Abstract
High mobility group box (HMGB) 1 and 2 are two abundant nonhistone nuclear proteins that have been found in association with chromatin. Previous studies based on immunofluorescence analysis indicated that HMGB1 dissociates from chromosomes during mitosis. In the present work, HMGB1 and 2 subcellular localization was reinvestigated in living cells by using enhanced green fluorescent protein- and Discosome sp. red fluorescent protein-tagged proteins. Contrary to previous reports, HMGB1 and 2 were shown to be present under two forms in mitotic cells, i.e., free and associated with the condensed chromatin, which rapidly exchange. A detailed analysis of HMGB2 interaction with mitotic chromosomes indicated that two sites encompassing HMG-box A and B are responsible for binding. Importantly, this interaction was rapidly inactivated when cells were permeabilized or exposed to chemical fixatives that are widely used in immunodetection techniques. A comparable behavior was also observed for two proteins of the HMG-nucleosome binding (HMGN) group, namely, HMGN1 and HMGN2.
INTRODUCTION
High mobility group box (HMGB) 1 and 2 proteins, formerly called HMG1 and 2 (Bustin, 2001), are two highly abundant nuclear nonhistone proteins that are almost identical in all mammals and have counterparts in all eukaryotes (Bianchi, 1995; Bustin and Reeves, 1996). They have been implicated in transcription regulation, DNA repair, recombination, differentiation, and extracellular signaling (Muller et al., 2001b; Thomas, 2001; Scaffidi et al., 2002). Despite their high sequence similarities, tertiary structure, and biochemical activities (Bustin, 1999; Bianchi and Beltrame, 2000; Thomas, 2001), HMGB1 and 2 are not functionally completely redundant (Calogero et al., 1999; Ronfani et al., 2001).
HMGB1/2 bind in a sequence-nonspecific manner and with a low affinity to single-stranded, linear duplex and supercoiled DNA, and exhibit a higher affinity for unusual DNA structures such as four-way junctions, cruciform DNA and cisplatin-modified DNA. In turn, these proteins induce structural modifications in the linear DNA they are bound to, such as bending and looping (Thomas, 2001).
Two nonhomologous but structurally related domains called HMG boxes A and B are responsible for HMGB DNA binding properties. Both DNA boxes bind and bend DNA, although they exhibit differences in their DNA binding and bending activities (Teo et al., 1995; Payet et al., 1999; Webb and Thomas, 1999). These activities are modulated, at least in vitro, by sequences that flank the HMG boxes (Sheflin et al., 1993; Stros et al., 1994; Payet and Travers, 1997; Ritt et al., 1998; Stros, 1998; Muller et al., 2001a).
HMGB1/2 interact with cellular (RAG1, p53, Oct, and Hox proteins, some steroid receptors, and TATA binding protein) and viral proteins (Rep78, Rep68 of the adeno-associated virus, and ZEBRA of the Epstein-Barr virus) (Ge and Roeder, 1994; Onate et al., 1994; Zwilling et al., 1995; Zappavigna et al., 1996; Costello et al., 1997; Boonyaratanakornkit et al., 1998; Jayaraman et al., 1998; Aidinis et al., 1999; Sutrias-Grau et al., 1999; Butteroni et al., 2000; Lu et al., 2000) and usually increases the ability of their partners to interact with DNA. Together, these observations lead to the proposal that HMGBs act as architectural facilitators in the assembly of nucleoprotein complexes.
HMGBs have been proposed to be important component of chromatin and to share some functions with histone H1. Notably, they have been purified in association with chromatin (Johns, 1982), and both H1 and HMGB bind to linker DNA (Schroter and Bode, 1982), to four-way junctions (Hill and Reeves, 1997) and to cis-platin–modified DNA (Yaneva et al., 1997). However, HMGB1 binding affinity is much lower than H1's, suggesting that HMGB may interact with the linker region only when histone H1 is absent or weakly expressed (Ura et al., 1996), such as during Xenopus (Dimitrov and Wolffe, 1996; Nightingale et al., 1996) or Drosophila development (Ner and Travers, 1994). In mammalian cells, HMGB1 attaches only loosely to chromatin (Falciola et al., 1997).
EBNA1, a nuclear protein encoded by the Epstein-Barr virus, interacts with cellular chromosomes during mitosis, most likely by the intermediate of one or several cellular protein(s) (Petti et al., 1990; Marechal et al., 1999; Shire et al., 1999). Preliminary results in one of our laboratories suggested that HMGB2 could recruit or be recruited by EBNA1 onto mitotic chromosomes. However, previous studies based on immunofluorescence analysis indicated that HMGB1, which is closely related to HMGB2, dissociates from mitotic chromosomes (Falciola et al., 1997).
In the present work, the use of enhanced green fluorescent protein (EGFP)- and DsRed-tagged proteins allowed to reinvestigate HMGB1/2 localization in the context of living cell, while preserving some of their biological properties. It is shown herein that HMBG1/2 interact with chromosomes in a highly dynamic manner in mitotic living cells and that the binding is mediated by two regions encompassing HMG boxes A and B. This interaction is abrogated in permeabilized or chemically fixed cells. A comparable behavior was also observed for two proteins of the HMG-nucleosome binding (HMGN) group, namely, HMGN1 and 2.
MATERIALS AND METHODS
Cells Lines
The Raji cell line (ATCC CCL 86) was grown in RPMI 1640 medium supplemented with 10% fetal calf serum. Human HeLa cells (ATCC CCL2) and mouse 3T3 fibroblasts were grown in DMEM supplemented with 10% fetal calf serum, streptomycin (105 U/liter), vancomycin (0.1 g/liter), and glutamine (2 mM).
Recombinant Plasmids
Plasmids pEGFP-N1, pEGFP-C1, pEGFP-C2, pDsRed-N1, and pDsRed1-C1 (BD Biosciences Clontech, Palo Alto, CA) encoded a variant of the green fluorescent protein with enhanced fluorescence (EGFP) and a red fluorescent protein (DsRed1). Human HMGB2 was expressed as a fusion protein to the N and the C termini of EGFP, the N and the C termini of DsRed, respectively, in pHMGB2-EGFP, pEGFP-HMGB2, pHMGB2-DsRed, and pDsRed-HMGB2. HMGB2 cDNA was cloned from HeLa cells. For this purpose, total RNA was extracted with High Pure RNA isolation kit (Roche Diagnostics, Mannheim, Germany) and subjected to a reverse transcription in the presence of oligo(dT) primers by using the Ready To Go You Prime First Strand Beads kit following the manufacturer's recommendations (Amersham Biosciences UK, Little Chalfont, Buckinghamshire, United Kingdom). The region encoding human HMGB2 was subsequently amplified from the cDNA by polymerase chain reaction (PCR) with HMGB2 upstream primer 5′-CCCAAGCTTGGGCCACCATGGGTAAAGGAGACCCCAACAAG-3′ and HMGB2 downstream primer 5′-CGCGGATCCCGTTCTTCATCTTCATCCTCTTCCTCC-3′. The resulting PCR products was digested by HindIII and BamHI, gel purified, and cloned into pEGFP-N1 and pDsRed-N1. Then, the HindIII/BamHI insert from pHMGB2-DsRed was subcloned into pDsRed-C1 and pEGFP-C1.
