Abstract
Synaptogenesis and synaptic plasticity depend crucially on the dynamic and locally specific regulation of the actin cytoskeleton. We identified an important component for controlled actin assembly, abelson interacting protein-1 (Abi-1), as a binding partner for the postsynaptic density (PSD) protein ProSAP2/Shank3. During early neuronal development, Abi-1 is localized in neurites and growth cones; at later stages, the protein is enriched in dendritic spines and PSDs, as are components of a trimeric complex consisting of Abi-1, Eps8 and Sos-1. Abi-1 translocates upon NMDA application from PSDs to nuclei. Nuclear entry depends on abelson kinase activity. Abi-1 co-immunoprecipitates with the transcription factor complex of Myc/Max proteins and enhances E-box-regulated gene transcription. Downregulation of Abi-1 by small interfering RNA results in excessive dendrite branching, immature spine and synapse morphology and a reduction of synapses, whereas overexpression of Abi-1 has the opposite effect. Data show that Abi-1 can act as a specific synapto-nuclear messenger and is essentially involved in dendrite and synapse formation.
Keywords: actin, ProSAP2, PSD, Shank3, synapse
Introduction
Communication in the central nervous system is mainly accomplished via highly specialized cellular contact sites called chemical synapses. Spinous excitatory synapses are highly dynamic and the morphology of dendritic spines can be altered within a few minutes (Hering and Sheng, 2001). These morphological alterations can be induced by synaptic activity and are thought to be the structural basis for synaptic plasticity underlying learning and memory formation (Yuste and Bonhoeffer, 2004). It is generally accepted that motility as well as dynamic changes of synaptic contacts is brought about by the reorganization of the actin and microtubule cytoskeleton via small GTPases. These GTPases are organized and regulated in micromolecular or macromolecular complexes, which localize to specific cellular microdomains (Wong et al, 2000; Ichigotani et al, 2002; Innocenti et al, 2003, 2004; Disanza et al, 2004).
One signaling complex that has been studied extensively is the multiprotein complex of the ubiquitously expressed non-receptor tyrosine kinase c-Abl, which is localized both at the cellular membrane and in the nucleus (Taagepera et al, 1998; Pendergast, 2002). In brain, non-receptor tyrosine kinases of the c-Abl family could be detected at the presynaptic membrane and in the postsynaptic density (PSD), where they regulate synaptic efficacy and synaptic plasticity, presumably through their ability to locally reorganize the actin-based cytoskeleton (Moresco and Koleske, 2003). One important interacting protein of c-Abl is the abelson tyrosine kinase substrate or abelson interactor 1, abelson interacting protein-1 (Abi-1). Abi-1 was found to be part of several macromolecular complexes, including a trimeric signaling complex, where it closely interacts with Eps8 and Sos-1. Abi-1 is essential, via the control of Rac activity, for the formation and activation of the WAVE2 signaling complex. In turn, the activated WAVE proteins lead to enhanced actin nucleation via the Arp2/3 complex (Ichigotani et al, 2002; Innocenti et al, 2003, 2004). The role and functional importance of the Abi-1 complex has been studied in various cellular systems, showing that Abi-1 proteins play a pivotal role in phosphorylation (Tani et al, 2003) and localization of protein complexes to cellular subcompartments and in actin reorganization, especially through the regulation of Rac-dependent pathways (Stradal et al, 2001; Leng et al, 2005). Recently, it has been reported that Abi-1, like c-Abl, shuttles to the nucleus in NIH 3T3 fibroblasts, pointing to a dual function of the protein in distinct cellular compartments (Echarri et al, 2004).
