [Google Scholar]
- 3.Leyden JJ, McGinley KJ, Nordstrom KM, Webster GF. J Invest Dermatol. 1987;88:65s–72s. doi: 10.1111/1523-1747.ep12468965. [DOI] [PubMed] [Google Scholar]
- 4.Roth RR, James WD. Annu Rev Microbiol. 1988;42:441–464. doi: 10.1146/annurev.mi.42.100188.002301. [DOI] [PubMed] [Google Scholar]
- 5.Noble WC. Br J Dermatol. 1998;139(Suppl 53):9–12. doi: 10.1046/j.1365-2133.1998.1390s3009.x. [DOI] [PubMed] [Google Scholar]
- 6.Akiyama H, Morizane S, Yamasaki O, Oono T, Iwatsuki K. J Dermatol Sci. 2003;32:193–199. doi: 10.1016/s0923-1811(03)00096-3. [DOI] [PubMed] [Google Scholar]
- 7.El Baze P, Thyss A, Caldani C, Juhlin L, Schneider M, Ortonne JP. Arch Dermatol. 1985;121:873–876. doi: 10.1001/archderm.121.7.873. [DOI] [PubMed] [Google Scholar]
- 8.Davies CE, Wilson MJ, Hill KE, Stephens P, Hill CM, Harding KG, Thomas DW. Wound Repair Regen. 2001;9:332–340. doi: 10.1046/j.1524-475x.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- 9.McCaig AE, Glover LA, Prosser JI. Appl Environ Microbiol. 1999;65:1721–1730. doi: 10.1128/aem.65.4.1721-1730.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tlaskalova-Hogenova H, Stepankova R, Hudcovic T, Tuckova L, Cukrowska B, Lodinova-Zadnikova R, Kozakova H, Rossmann P, Bartova J, Sokol D, et al. Immunol Lett. 2004;93:97–108. doi: 10.1016/j.imlet.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Amann RI, Ludwig W, Schleifer KH. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hugenholtz P, Pace NR. Trends Biotechnol. 1996;14:190–197. doi: 10.1016/0167-7799(96)10025-1. [DOI] [PubMed] [Google Scholar]
- 13.Pace NR. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 14.Zoetendal EG, Vaughan EE, de Vos WM. Mol Microbiol. 2006;59:1639–1650. doi: 10.1111/j.1365-2958.2006.05056.x. [DOI] [PubMed] [Google Scholar]
- 15.Schloss PD, Handelsman J. Microbiol Mol Biol Rev. 2004;68:686–691. doi: 10.1128/MMBR.68.4.686-691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams MM, Domingo JW, Meckes MC, Kelty CA, Rochon HS. J Appl Microbiol. 2004;96:954–964. doi: 10.1111/j.1365-2672.2004.02229.x. [DOI] [PubMed] [Google Scholar]
- 17.Shivaji S, Reddy GS, Aduri RP, Kutty R, Ravenschlag K. Cell Mol Biol. 2004;50:525–536. [PubMed] [Google Scholar]
- 18.Smit E, Leeflang P, Gommans S, van den BJ, van Mil S, Wernars K. Appl Environ Microbiol. 2001;67:2284–2291. doi: 10.1128/AEM.67.5.2284-2291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroes I, Lepp PW, Relman DA. Proc Natl Acad Sci USA. 1999;96:14547–14552. doi: 10.1073/pnas.96.25.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pei Z, Bini EJ, Yang L, Zhou M, Francois F, Blaser MJ. Proc Natl Acad Sci USA. 2004;101:4250–4255. doi: 10.1073/pnas.0306398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. Proc Natl Acad Sci USA. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X, Bent SJ, Schneider MG, Davis CC, Islam MR, Forney LJ. Microbiology. 2004;150:2565–2573. doi: 10.1099/mic.0.26905-0. [DOI] [PubMed] [Google Scholar]
- 26.Harris KA, Hartley JC. J Med Microbiol. 2003;52:685–691. doi: 10.1099/jmm.0.05213-0. [DOI] [PubMed] [Google Scholar]
- 27.Saglani S, Harris KA, Wallis C, Hartley JC. Arch Dis Child. 2005;90:70–73. doi: 10.1136/adc.2003.042176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dekio I, Hayashi H, Sakamoto M, Kitahara M, Nishikawa T, Suematsu M, Benno Y. J Med Microbiol. 2005;54:1231–1238. doi: 10.1099/jmm.0.46075-0. [DOI] [PubMed] [Google Scholar]