Deletion mutants were generated from pHMGB2-DsRed by PCR by using the indicated primers: A, 5′-CGCGGATCCCGACCTTTGGGAGGAACGTAATTTTTC-3′; AN, 5′-CGCGGATCCCGCTTTTTCTTCCCCTTCTTATCACC-3′; NB, 5′-CCCAAGCTTGGGCCACCATGGATAAGAAGGGAAAGAAAAAGGAC-3′; B1, 5′-GCTCAAGCTTCACCATGGGTGACCCCAATGCTCCTAAAAGG-3′; and B2, 5′-CGCGGATCCCGGCCCTTGGCACGATATGCAGC-3′. These primers were designed to contain HindIII or BamHI restriction sites to allow cloning in pEGFP-N1. Mutant A was generated with primers HMGB2 upstream and A; mutant AN, with primers HMGB2 upstream and AN; mutant B, with primers B1 and B2; mutant NB, with primers NB and B2; mutant ANB, with primers HMGB2 upstream and B2; mutant BC, with primers B1 and HMGB2 downstream; mutant NBC, with primers NB and HMGB2 downstream. Mutant N was obtained by cloning the HindIII-BamHI linker obtained by hybridizing oligonucleotides Nfor 5′-AGCTTGGGCCACCATGGATAAGAAGGGGAAGAAAAAGCGG-3′ and Nrev 5′-GATCCCGCTTTTTCTTCCCCTTCTTATCCATGGTGGCCCA-3′.
pHMGB1-EGFP and pHMGB1-DsRed encode human HMGB1 fused to the N terminus of EGFP and DsRed, respectively. The region encoding human HMGB1 was generated by PCR from reverse transcribed polyadenylated RNA by using HMGB1 upstream primer 5′-GGAAGATCTTCGCCACCATGGGCAAAGGAGATCCTAAGAAGC-3′ and HMGB1 downstream primer 5′-CCGGAATTCCTTCATCATCATCATCTTCTTCTTCATC-3′. The PCR product was digested by BglII and EcoRI, gel purified and inserted into pEGFP-N1 and pDsRed-N1 between the BglII and EcoRI sites. Using HMGB1.2 upstream primer 5′-GGAAGATCTTTCGCCACCATGGGCAAAGGAGATCCTAAGAAGC-3′ and HMGB1.2 downstream primer 5′-CCGGAATTCCTTATTCATCATCATCATCTTCTTCTTCATC-3′, a PCR product containing the HMGB1 coding region was generated from pHMGB1-DsRed, digested with BglII and EcoRI, and inserted within pDsRed-C1 and pEGFP-C1 between the BglII and EcoRI restriction sites.
The regions encoding HMGN1 and HMGN2 were generated by PCR from purified human fibroblast DNA by using primers HMG14/17for 5′-GGGGGAAGCTTCCGCCGCCACCATGCCCAAGAG-3′ and HMG14rev 5′-GGGGGATCCGACTTGGCTTCTTTCTCTCC-3′, and HMG14/17for and HMG17rev 5′-GGGGGATCCTTGGCATCTCCAGCACCTTC-3′, respectively. The PCR products digested by HindIII and BamHI were cloned into pEGFP-N1 and pDsRed-N1, which resulted in vectors pHMGB14-EGFP, pHMGB14-DsRed, pHMGB17-EGFP, and pHMGB17-DsRed, respectively.
The cDNA encoding human histone H1 was obtained by reverse transcription on total human RNA extracted from Raji cells by using primer H1.F3C 5′-GGGAATTCTCACTTTTTCTTCGGAGCTGCCTTCTTTGC-3′. The region encoding H1 was produced from the cDNA by PCR by using primers H1.F3N 5′-GGCGGGATCCTGTCGGAGACTGCTCCACTTGCTCCTAC-3′ and H1.F3C. The PCR product was gel purified, digested with BamHI and EcoRI, and cloned into pEGFP-C2 (BD Biosciences Clontech) digested by BglII and EcoRI. The resulting plasmid was named pEGFP-H1. Plasmid DNA was purified using the QIAGEN-plasmid maxi kit (QIAGEN, Hilden, Germany). DNA sequencing was performed by automated sequencing using the dideoxynucleotide chain termination method accordingtothemanufacturer'srecommendations(ABIPrismdRhodamine Terminator Cycle Sequencing Ready Mix; Applied Biosystems, Foster City, CA). Plasmids pTHCR, pSGD9, and pHMG1 were described previously (Zappavigna et al., 1996).
Transfections
HeLa cells were grown in six-well plates until they reached ∼80% confluence. Plasmids were transfected with the FuGENE 6 transfection reagent (Roche Diagnostics) according to manufacturer's recommendations. Briefly, 1 μg of purified plasmid DNA was mixed with 100 μl of antibiotic-free culture medium complemented with 10% fetal calf serum and 4 μl of FuGENE 6. The DNA–FuGENE complex was incubated for 30 min at room temperature and then added dropwise into the wells containing 2 ml of antibiotic-free DMEM complemented with 10% fetal calf serum. The cells were incubated at 37°C for 16 h.
For stable cell lines, one-fifth of the cells from a well were plated on a 78-cm2 culture dish 16 h after transfection, and grown in the presence of 400 μg/ml geneticin (Sigma-Aldrich, St. Louis, MO) for 3 wk. Fluorescent foci were then cloned, plated in a six-well plate, and grown in the absence of selection until ∼80% confluence. Subcloning was repeated as described above from three to up to five times.
Fluorescence Microscopy
Unless otherwise indicated, fluorescent microscopy was performed on living cells 24–48 h after transfection. Briefly, one-fourth of the transfected cells were plated in a single-well chamber culture slide (Falcon Plastics, Oxnard, CA) and incubated for 48 h. The cells were incubated for an additional 10 min at 37°C in culture medium containing 0.2 μg/ml Hoechst 33342, washed once, and observed by fluorescence microscopy in the presence of prewarmed culture medium either at 365 nm (Hoechst), at 488 nm (EGFP and enhanced yellow fluorescent protein), or at 558 nm (DsRed).
For the preparation of chromosome spreadings, transfected HeLa cells were washed once in prewarmed culture medium, incubated for an additional 16 h in the presence of 0.1 μg/ml colcemid (Sigma-Aldrich), and then stained for 10 min with Hoechst 33342 (0.2 μg/ml) at 37°C. Mitotic cells were then collected by gentle pipetting, washed once, and resuspended in 75 mM KCl. After 10 min at room temperature, swelling cells were cytocentrifuged on slides (3 min at 500 rpm; Cytospin 3; Shandon Scientific, Cheshire, England, United Kingdom). The slides were then observed by epifluorescence microscopy either immediately or after the addition of phosphate-buffered saline (PBS) containing 20% glycerol.
For the analysis of fixed cells, transfected HeLa cells were transferred on glass coverslides 16 h after transfection and grown for an additional 24–48 h. Then, the cells were washed once in prewarmed PBS and treated at room temperature either by 4% paraformaldehyde in PBS or by 1% formaldehyde in PBS for 10 min.
Fluorescence Loss in Photobleaching (FLIP) Experiments
FLIP experiments were carried out on a TCS-SP confocal microscope (Leica, Wetzlar, Germany) by using the 488-nm line of an Ar laser (20 mW nominal output, beam width at specimen 0.2 μm, detection 500–575 nm) as described previously (Phair and Misteli, 2000). In brief, five single scans were acquired, followed by a series of bleach pulses of 200–500 ms by using a spot of 1 μm in radius followed by imaging scanning. For imaging, the laser power was attenuated to 1% of the bleach intensity. The bleach/scanning iterations were separated by 6-s intervals. Relative loss of fluorescence was determined as described previously (Phair and Misteli, 2000).
Western Blot Analysis
Western blot analyses were performed 24–48 h after transfection as described previously (Marechal et al., 1999). EGFP fused proteins were detected with a 1:1000 dilution of the mouse JL-8 monoclonal antibody (BD Biosciences Clontech). Human HMGB1 was detected with a 1:3000 dilution of a rabbit antiserum (Falciola et al., 1997). Detection of the primary antibodies was performed with a 1:10,000 dilution of a peroxidase-conjugated anti mouse or anti rabbit IgG polyclonal antibody (Amersham Biosciences UK). Proteins were detected by chemiluminescence according to the manufacturer's recommendations (ECL Western blotting detection reagents; Amersham Biosciences UK).
Cell Permeabilization Assay
Cells were cultured on a glass coverslip in a 6-cm dish; the coverslip was inserted into a coverslip dish assembly (Harvard/Medical Systems, Holliston, MA) containing 500 μl of PBS. Cells were permeabilized by adding NP-40 to a final concentration of 0.15% while imaged on an inverted microscope (Axiovert 135 M; Carl Zeiss), and sequential images were collected.
For Western blot analysis of the soluble and insoluble fractions, the cells were transfected in a 6-cm dish. Twenty-four hours after the transfection, the cells were washed once in PBS and incubated for 5 min in 500 μl of PBS containing 0.1% NP-40. The soluble and insoluble fractions were separated by centrifugation (20,000 × g; 10 min; 4°C). The pellet was resuspended in 500 μl of denaturing solution (50 mM Tris-HCl, 2% SDS, 2% β-mercaptoethanol) and incubated at 90°C for 3 min. Ten micrograms of soluble proteins and a comparable volume of the insoluble fraction were subjected to SDS-PAGE and Western blot analysis. The membrane was first probed with the antibody directed against EGFP. After detection, the primary and secondary antibodies were completely removed by incubating the membrane in a stripping solution (100 mM β-mercaptoethanol, 2% SDS, 62.3 mM Tris-HCl, pH 6.7) for 30 min at 50°C. The membrane was washed in PBS, blocked for 1 h, and reprobed with a rabbit antiserum directed against human HMGB1.