Spines and synapses are neuronal cell membrane protrusions that are characterized by constant remodeling processes. Several molecules can induce or influence the shape and maturation of these highly specialized contact sites (Hering and Sheng, 2001) mostly by binding and/or by regulating small GTPases (Pak et al, 2001; Ma et al, 2003). Interestingly, members of the ProSAP/Shank family of PSD scaffolding molecules can also effectively influence spine shape and size (Sala et al, 2001), and are even able to induce spines in normally aspiny neurons (Roussignol et al, 2005). Recently, it has been shown that multimers of the ProSAP/Shank adapter proteins are organized as huge macromolecular platforms in parallel to the postsynaptic membrane (Baron et al, 2006), which organize higher order receptor clustering, actin binding and reorganization as well as regulated signal transduction upon receptor stimulation (Sheng and Kim, 2000; Boeckers et al, 2002).
Here, we report that in the central nervous system Abi-1 is a synaptic molecule that is tightly bound to the PSD matrix by interaction with ProSAP2/Shank3. Interestingly, Abi-1 is not confined to the PSD but shuttles to the nucleus upon NMDA application. Moreover, we found that Abi-1 is part of a Myc/Max complex of transcription factors. The entry into the nucleus is dependent on phosphorylation at tyrosine 53. Downregulation of Abi-1 during synaptogenesis by small interfering RNAs results in increase in the number of dendrites and a decrease in synaptic sites. These synapses display a significant higher percentage of immature morphology. These results point to an essential role of the Abi-1 complex in the establishment and structural reorganization of dendrites, spines and synapses. As Abi-1 proteins that are resistant to tyrosine 53 phosphorylation and unable to enter the nucleus cannot rescue the small interference RNA (RNAi) phenotype, it is tempting to speculate that the regulation of dendritic outgrowth and synapse maturation is at least in part dependent upon the nuclear role of Abi-1 interacting with the Myc/Max transcriptional complex.
Results
Abi-1 interacts with ProSAP2/Shank3 via the C-terminal SH3 domain
In a yeast two-hybrid (YTH) screen using a conserved proline-rich region of ProSAP2/Shank3, we identified Abi-1 as a binding partner. Abi-1 is a 476-aa proline-rich protein comprising an N-terminal WAB (Wave binding) and SNARE (syntaxin binding) domain, a homeobox homology region (HHR domain) and a C-terminal SH3 domain (Figure 1A–C). In a detailed YTH assay, we identified a proline-rich consensus motif in ProSAP2/Shank3, which is responsible for the interaction with the Abi-1 SH3 domain (Figure 1A and D). A similar motif at a conserved, identical site in the protein is present in ProSAP1/Shank2 but not Shank1 (Figure 1A). The interaction via these domains could be confirmed by cotransfection experiments in Cos-7 cells using ProSAP2/Shank3, ProSAP1/Shank2 and Abi-1 deletion constructs (Figure 1E). The proline-rich segment of ProSAP2/Shank3 was recruited to Abi-1 or Abi-SH3 domain clusters upon coexpression (Figure 1E(II,III and VI)). In contrast, Shank1A coexpression, removal of the proline-rich domain of ProSAP2/Shank3 or the SH3 domain of Abi-1 abolished colocalization (Figure 1E(IV,V and VII)). To test whether the interaction between ProSAP2/Shank3 and Abi-1 is direct and occurs in vivo, we confirmed the interaction in a GST pull-down assay (Figure 1F). Finally, we could demonstrate the co-immunoprecipitation of Abi-1 from rat brain lysate using ProSAP2/Shank3 antibodies and vice versa. Moreover, the Abi-1 precipitate was found to contain ProSAP1/Shank2 and proteins of the Abi-1 complex (i.e. Eps8 and WAVE) but not MAP2 (Figure 1F).
Figure 1.