- 29.Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJ. Appl Environ Microbiol. 2001;67:4399–4406. doi: 10.1128/AEM.67.10.4399-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaughan EE, Schut F, Heilig HG, Zoetendal EG, de Vos WM, Akkermans AD. Curr Issues Intest Microbiol. 2000;1:1–12. [PubMed] [Google Scholar]
- 31.Breitkopf C, Hammel D, Scheld HH, Peters G, Becker K. Circulation. 2005;111:1415–1421. doi: 10.1161/01.CIR.0000158481.07569.8D. [DOI] [PubMed] [Google Scholar]
- 32.Colwell RR. In: Nonculturable Microorganisms in the Environment. Colwell RR, Grimes DJ, editors. Washington, DC: Am Soc Microbiol; 2000. pp. 325–342. [Google Scholar]
- 33.Chiller K, Selkin BA, Murakawa GJ. J Investig Dermatol Symp Proc. 2001;6:170–174. doi: 10.1046/j.0022-202x.2001.00043.x. [DOI] [PubMed] [Google Scholar]
- 34.Berlau J, Aucken H, Malnick H, Pitt T. Eur J Clin Microbiol Infect Dis. 1999;18:179–183. doi: 10.1007/s100960050254. [DOI] [PubMed] [Google Scholar]
- 35.Kloos WE, Musselwhite MS. Appl Microbiol. 1975;30:381–385. doi: 10.1128/am.30.3.381-395.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brook I. Anaerobe. 2006;12:5–12. doi: 10.1016/j.anaerobe.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Ehlers C, Ivens UI, Moller ML, Senderovitz T, Serup J. Skin Res Technol. 2001;7:90–94. doi: 10.1034/j.1600-0846.2001.70206.x. [DOI] [PubMed] [Google Scholar]
- 38.Paulino LC, Tseng CH, Strober BE, Blaser MJ. J Clin Microbiol. 2006;44:2933–2941. doi: 10.1128/JCM.00785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayashi H, Sakamoto M, Benno Y. Microbiol Immunol. 2002;46:819–831. doi: 10.1111/j.1348-0421.2002.tb02769.x. [DOI] [PubMed] [Google Scholar]
- 40.Kazor CE, Mitchell PM, Lee AM, Stokes LN, Loesche WJ, Dewhirst FE, Paster BJ. J Clin Microbiol. 2003;41:558–563. doi: 10.1128/JCM.41.2.558-563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, Dore J. Appl Environ Microbiol. 1999;65:4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schloss PD, Handelsman J. Appl Environ Microbiol. 2006;72:6773–6779. doi: 10.1128/AEM.00474-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edwards U, Rogall T, Blocker H, Emde M, Bottger EC. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagashima K, Hisada T, Sato M, Mochizuki J. Appl Environ Microbiol. 2003;69:1251–1262. doi: 10.1128/AEM.69.2.1251-1262.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maidak BL, Cole JR, Lilburn TG, Parker CT, Jr, Saxman PR, Farris RJ, Garrity GM, Olsen GJ, Schmidt TM, Tiedje JM. Nucleic Acids Res. 2001;29:173–174. doi: 10.1093/nar/29.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huber T, Faulkner G, Hugenholtz P. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- 47.DeSantis TZ, Jr, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, et al. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar S, Tamura K, Nei M. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 50.Jukes TH, Cantor CR. In: Mammalian Protein Metabolism. Munro HN, editor. New York: Academic; 1969. pp. 21–132. [Google Scholar]
- 51.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 52.Pavoine S, Dufour AB, Chessel D. J Theor Biol. 2004;228:523–537. doi: 10.1016/j.jtbi.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 53.Lozupone C, Hamady M, Knight R. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin AP. Appl Environ Microbiol. 2002;68:3673–3682. doi: 10.1128/AEM.68.8.3673-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.