HoxD9 Transactivation Assay
Cells (200,000) were transfected with 1.5 μg of reporter plasmid (pTHCR), 1 μg of pSGD9, increasing amounts (0–2 μg) of constructs expressing HMGB1 or HMGB1-GFP (pHMGB1 or pEGFP-HMGB1), and 300 ng of pRLnull as an internal control. Transfection was carried out in triplicate batches. Forty-eight hours after transfection, cells were harvested and luciferase activities were measured using the dual-luciferase reporter assay system (Promega, Madison, WI) and Lumino luminometer (Stratec Biomedical Systems, Birkenfeld, Germany).
RESULTS
HMGB1/2 Fused to EGFP Interacts with Mitotic Chromosomes in Living Cells
EBNA1 is a viral protein that interacts with cellular chromosomes during interphase and mitosis (Petti et al., 1990). Importantly, its chromosome binding domains (CBDs) are independent from the DNA binding domain (DBD), which suggests that EBNA1 may be recruited onto chromosomes by one or several cellular proteins (Marechal et al., 1999; Shire et al., 1999). Preliminary results from a double-hybrid screening identified HMGB2 as a potential cellular partner of EBNA1, raising the possibility that HMGB2 could recruit EBNA1 onto mitotic chromatin. However, the association of HMGB2 itself with mitotic chromosomes was unknown, and results from one of our laboratory indicated that the closely related protein HMGB1 was not associated with mitotic chromosomes (Falciola et al., 1997). To examine this issue in greater detail, we analyzed the localization of HMGB proteins fused to EGFP in living HeLa cells.
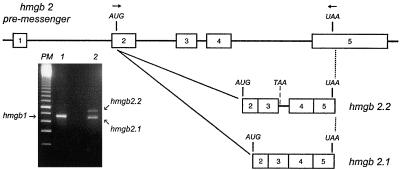
For this purpose, cDNAs encompassing human HMGB2 were generated by reverse transcription from polyadenylated RNAs and amplified by PCR. Two cDNAs were amplified, cloned into pEGFP-N1 and sequenced (Figure 1). Sequence analysis indicated that hmgb2.1 resulted from a mature mRNA, whereas hmgb2.2 contained an intron between exons 3 and 4. After transfection in HeLa cells, both expression vectors gave rise to fusion proteins of comparable fluorescence, cellular localization and mobility on SDS-PAGE (our unpublished data), indicating that the intron had been spliced from primary transcripts originating from the longer expression construct. Therefore, only the construct containing hmgb2.1 was used for further experiments.
Figure 1.
Cloning of human HMGB cDNAs. HMGB1 and HMGB2 cDNAs were amplified by PCR from reverse transcribed polyadenylated RNAs. The position of the primers used to amplify HMGB2 is indicated by arrows on the premessenger. Numbered open boxes represent exons. Whereas a single cDNA was detected for HMGB1 (lane 1), two cDNAs (hmgb 2.1 and hmgb 2.2) were amplified for HMGB2 (lane 2). Sequence analysis indicated that hmgb1 and hmgb 2.1 corresponded to the expected mature mRNA, whereas hmgb 2.2 arose from a partly spliced RNA which contained an intron between exon 3 and 4 (open boxes).
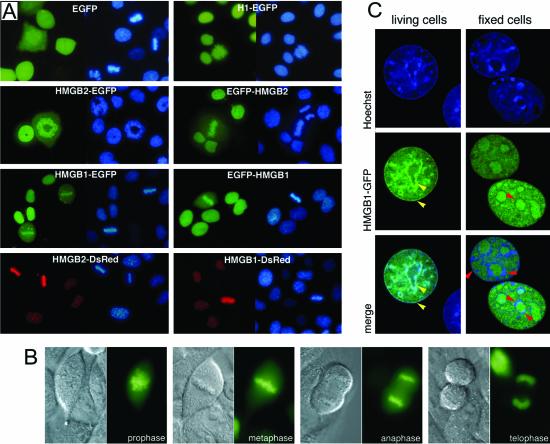
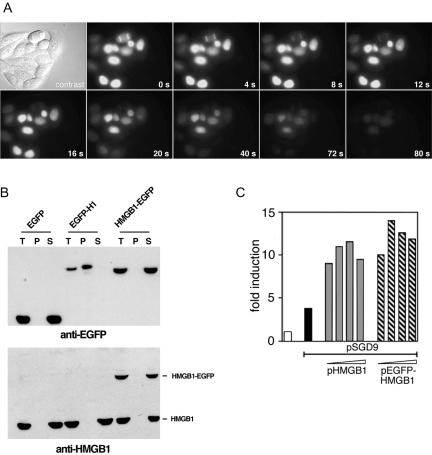
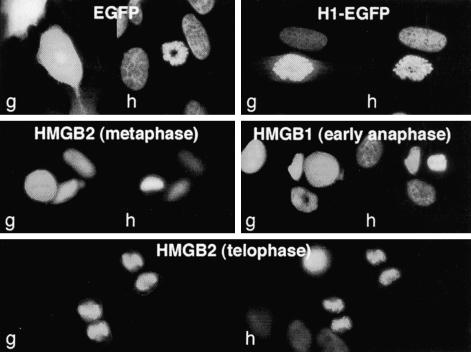
The subcellular localization of HMGB2 fused to EGFP was assessed by fluorescence microscopy in living cells from 48–72 h after transfection. In interphase cells, HMGB2-EGFP localized in the nucleus (Figures 2 and 3). In mitotic cells, HMGB2-EGFP colocalizes with the condensed chromatin, although some fluorescence was also visible in the cytoplasm (Figure 2). Western blot analysis indicated that the cytoplasmic fluorescence was not attributable to free EGFP (Figure 6C). This localization contrasts with that of unfused EGFP, which did not interact with mitotic chromosomes, and with that of histone H1-EGFP, which was exclusively localized to the chromatin both in interphase and in mitotic cells (Figure 2A). To evaluate the effect of the fluorescent tag and of its position in the fusion protein, additional vectors were constructed that encoded HMGB2 fused to the C termini of EGFP and to the C and N terminus of the DsRed protein. As shown on Figure 2A, HMGB2 binding to mitotic chromosomes was not affected by the presence of EGFP at its N terminus. Similarly, DsRed-tagged HMGB2 was also capable of interacting with mitotic chromosomes. However, DsRed protein is known to oligomerize to form tetramers, which may be not compatible with the intrinsic properties of the proteins to which it is fused (Baird et al., 2000). For this reason, subsequent experiments were mainly done with EGFP-fused proteins.
Figure 2.
HMGB proteins interact with human and mouse chromosomes. (A) Living HeLa cells expressing EGFP or histone H1, HMGB2, HMGB1 fused to EGFP (green) or to DsRed (red) were observed by fluorescent microscopy. DNA was stained with Hoechst 33342 (blue). Whereas EGFP does not interact with mitotic chromosomes, H1-EGFP strictly colocalized with condensed chromatin during mitosis. HMGB2 binding to mitotic chromosomes was observed independently of the N- or C-terminal position of EGFP. HMGB2 binding was also observed in the context of DsRed fusions. Similar observations were done for HMGB1 tagged with either EGFP or DsRed to its N or C termini. (B) HeLa cells were transfected by an expression vector encoding HMGB1-EGFP. Binding was observed during early to late phases of mitosis. Similar observations were done for HMGB2 (our unpublished data). (C) Mouse 3T3 cells were transfected by an expression vector encoding HMGB1-EGFP (green). DNA was stained by Hoechst 33342 (blue). In living cells, HMGB1 localized to the nucleus and was not excluded from the nucleolus. It colocalized to most heterochromatin containing regions (yellow arrowheads). In paraformaldehyde-fixed cells, HMGB1-EGFP detached from Hoechst-bright spots (red arrowheads) and remained in the nucleolus.
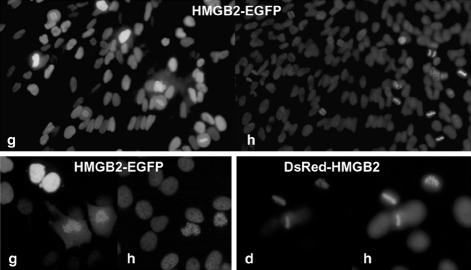
Figure 3.