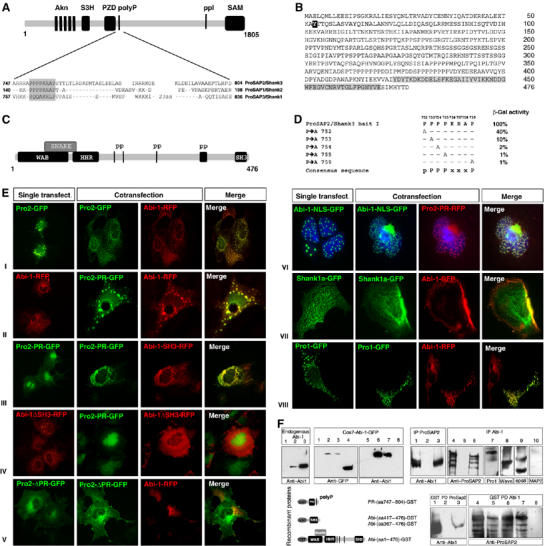
Abi-1 interacts with a proline-rich region of ProSAP2/Shank3. (A) Schematic representation of the ProSAP2/Shank3 protein showing the localization and sequence of a proline-rich region (bait) flanking the PDZ domain (conserved in ProSAP1/Shank2 and ProSAP2/Shank3). (B, C) Characteristic Abi-1 protein/protein interaction domains. The C-terminal SH3 domain is marked in gray. The tyrosine phosphorylation site (tyrosine 53) of Abi-1 is marked in black. The 476-aa Abi-1 protein is coding for proline-rich regions (PP), an N-terminal WAB and SNARE domain, a conserved Abi-1 HHR and a C-terminal SH3 domain. (D) Yeast two-hybrid assay showing that point mutations of the ProSAP2/Shank3 bait lead to a partial or complete loss of protein/protein interaction with Abi-1; pPPPxxxP was found as a consensus sequence. (E) GFP- and RFP-fusion proteins coding for ProSAP1,2/Shank2,3 or Abi-1 that are cotransfected in Cos7 cells colocalize in clusters when coding for the above-described interacting domains (I–IV and VIII). Interaction is abolished when the Abi-1 SH3 domain or the ProSAP2 proline-rich domain (PR) is deleted (IV and V); Shank1a does not cocluster with Abi-1 (VII). Permanent nuclear localization of Abi-1 by adding a nuclear localization signal (Abi-1-NLS-GFP) results in the recruitment of the ProSAP2/Shank3 proline-rich domain into the nucleus (VI). (F) Characterization of the monoclonal Abi-1 antibody used in the study. The antibody recognizes the endogenous protein in HeLA cells (lane 2), in hippocampal neurons (lane 3) but not in Cos7 cells (lane 1). Expression of different GFP proteins shows that the antibody is directed against the N-terminal part of the protein (lanes 1 and 5, untransfected Cos7 cells; lanes 2 and 6, Abi-1-GFP transfected; lanes 3 and 7, Abi-1ΔSH3-GFP transfected; lanes 4 and 8, Abi-1-SH3-GFP transfected). Co-immunoprecipitation (Co-IP) and GST pull-down experiments (PD) from rat brain lysate (5 μg of protein has been loaded) were performed. Rat brain extracts (input,1 and 4) were immunoprecipitated with Abi-1 (lanes 6–10), ProSAP2/Shank3 (lane 3) or control (IgG, lanes 2 and 5). Subsequently, immunoprecipitates were blotted for Abi-1 (lane 3) and ProSAP2/Shank3 (lane 6). These Co-IPs experiments from brain lysates show that both proteins interact in rat brain. Moreover, ProSAP1, Wave and Eps8 but not Map2 are detected in the Abi-1 precipitate (lanes 7–10). PD experiments show the direct interaction of the proline-rich region of ProSAP2 with Abi-1 in both directions.