HMGB2 fused to EGFP or to DsRed interact with mitotic chromosomes 8 wk after transfection. HeLa cells were transfected by pHMGB2-EGFP or pDsRed-HMGB2, grown for 3 wk in the presence of geneticin and then for 5 wk in the absence of selection. Fluorescent foci were cloned from 3 to 5 times. Positive foci contained cells expressing undetectable to high level of the fluorescent protein (top, low magnification). In all instances, HMGB2-EGFP (g) interacts with mitotic chromosomes. Similar results were obtained with cloned HeLa cells expressing DsRed-HMGB2 (d) (bottom, high magnification). DNA was stained with Hoechst 33342 (h). Note that DsRed-HMGB2 colocalized exclusively with the condensed chromosomes, whereas HMGB2-EGFP was also detected in the cytoplasm.
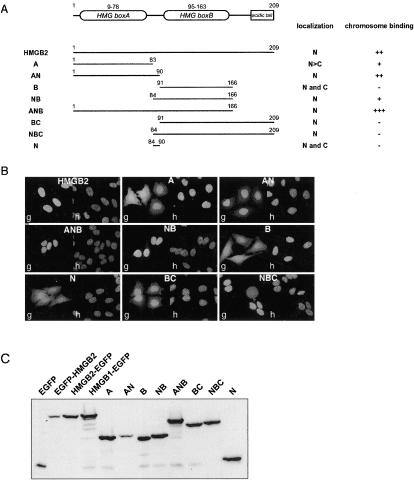
Figure 6.
Expression and localization of HMGB deletion mutants. (A) Schematic representation of HMGB2 and truncated forms. The nuclear (N) or cytoplasmic localization (C) as well as their binding to mitotic chromosomes is indicated. (B) Cellular localization of HMGB2 and truncated forms fused to EGFP (g) in HeLa cells. Cellular DNA was counterstained with Hoechst 33342 (h). (C) Expression of HMGB2, HMGB1 and various deletion mutants of HMGB2 fused to EGFP. Ten micrograms of total protein extract was subjected to SDS-PAGE and analyzed by Western blot with an antibody directed against EGFP.
HMGB1 is highly homologous to HMGB2 but may not be completely redundant to it functionally (Calogero et al., 1999; Ronfani et al., 2001). It was therefore important to test whether binding to condensed chromosomes was a common property of HMGB proteins. A unique cDNA, whose sequence corresponded to the expected mature HMGB1 mRNA, was obtained from polyadenylated RNA by reverse PCR (Figure 1) and cloned into EGFP and DsRed expression vectors. As shown on Figure 2, transfections of vectors encoding HMGB1 fused either to EGFP or to DsRed confirmed that HMGB1 associates to condensed chromosomes as well; this property was independent of the position and of the nature of the fluorophore. Importantly, HMGB1 and HMBG2 interact with condensed chromosomes from early to late phases of mitosis (Figure 2B; our unpublished data). Experiments were also performed in mouse 3T3 cells, where it is possible to analyze subnuclear compartments in more detail.
Figure 2C shows mouse fibroblast fixed with PFA (right): HMGB1-GFP is almost excluded from heterochromatic blocks close to nucleoli and to the nuclear membrane (red arrowheads). In living cells (left), the distribution of HMGB1 overlaps rather completely the Hoechst staining, confirming that the protein roams the whole nucleus, including heterochromatic AT-rich regions (yellow arrowheads). In contrast, HMGB1 seems to be less concentrated in nucleoli of living cells, compared with fixed ones. These results would suggest that PFA is able to fix better HMGB1 on chromatin where it is less compacted, such as euchromatin. However, in vivo HMGB1 seems to be distributed both in euchromatin and heterochromatin.
HMGB Proteins Fused to EGFP or to DsRed Associate with Mitotic Chromosomes in Stably Transfected Cell Lines
The present findings conflict in part with the previous observation that HMGB1 could not be detected on condensed chromosomes during mitosis (Falciola et al., 1997). However, the present study was performed in living cells that were transiently transfected with HMGB proteins fused to fluorescent proteins, whereas the previous study was performed on endogenous HMGB1 in fixed cells.
In a first attempt to solve this discrepancy, we wondered whether transient transfections might induce an abnormal interaction of HMGB proteins with mitotic chromosomes. For example, if the binding of endogenous HMGB proteins to mitotic chromosomes is normally prevented by posttranslational modification(s), transient transfections may result in the rapid accumulation of an unproperly modified protein that, in contrast to its endogenous counterpart, might be able to interact with mitotic chromosomes. If true, binding of HMGB2 (or HMGB1) should not be observed in stable cell lines, or when the proteins are expressed at a low level.
To test this hypothesis, HeLa cells were transfected with pHMGB2-EGFP or with pHMGB2-DsRed and grown for 3 wk in the presence of geneticin. Fluorescent foci were then cloned and grown for five additional weeks in the absence of geneticin. Although the positive foci were repeatedly subcloned, a large cell-to-cell variation in the fluorescence level was noted, which likely reflects cell-to-cell fluctuation in the expression of the fusion protein (Figure 3). Because stable clones expressing EGFP homogeneously were obtained in the same experiment, this suggests that the cells contain integrated vectors, but that the expression of the fusion proteins may be modulated by epigenetic events and/or counterselected. Nonetheless, a specific association of HMGB2 with mitotic chromosomes was still observed, and this interaction was not dependent on the expression level of the fusion protein. Similar results were obtained for HeLa clones expressing EGFP- and DsRed-tagged HMGB1 for >2 mo (our unpublished data). Therefore, we concluded that the interaction of HMGB proteins with mitotic chromosomes is not linked to their transient expression.
EGFP Does Not Significantly Alter Known Biological Properties of HMGB Proteins
We next examined whether EGFP-tagged HMGB proteins behave like their endogenous counterparts. Previous work had shown that endogenous HMGB1 interacts only weakly with mitotic and interphase chromatin. As a consequence, HMGB1 rapidly diffuses in the extracellular environment when the cellular membranes are permeabilized (Falciola et al., 1997). HeLa cells transfected with an expression vector encoding EGFP-fused HMGB2 or HMGB1 were exposed to a low level of NP-40. After the addition of detergent, both EGFP-tagged HMGB1 (Figure 4A) and HMGB2 (our unpublished data) rapidly diffuse out of interphase and mitotic cells, as previously observed for endogenous HMGB1.
Figure 4.
EGFP does not alter known properties of HMGB1. (A) Leakage of HMGB1-EGFP from permeabilized cells. HeLa cells expressing HMGB1-EGFP were permeabilized with NP-40 directly on the microscope, and sequential images were collected before and after detergent addition. Less than 1.5 min is sufficient to lose the entire complement of HMGB1 upon permeabilization. (B) HMGB1 and HMGB1-EGFP diffuse similarly upon cell permeabilization. HeLa cells expressing EGFP, EGFP-H1, or HMGB1-EGFP were incubated in the presence of PBS plus NP40 for 5 min. An equivalent volume of soluble (S) and insoluble (chromatin bound) (P) proteins was analyzed by Western blot by using an antibody directed against EGFP. The membrane was then reprobed with an antibody directed against human HMGB1. A comparable amount of total proteins was also analyzed (T). HMGB1 and HMGB1-EGFP were exclusively detected in the soluble fraction, indicating that both proteins similarly diffuse out of the cells.(C) The fusion to EGFP does not impair the transactivational activity of HMGB1. HeLa cells were transfected with fixed amounts of the reporter plasmid pTHCR and pSGD9, and increasing amounts of either pHMGB1 or pEGFP-HMGB1. Both HMGB1 and HMGB1-EGFP enhance HOXD9 transcriptional activity in transient cotransfection assays.
This was further confirmed by comparing the diffusion of the EGFP-tagged and endogenous HMGB in a permeabilization assay. Briefly, living HeLa cells expressing HMGB1-EGFP were incubated in the presence of NP-40 for 5 min. Then the soluble and insoluble fractions were separated by centrifugation and their protein content was analyzed by Western blot (Figure 4B). In contrast to EGFP-H1, which was detected in the insoluble fraction only, HMGB1-EGFP as well as the endogenous HMGB1 was exclusively detected in the soluble fraction. Therefore, HMGB1 and HMGB1-EGFP diffuse similarly in a permeabilization assay. Complementary experiments indicated that HMGB2 and HMGB2-EGFP behaved similarly and that this was not dependent of the salt concentration in the permeabilization buffer (our unpublished data).
Another experiment was performed in order to test whether EGFP affected the ability of HMGB1 to enhance HOXD9-controlled transcriptional activation. Figure 4C shows that EGFP-HMGB1 can enhance the transcriptional activity from an HOXD9-dependent promoter like the untagged HMGB1 protein.
Together, these experiments provide evidences that EGFP does not significantly alter known biological properties of HMGB proteins.
HMGB Proteins Rapidly Exchange between Chromosome and Cytoplasm during Mitosis
A significant amount of fluorescence was detected in the cytoplasm of mitotic cells expressing EGFP-fused HMGB1 or HMGB2. This could be observed both after short- and long-term expression of the fusion protein. Western blots did not indicate the presence of major EGFP-tagged breakdown products, and we concluded that during mitosis HMGB1 and HMGB2 exchange between a free and a chromosome-associated form. To prove this, we performed FLIP experiments.
Photobleaching techniques are noninvasive microscopy methods that reveal the dynamics underlying the steady-state distribution of a fluorescently tagged protein in living cells. A fluorophore within a small volume of the cell is irreversibly destroyed with a high-intensity laser pulse. After bleaching, the labeled protein is photochemically altered, so that it no longer fluoresces, but otherwise retains completely its biological activity (Tsien and Waggoner, 1995). The exchange between the bleached and unbleached populations of fluorophores is then monitored and used as an indicator of the overall mobility of the protein.
In FLIP, a region of interest is repeatedly bleached, and the loss of fluorescence from outside the bleached region is monitored by imaging after each bleach pulse. FLIP studies are particularly useful when the protein is not uniformly distributed but concentrated in defined intracellular sites, because it visualizes the flux between populations of fluorophores localized in different regions. In mitotic cells, HMGB1 is mostly associated with chromosomes, but it is also clearly visible in the cytoplasm. In this situation, it is possible to discriminate between soluble and chromatin-bound molecules. We performed FLIP experiments by targeting either the condensed chromosomes, or the cytoplasm. In both cases, we recorded images of the whole cell (Figure 5A) and estimated the residual amount of fluorescence remaining both on chromosomes and in the cytoplasm (Figure 5B). We found that in both cases (chromosomes and cytoplasm) repeated photobleaching led to a rapid and complete loss of fluorescence both from condensed chromosomes and from the cytoplasm, with comparable kinetics. This result indicates that all HMGB1 molecules are highly dynamic and rapidly shuttle between the cytoplasm and chromosomes. The concentration of HMGB1 on chromosomes therefore is not due to a static binding, but rather is the result of a steady state in which HMGB1 rapidly and continuously associates and dissociates on DNA.
Figure 5.
The cytoplasmic and chromosome associated forms of HMGB1 can rapidly exchange. (A) FLIP on a cytoplasmic region of a mitotic cell expressing HMGB1-EGFP (a). The area indicated by a circle was repeatedly bleached for 200 ms, and cells were imaged between bleach pulses. The loss of fluorescence was fast and complete in the entire cell, including the condensed chromosomes. There is interchange between chromosomal and cytoplasmic pools of HMGB1-EGFP, and a rapid dissociation of the protein from condensed chromatin. FLIP on mitotic cells expressing EGFP-H1 (b). A cytoplasmic region (cell on the left) and a region on condensed chromosomes (cell on the right) of two mitotic cells were repeatedly bleached for 200 ms, and cells were imaged between bleach pulses. Bleaching in the cytoplasm did not lead to loss of fluorescence on chromosomes, whereas bleaching on chromosomes lead to a slow loss of fluorescence in the regions near the bleached spot. (B) FLIP quantitation in living HeLa cells expressing EGFP-H1 or HMGB1-EGFP.
Control experiments using H1-EGFP confirmed that histone H1 is stably associated to condensed mitotic chromosomes, and exchanges very slowly with the surrounding cytoplasm.
The Chromosome Binding Regions of HMGB2 Encompass HMG Boxes A and B
In a first attempt to investigate the molecular details of HMGB interaction with mitotic chromatin, we evaluated the ability of various truncated forms of HMGB2 to address EGFP onto mitotic chromosomes. A schematic representation of these proteins is shown in Figure 6A. Western blot analysis indicated that these proteins were expressed in HeLa cells with minor amounts of breakdown products (Figure 6C). Therefore, an apparent absence of binding was not attributable to the presence of free EGFP.
Recent work in one of our laboratories suggested that some proteins need to reach the nucleus during interphase to be able to interact with chromatin during mitosis (Piolot et al., 2001). As shown on Figure 6B, HMGB2-EGFP–truncated forms were either strictly or partly nuclear and therefore competent to bind to chromosomes during mitosis.
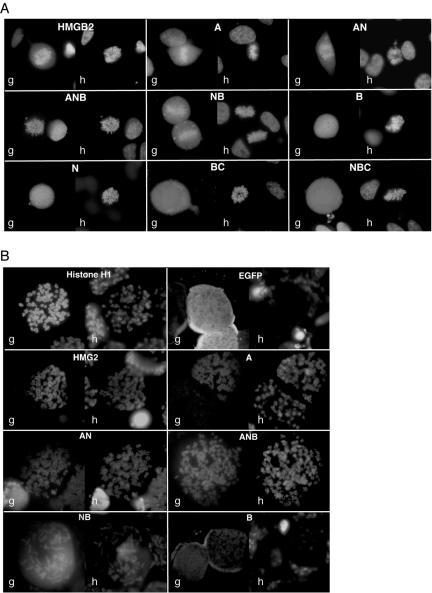
In a first set of experiments, the chromosome binding properties of these proteins were assessed in living HeLa cells 48–72 h after transient transfection. As indicated in Figure 6A and illustrated on Figure 7A, binding was observed for truncation A, which encompasses the HMG box A. Binding was notably increased by the addition of regions N and B and truncation ANB exhibited a chromosome binding activity that was even stronger than that of the wild-type protein, as estimated from the relative level of fluorescence on the chromosomes and in the cytoplasm. Surprisingly, HMG box B did not significantly target EGFP onto mitotic chromosomes, except when the N region was also present. Therefore, although amino acids 84–90 did not relocate EGFP to the mitotic chromosomes, they seemed 1) to be necessary for the binding of box B and 2) to increase binding of box A. The apparent lack of binding observed for mutant NBC argued for a negative effect of the acidic tail, at least in the context of EGFP-fused proteins.
Figure 7.
Two regions are responsible for HMGB2 binding to mitotic chromosomes. (A) Analysis of the chromosome binding properties of HMGB2 truncated forms fused to EGFP (g) in living HeLa cells. Cells were observed in culture medium 72 h after transfection. DNA was counterstained by Hoechst 33342 (h). (B) Analysis of the chromosome binding properties of HMGB2 derivatives fused to EGFP (g) after chromosome spread. HeLa cells were arrested in mitosis by a 16-h treatment in the presence of colchicine. DNA was counterstained by Hoechst 33342 (h). Mitotic cells were then subjected to a hypotonic shock and cytocentrifuged on microscopic slides. Binding to mitotic chromosomes was assessed immediately either in the absence of mounting medium (EGFP and mutant B) or in the presence of PBS plus 20% glycerol. Note that Hoechst staining was barely detectable in the absence of mounting medium. Unbound proteins rapidly diffuse in the mounting medium, whereas bound proteins were observed for up to 1 h.
This assay was convenient to assess the possible interaction of HMGB2 mutants with mitotic chromosomes. However, it may not be able to detect weak binding, such as in the case of mutants B, BC, NBC, and N. To circumvent this possible limit, complementary studies were performed on mitotic cells subjected to a mild chromosome spreading that does not involve fixation. Briefly, HeLa cells were transfected as described above and incubated for 16 h in the presence of colcemid. Mitotic cells were collected by gentle pipetting, subjected to a hypotonic shock, and centrifuged onto glass slides. Chromosome binding was then immediately assessed either in the absence of any mounting medium or in the presence of PBS containing 20% glycerol. Figure 7B shows that this treatment did not affect H1–EGFP interaction with metaphase chromosomes. EGFP was detected in the cytoplasm of the transfected cells in the absence of mounting medium, but rapidly diffused away from most cells when the mounting medium was added. Overall, the observations made with the various truncated forms of HMGB2 were in agreement with the results obtained in living cells. Notably, it was confirmed that mutants N, B, NBC, and BC did not significantly interact with chromatin in mitotic cells.
As a conclusion, HMGB2 binding to mitotic chromosomes is mediated by two regions that encompass but are not strictly identical to HMG box A and B. Because HMGB1 and HMGB2 are highly homologous, it is very likely that corresponding regions are responsible for HMGB1 chromosome binding activity.