Abi-1 and interacting proteins are widely expressed in brain and are components of the postsynaptic density complex
The expression profile of Abi-1 mRNA and protein in rat brain was analyzed by in situ hybridization and immunohistochemistry. During all stages of development, Abi-1 transcripts as well as Abi-1 protein were widely expressed in rat brain. High levels were detected in cortex, hippocampus (HC) and cerebellum (CE) (Figure 2A(I–III)). Higher magnification (Figure 2A(III)) revealed a dendritic and a punctate labeling of neuropil in all brain areas. Immunoelectron microscopy showed that Abi-1 can be detected in dendrites and at PSDs of excitatory synapses (Figure 2A(IV), inset, arrowheads). In addition, the antigen could also be identified in some cells close to the nuclear membrane and in the nucleus (Figure 2A(IV)). Postsynaptic staining of Abi-1 was confirmed by immunofluorescence staining of cultured hippocampal neurons where it completely overlaps with ProSAP2/Shank3 and colocalized with the presynaptic marker protein Bassoon (Figure 2B). Staining with antibodies directed against other components of the Abi-1 complex (Eps8, Sos-1 and WAVE) revealed that these molecules are also enriched in spines and/or PSDs (Figure 2B). After subcellular fractionation of brain tissue, ProSAP2/Shank3, Abi-1, c-Abl and WAVE were enriched within the PSD fraction, whereas Eps8 showed only a weak band in the PSD fraction. Sos-1 is not enriched within the cytoskeletal matrix of the PSD (see Figure 2C). The analysis of the spatial and temporal pattern of Abi-1 expression showed that the mRNA expression only slightly increases between days 0 and 14. Protein expression, however, rises significantly between days 1 and 3. Moreover, additional bands at higher molecular weight, reflecting phosphorylated Abi-1 (Courtney et al, 2000), are seen at later stages of neuronal development starting from day 10 onwards (Figure 2C). Immunohistochemical staining shows Abi-1 enriched in growth cones of young neurons (days 1 and 3; arrowheads). At later developmental stages (days 10 and 14), Abi-1 is finally localized in spines and synapses as shown (Figure 2B and C).
Figure 2.
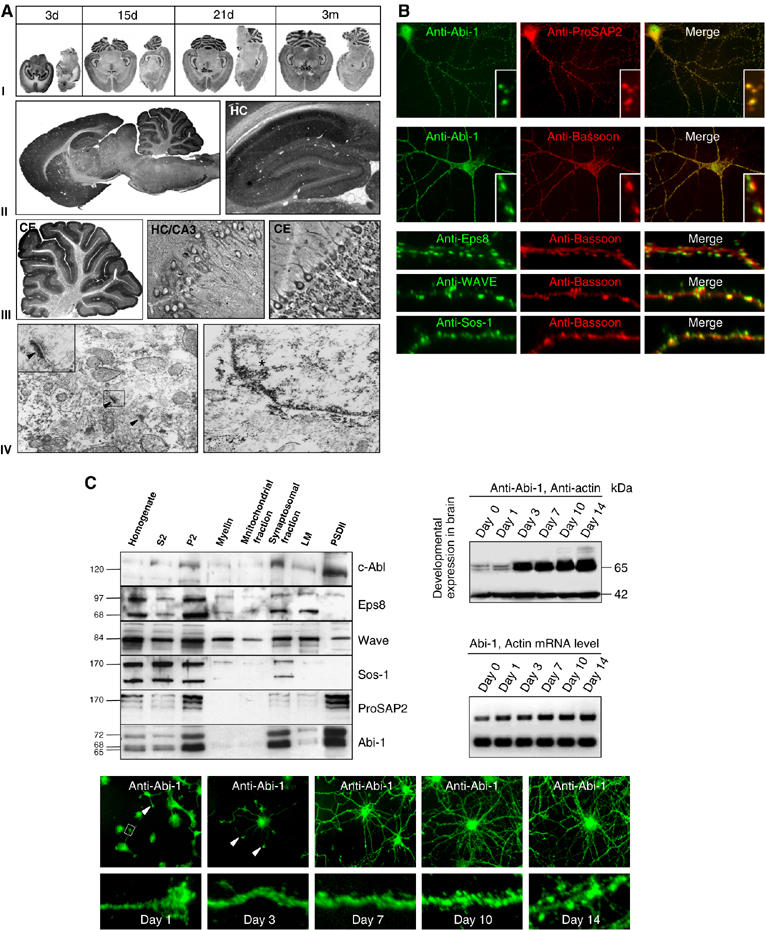
Spatial and temporal localization of endogenous Abi-1 in hippocampal neurons and rat brain. (A) In situ hybridization and immunohistochemistry during rat brain development reveals high expression levels of Abi-1, especially in cortex, hippocampus (HC) and cerebellum (CE, I–III). In the hippocampal CA3 region, an intense punctate labeling of the stratum oriens and stratum radiatum is seen. In cerebellum, granule cells are immunopositive and some nuclei are stained (III). Immunoelectron microscopy (magnification × 37 800) revealed that Abi-1 can be detected in dendrites and at PSDs (arrowheads, inset). In some neurons, the DAB/silver/gold precipitate could also be identified close to