HMGB Protein Interaction with Mitotic Chromosomes Is Abrogated by Cross-linking Fixatives
Because the previous experiments established that HMGB1 and HMGB2 interact with mitotic chromosomes in living cells, we suspected that the previously reported release of HMGB1 from mitotic chromosomes might be due to the procedure that was used, i.e., immunofluorescence on paraformaldehyde-treated cells. Because paraformaldehyde does not abrogate EGFP fluorescence, its effect on HMGB protein binding could be investigated. HeLa cells were transfected by vectors encoding HMGB1- or HMGB2-EGFP, grown on glass coverslides for 48 h, and incubated in the presence of PBS containing 4% paraformaldehyde while being observed under the microscope. As early as 1 min after paraformaldehyde addition, HMGB1- and HMGB2-EGFP mostly diffused away from chromosomes of mitotic cells. This resulted in the absence of any detectable signal on the chromosomes after a 10-min incubation (Figure 8). Nevertheless, it should be noticed that HMGB were detected in the vicinity of condensed chromosomes in late phases of mitosis, such as in telophase (Figure 8). In contrast, H1-EGFP binding was remarkably stable. Similar results were obtained when the cells were treated with 1% formaldehyde (FA) (our unpublished data), a cross-linking fixative that is broadly used to investigate protein–protein and protein–DNA interactions. Paraformaldehyde was also shown to alter the distribution of HMGB proteins in interphase cells, where it induces nucleolar concentration and loss of colocalization with some heterochromatin rich regions (Figure 2C).
Figure 8.
HMGB interaction with mitotic chromosomes is disrupted by paraformaldehyde. HeLa cells expressing EGFP or histone H1, HMGB1, and HMGB2 fused to EGFP were incubated for 10 min in the presence of PBS containing 4% paraformaldehyde. As soon as 1 min after the beginning of the treatment, HMGB proteins dissociate from the condensed chromatin, whereas H1-EGFP stayed stably associated even after prolonged incubation. Note that HMGB proteins were detected in the vicinity of telophase chromosomes even in fixed cells.
Thus, we concluded from these experiments that 1) HMGB proteins interact with mitotic chromosomes in living cells and 2) this interaction is highly sensitive to cross-linking agents commonly used to investigate the subcellular localization of proteins or their interaction with DNA.
Other HMG Proteins Interact with Mitotic Chromosomes in Living Cells but Not in Paraformaldehyde-fixed Cells
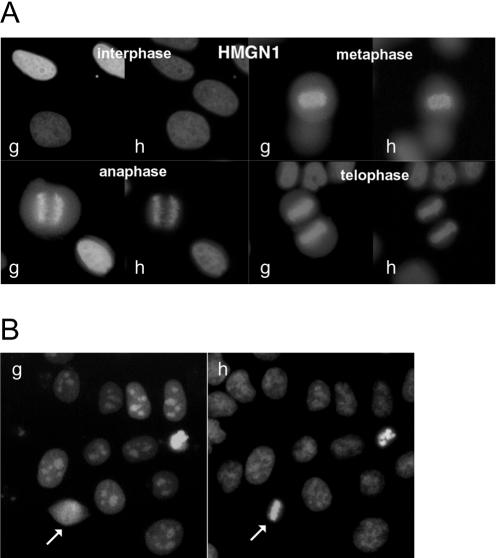
The HMGA group of proteins do interact with metaphase chromosomes (Disney et al., 1989; Saitoh and Laemmli, 1994). Conversely, proteins of the HMGN group have not been detected on mitotic chromosomes (Hock et al., 1998). Because these studies were also done by immunofluorescence on paraformaldehyde-fixed cells, we wondered whether fusion to EGFP or DsRed might indeed reveal that HMGN proteins are associated with mitotic chromosomes in living cells. As illustrated by Figure 9, HMGN1-EGFP was detected in association with the chromosomes at every stage of mitosis. This behavior was also observed for HMGN2-EGFP and did not depend either on the position or on the nature of the fluorophore (our unpublished data). In contrast to HMGB proteins, only minor amounts of protein were detected in the cytoplasm during mitosis. The effect of paraformaldehyde on HMGN proteins was also investigated and proved to induce the release of most if not all the protein from the chromosomes (Figure 9B).
Figure 9.
HMGN1-EGFP binding to mitotic chromosomes is observed in living cells, not in paraformaldehyde-treated cells. (A) HeLa cells were transfected by an expression vector encoding HMGN1 fused to the N terminus of EGFP (g). DNA was counterstained by Hoechst 33342 (h). Living cells were observed 72 h after transfection by fluorescent microscopy. HMGN1 was associated with mitotic chromosomes from early to late phases of mitosis. (B) Binding of HMGN1 to mitotic chromosomes was disrupted when the cells were treated for 10 min by PBS containing 4% paraformaldehyde (white arrow). Note that HMGN1 concentrated in nucleolar areas in paraformaldehyde-treated cells.
DISCUSSION
Ongoing experiments in one of our laboratories suggest that EBNA1, a protein encoded by the Epstein-Barr virus that is known to associate with cellular chromatin during interphase and mitosis, interacts with HMGB2. We then suspected that HMGB2 might contribute to EBNA1 interaction with mitotic chromosomes. To test this hypothesis, HMGB1 and 2 subcellular localization was investigated in living HeLa cells by using EGFP- and DsRed-tagged proteins. HMGB2 turned out to be a component of mitotic chromosomes, although it exchanges rapidly between a soluble and a chromatin-bound form. This is all the more significant because HMGB1, which is highly homologous to HMGB2, was described to interact weakly with chromatin and was not detected on mitotic chromosomes. We also show herein that HMGB1 behaves indistinguishably from HMGB2 and that previous conclusions were based on paradoxical effects of cross-linking agents such as PFA. Finally, we show that the artifacts created during PFA cross-linking might lead to the conclusion that proteins of the HMGN family dissociate from mitotic chromosomes, whereas HMGN-EGFP fusions clearly indicate that HMGN1 and HMGN2 are tightly chromosome associated.
During interphase, HMGB1- and HMGB2-EGFP fusions were fairly homogeneously distributed in the cell nucleus of human cells, including the nucleolus, and occasionally colocalized with DNA-rich regions. In contrast with human cells where the border between euchromatin and heterochromatin is not very clear, heterochromatin occurs in the form of well defined chromatin blobs with a high concentration of DNA in mouse cells. The use of an EGFP-tagged protein in living mouse fibroblasts indeed confirmed that HMGB protein localizes with both euchromatin and heterochromatin. Whether binding to certain heterochromatic regions reflects the ability of some HMGB proteins to form a complex with SP100B and HP-1, a protein that is also found predominantly in heterochromatin, has still to be determined (Lehming et al., 1998). During mitosis, both human HMGB1 and 2 associate with condensed chromosomes in living HeLa cells. In contrast to histone H1, which is stably and exclusively associated with chromosomes, HMGB1 and 2 were also detected in the cytoplasm, and evidence is provided herein that the bound and unbound forms of HMGB1/2 can rapidly exchange. This high mobility of HMGB1 has also recently been observed in interphase cells (Scaffidi et al., 2002)
Two CBDs have been identified in HMGB2. The first one encompasses HMG box A (aa 1–83), but amino acids 84–90 are required to confer a chromosome binding activity comparable with the entire protein. The second CBD mapped between amino acids 84–166. It comprises HMG box B (aa 91–166) but was strictly dependent on the presence of amino acids 84–90 for binding. The HMG boxes are defined as the DBDs of HMGB proteins; therefore, DBDs and CBDs do not coincide exactly. In vitro, the regions immediately C-terminal to the HMG boxes have been shown to increase the DNA-binding activity to some forms of DNA, including supercoiled DNA (Stros, 2001). Importantly, we also noticed that the C-terminal acidic tail of HMGB2 exerts a negative effect on chromosome binding, either in association with box B or in the context of the full-length protein (compare NB with NBC and ANB to full-length protein on Figure 6). Together, these results are reminiscent of a recent investigation of HMGB1 interaction with linear duplex DNA where it was shown that 1) box A has a higher affinity than box B for double-stranded linear DNA in solution, 2) box A and B behave as independent domains, and 3) the acidic tail reduces DNA binding affinity, possibly by interacting with box B (Muller et al., 2001a).
Together, these results indicate that HMGB proteins directly interact with chromatin in mitotic chromosomes. Although the nature of this interaction is still elusive, indirect evidence suggests that HMGB and histone H1 may share a common role in organizing higher order chromatin structure (Nightingale et al., 1996; Bustin, 1999). Notably, both proteins bind to linker DNA (Ura et al., 1996), four-way junctions or cis-platined DNA in vitro (Hill and Reeves, 1997; Yaneva et al., 1997). However, the affinity of histone H1 for these structures is much higher than HMGB in vitro, suggesting that the binding of HMGB proteins may occur in vivo only where histone H1 is absent. Accordingly, HMGB proteins have been shown to interact with mitotic chromosomes during Xenopus laevis early embryogenesis (Dimitrov et al., 1993; Dimitrov et al., 1994) until histone H1 accumulates. A related protein, HMG-D, is also expressed early in Drosophilia melanogaster development (Ner and Travers, 1994) and associates with mitotic chromosomes only until histone H1 first accumulates in the dividing cells. In differentiated mammalian cells the concentration of histone H1 is ∼10 times higher than that of HMGB1 (Einck and Bustin, 1985). Therefore, H1 should efficiently outcompete HMGB for binding to linker DNA, and our results might seem surprising. Two nonmutually exclusive hypotheses can be proposed: 1) HMGBs interact with linker DNA regions that remain free after all histone H1 has occupied its preferred binding sites, and 2) HMGB interaction with mitotic chromosomes is mediated through other proteins bound to nonlinker DNA. The first hypothesis rests on the observation that histone H1 is substantially substoichiometric in relation to core histones, so that a large amount of nucleosomes remain H1-free. The second possibility is supported by the fact that HMGB proteins interact with several chromatin-associated proteins, including the TATA binding protein (TBP) (Ge and Roeder, 1994; Sutrias-Grau et al., 1999). TBP is associated with condensed chromosomes during mitosis (Chen et al., 2002; Christova and Oelgeschlager, 2002), and might “bookmark” previously active genes and promote their rapid reactivation after mitosis. We currently do not have evidence for a direct binding between HMGB proteins and TBP on mitotic chromosomes, but this assumption is compatible with our current knowledge of TBP–HMGB interaction. Indeed, HMGB1 interacts with the core domain of TBP through multiple regions, including HMG box A, and this results in a HMGB1/TBP/TATA complex that can modulate RNA pol II transcription (Sutrias-Grau et al., 1999).
The discrepancy between the present work and previous ones with regard to the presence of HMGB proteins on mitotic chromosomes is most likely due to the paradoxical effect of paraformaldehyde and formaldehyde. Second, we could observe chromosome associated EGFP-tagged HMGB for >10 min in cells incubated in a methanol/acetone mixture. However, the fluorescence was lost when the cells were dried and incubated in PBS, suggesting that HMGB proteins were not properly fixed by this procedure. Paraformaldehyde is extensively used to cross-link proteins to chromatin. Because formaldehyde can efficiently cross-link HMGB1/2 on nucleosomes reconstituted in vitro (Stros et al., 1985), this treatment is unlikely to directly alter preformed complexes. Rather, PFA or FA may alter the accessibility of HMGB to their target(s) by modifying the overall structure of the mitotic chromosomes, and/or inactivate the free form of HMGB1/2. HMGBs contain >40 lysine residues, and a lot of these are expected to interact directly with the charged phosphate backbone of DNA. PFA reacts with the ε amino group of lysines, and the reaction product is a Schiff base. This is still charged, but both hydrogen bonding and van der Waals contacts of the lysine residue will be disrupted.
The deleterious effect of PFA has also been demonstrated for other members of the HMG protein family. Indeed, it was initially considered that HMGN1 and 2 were released from mitotic chromosomes when cells enter mitosis (Hock et al., 1998). Actually, EGFP-HMGN1 and 2 do not dissociate from condensed chromosomes in living cells, but they are rapidly released in the cytoplasm upon treatment by PFA and FA. Altogether, our results indicate that many members of the HMG protein family, including HMGB, HMGN, and HMGI/Y, interact with chromatin during interphase and mitosis. Our results justify a careful reexamination of nuclear protein interactions with mitotic chromosomes.
Acknowledgments
We are grateful to Corinne Dutreuil for sequencing and to Emmanuelle Lepin for technical help. This work was supported by the Universite Pierre et Marie Curie (UPRES EA 3500), the Program deRecherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires, the Association pour la Recherche sur le Cancer, and the Associazione Italiana per la Ricerca sul Cancro.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02-09-0581. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-09-0581.
References
- Aidinis, V., Bonaldi, T., Beltrame, M., Santagata, S., Bianchi, M.E., and Spanopoulou, E. (1999). The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Mol. Cell. Biol. 19, 6532–6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird, G.S., Zacharias, D.A., and Tsien, R.Y. (2000). Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. USA 97, 11984–11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi, M.E. (1995). The HMG-box domain. In: DNA-Protein: Structural Interactions, ed. D. Lilley, Oxford: Oxford University Press, 177–200.
- Bianchi, M.E., and Beltrame, M. (2000). Upwardly mobile proteins. Workshop: the role of HMG proteins in chromatin structure, gene expression and neoplasia. EMBO Rep. 1, 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonyaratanakornkit, V., Melvin, V., Prendergast, P., Altmann, M., Ronfani, L., Bianchi, M.E., Taraseviciene, L., Nordeen, S.K., Allegretto, E.A., and Edwards, D.P. (1998). High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol. Cell. Biol. 18, 4471–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin, M. (1999). Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 19, 5237–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin, M. (2001). Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem. Sci. 26, 152–153. [DOI] [PubMed] [Google Scholar]
- Bustin, M., and Reeves, R. (1996). High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol. 54, 35–100. [DOI] [PubMed] [Google Scholar]
- Butteroni, C., De Felici, M., Scholer, H.R., and Pesce, M. (2000). Phage display screening reveals an association between germline-specific transcription factor Oct-4 and multiple cellular proteins. J. Mol. Biol. 304, 529–540. [DOI] [PubMed] [Google Scholar]
- Calogero, S., Grassi, F., Aguzzi, A., Voigtlander, T., Ferrier, P., Ferrari, S., and Bianchi, M.E. (1999). The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat. Genet. 22, 276–280. [DOI] [PubMed] [Google Scholar]
- Chen, D., Hinkley, C.S., Henry, R.W., and Huang, S. (2002). TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes. Mol. Biol. Cell 13, 276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christova, R., and Oelgeschlager, T. (2002). Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat. Cell Biol. 4, 79–82. [DOI] [PubMed] [Google Scholar]
- Costello, E., Saudan, P., Winocour, E., Pizer, L., and Beard, P. (1997). High mobility group chromosomal protein 1 binds to the adeno-associated virus replication protein (Rep) and promotes Rep-mediated site-specific cleavage of DNA, ATPase activity and transcriptional repression. EMBO J. 16, 5943–5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov, S., Almouzni, G., Dasso, M., and Wolffe, A.P. (1993). Chromatin transitions during early Xenopus embryogenesis: changes in histone H4 acetylation and in linker histone type. Dev. Biol. 160, 214–227. [DOI] [PubMed] [Google Scholar]
- Dimitrov, S., Dasso, M.C., and Wolffe, A.P. (1994). Remodeling sperm chromatin in Xenopus laevis egg extracts: the role of core histone phosphorylation and linker histone B4 in chromatin assembly. J. Cell Biol. 126, 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov, S., and Wolffe, A.P. (1996). Remodeling somatic nuclei in Xenopus laevis egg extracts: molecular mechanisms for the selective release of histones H1 and H1(0) from chromatin and the acquisition of transcriptional competence. EMBO J. 15, 5897–5906. [PMC free article] [PubMed] [Google Scholar]
- Disney, J.E., Johnson, K.R., Magnuson, N.S., Sylvester, S.R., and Reeves, R. (1989). High-mobility group protein HMG-I localizes to G/Q- and C-bands of human and mouse chromosomes. J. Cell Biol. 109, 1975–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einck, L., and Bustin, M. (1985). The intracellular distribution and function of the high mobility group chromosomal proteins. Exp. Cell Res. 156, 295–310. [DOI] [PubMed] [Google Scholar]
- Falciola, L., Spada, F., Calogero, S., Langst, G., Voit, R., Grummt, I., and Bianchi, M.E. (1997). High mobility group 1 protein is not stably associated with the chromosomes of somatic cells. J. Cell Biol. 137, 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, H., and Roeder, R.G. (1994). The high mobility group protein HMG1 can reversibly inhibit class II gene transcription by interaction with the TATA-binding protein. J. Biol. Chem. 269, 17136–17140. [PubMed] [Google Scholar]
- Hill, D.A., and Reeves, R. (1997). Competition between HMG-I(Y), HMG-1 and histone H1 on four-way junction DNA. Nucleic Acids Res. 25, 3523–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock, R., Scheer, U., and Bustin, M. (1998). Chromosomal proteins HMG-14 and HMG-17 are released from mitotic chromosomes and imported into the nucleus by active transport. J. Cell Biol. 143, 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman, L., Moorthy, N.C., Murthy, K.G., Manley, J.L., Bustin, M., and Prives, C. (1998). High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 12, 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns, E.W. (1982). The HMG Chromosomal Proteins, London: Academic Press.
- Lehming, N., Le Saux, A., Schuller, J., and Ptashne, M. (1998). Chromatin components as part of a putative transcriptional repressing complex. Proc. Natl. Acad. Sci. USA 95, 7322–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, W., Peterson, R., Dasgupta, A., and Scovell, W.M. (2000). Influence of HMG-1 and adenovirus oncoprotein E1A on early stages of transcriptional preinitiation complex assembly. J. Biol. Chem. 275, 35006–35012. [DOI] [PubMed] [Google Scholar]
- Marechal, V., Dehee, A., Chikhi-Brachet, R., Piolot, T., Coppey-Moisan, M., and Nicolas, J.C. (1999). Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 73, 4385–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, S., Bianchi, M.E., and Knapp, S. (2001a). Thermodynamics of HMGB1 interaction with duplex DNA. Biochemistry 40, 10254–10261. [DOI] [PubMed] [Google Scholar]
- Muller, S., Scaffidi, P., Degryse, B., Bonaldi, T., Ronfani, L., Agresti, A., Beltrame, M., and Bianchi, M.E. (2001b). New EMBO members'review: the double life of HMGB1 chromatin protein: architectural factor and extracellular signal. EMBO J. 20, 4337–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ner, S.S., and Travers, A.A. (1994). HMG-D, the Drosophila melanogaster homologue of HMG 1 protein, is associated with early embryonic chromatin in the absence of histone H1. EMBO J. 13, 1817–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale, K., Dimitrov, S., Reeves, R., and Wolffe, A.P. (1996). Evidence for a shared structural role for HMG1 and linker histones B4 and H1 in organizing chromatin. EMBO J. 15, 548–561. [PMC free article] [PubMed] [Google Scholar]
- Onate, S.A., Prendergast, P., Wagner, J.P., Nissen, M., Reeves, R., Pettijohn, D.E., and Edwards, D.P. (1994). The DNA-bending protein HMG-1 enhances progesterone receptor binding to its target DNA sequences. Mol. Cell. Biol. 14, 3376–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payet, D., Hillisch, A., Lowe, N., Diekmann, S., and Travers, A. (1999). The recognition of distorted DNA structures by HMG-D: a footprinting and molecular modelling study. J. Mol. Biol. 294, 79–91. [DOI] [PubMed] [Google Scholar]
- Payet, D., and Travers, A. (1997). The acidic tail of the high mobility group protein HMG-D modulates the structural selectivity of DNA binding. J. Mol. Biol. 266, 66–75. [DOI] [PubMed] [Google Scholar]
- Petti, L., Sample, C., and Kieff, E. (1990). Subnuclear localization and phosphorylation of Epstein-Barr virus latent infection nuclear proteins. Virology 176, 563–574. [DOI] [PubMed] [Google Scholar]
- Phair, R.D., and Misteli, T. (2000). High mobility of proteins in the mammalian cell nucleus. Nature 404, 604–609. [DOI] [PubMed] [Google Scholar]
- Piolot, T., Tramier, M., Coppey, M., Nicolas, J.C., and Marechal, V. (2001). Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75, 3948–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritt, C., Grimm, R., Fernandez, S., Alonso, J.C., and Grasser, K.D. (1998). Basic and acidic regions flanking the HMG domain of maize HMGa modulate the interactions with DNA and the self-association of the protein. Biochemistry 37, 2673–2681. [DOI] [PubMed] [Google Scholar]
- Ronfani, L., Ferraguti, M., Croci, L., Ovitt, C.E., Scholer, H.R., Consalez, G.G., and Bianchi, M.E. (2001). Reduced fertility and spermatogenesis defects in mice lacking chromosomal protein Hmgb2. Development 128, 1265–1273. [DOI] [PubMed] [Google Scholar]
- Saitoh, Y., and Laemmli, U.K. (1994). Metaphase chromosome structure: bands arise from a differential folding path of the highly AT-rich scaffold. Cell 76, 609–622. [DOI] [PubMed] [Google Scholar]
- Scaffidi, P., Misteli, T., and Bianchi, M.E. (2002). Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 418, 191–195. [DOI] [PubMed] [Google Scholar]
- Schroter, H., and Bode, J. (1982). The binding sites for large and small high-mobility-group (HMG) proteins. Studies on HMG-nucleosome interactions in vitro. Eur. J. Biochem. 127, 429–436. [DOI] [PubMed] [Google Scholar]
- Sheflin, L.G., Fucile, N.W., and Spaulding, S.W. (1993). The specific interactions of HMG 1 and 2 with negatively supercoiled DNA are modulated by their acidic C-terminal domains and involve cysteine residues in their HMG 1/2 boxes. Biochemistry 32, 3238–3248. [DOI] [PubMed] [Google Scholar]
- Shire, K., Ceccarelli, D.F.J., Avolio-Hunter, T.M., and Frappier, L. (1999). EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J. Virol. 73, 2587–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stros, M. (1998). DNA bending by the chromosomal protein HMG1 and its high mobility group box domains. Effect of flanking sequences. J. Biol. Chem. 273, 10355–10361. [PubMed] [Google Scholar]
- Stros, M. (2001). Two mutations of basic residues within the N-terminus of HMG-1 B domain with different effects on DNA supercoiling and binding to bent DNA. Biochemistry 40, 4769–4779. [DOI] [PubMed] [Google Scholar]
- Stros, M., Shick, V.V., Belyavsky, A.V., and Mirzabekov, A.D. (1985). Interaction of high mobility group proteins HMG 1 and HMG 2 with nucleosomes studied by gel electrophoresis. Mol. Biol. Rep. 10, 221–226. [DOI] [PubMed] [Google Scholar]
- Stros, M., Stokrova, J., and Thomas, J.O. (1994). DNA looping by the HMG-box domains of HMG1 and modulation of DNA binding by the acidic C-terminal domain. Nucleic Acids Res. 22, 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutrias-Grau, M., Bianchi, M.E., and Bernues, J. (1999). High mobility group protein 1 interacts specifically with the core domain of human TATA box-binding protein and interferes with transcription factor IIB within the pre-initiation complex. J. Biol. Chem. 274, 1628–1634. [DOI] [PubMed] [Google Scholar]
- Teo, S.H., Grasser, K.D., and Thomas, J.O. (1995). Differences in the DNA-binding properties of the HMG-box domains of HMG1 and the sex-determining factor SRY. Eur. J. Biochem. 230, 943–950. [DOI] [PubMed] [Google Scholar]
- Thomas, J.O. (2001). HMG1 and 2, architectural DNA-binding proteins. Biochem. Soc. Trans. 29, 395–401. [DOI] [PubMed] [Google Scholar]
- Tsien, R., and Waggoner, A. (1995). Handbook of Confocal Fluorescence Microcopy, Vol. 10, ed. J. Pawley, New York: Plenum Press.
- Ura, K., Nightingale, K., and Wolffe, A.P. (1996). Differential association of HMG1 and linker histones B4 and H1 with dinucleosomal DNA: structural transitions and transcriptional repression. EMBO J. 15, 4959–4969. [PMC free article] [PubMed] [Google Scholar]
- Webb, M., and Thomas, J.O. (1999). Structure-specific binding of the two tandem HMG boxes of HMG1 to four-way junction DNA is mediated by the A domain. J. Mol. Biol. 294, 373–387. [DOI] [PubMed] [Google Scholar]
- Yaneva, J., Leuba, S.H., van Holde, K., and Zlatanova, J. (1997). The major chromatin protein histone H1 binds preferentially to cisplatinum-damaged DNA. Proc. Natl. Acad. Sci. USA 94, 13448–13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappavigna, V., Falciola, L., Helmer-Citterich, M., Mavilio, F., and Bianchi, M.E. (1996). HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J. 15, 4981–4991. [PMC free article] [PubMed] [Google Scholar]
- Zwilling, S., Konig, H., and Wirth, T. (1995). High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 14, 1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]