the nuclear membrane and in the nucleus (star, IV). (B) Abi-1 at synapses in hippocampal neurons. DIV21 staining of Abi-1 and ProSAP2/Shank3 reveals colocalization of these proteins at excitatory synapses. Double staining of Abi-1 (green) and Bassoon (red) shows a pattern of juxtaposed proteins at synapses. Double staining of other Abi-1 complex-forming proteins (Eps8, Sos-1 and WAVE; green) with the presynaptic marker Bassoon (red) shows the localization of these proteins at spines/PSDs. (C) Subcellular fractionation indicates the strong enrichment of c-Abl, ProSAP2/Shank3 and Abi-1 within the PSD preparation, whereas WAVE and Eps8 are only moderately attached to the PSD cytoskeleton. Sos-1 is not found in the PSD matrix. Developmental analysis of Abi-1 mRNA and protein expression in hippocampal culture shows a significant increase of mRNA and protein levels between days 1 and 3 (actin mRNA and protein are detected as internal control). Abi-1 staining of developing neurons reveals the localization of Abi-1 in dendrites and growth cones (arrowheads); from day 10 onwards the protein is localized to spines and synapses.
Abi-1 is also a nuclear protein in neurons and shuttles from the PSD to the nucleus upon NMDA application
Endogenous Abi-1 was found in the nucleus after treatment with the nuclear export inhibitor leptomycinB in NIH cells as well as in hippocampal neurons (Figure 3A), indicating that Abi-1 shuttles between cytoplasm and nucleus. As Abi-1 is concentrated in PSDs, we tested whether a translocation of Abi-1 can be induced by NMDA application. NMDA treatment for 3 min resulted in nuclear staining and a decrease of dendritic Abi-1 labeling that could already be observed 3 min after changing the media (Figure 3B(I)). Nuclear staining was substantiated by confocal analysis (Figure 3B(II)) and significantly increased with highest levels after 60 min. Moreover, it was found to be completely reversible, as after 2 h the neuronal staining pattern was identical to nontreated controls (Figure 3B(I)). The NMDA-induced translocation of Abi-1 could also be observed in acute slice preparations from rat brain. In contrast to controls, neurons of the NMDA-treated slices exhibited an obvious nuclear staining for Abi-1 (Figure 3C). The accumulation of Abi-1 in the nucleus could be verified by Western blot analysis of the cytoplasmic and the nuclear/pellet fraction of NMDA-stimulated neurons (Figure 3C). Moreover, arrowheads indicate the appearance of slightly larger Abi-1 bands in the nuclear/pellet fractions that have already been characterized as phosphorylated Abi-1 proteins (Juang and Hoffmann, 1999; Courtney et al, 2000). In vitro phosphorylation experiments of wild-type and mutated Abi-1-GFP proteins with a recombinant abelson kinase revealed that the mutation at tyrosine 53 results in a nearly complete loss of abelson kinase-mediated tyrosine phosphorylation.
Figure 3.
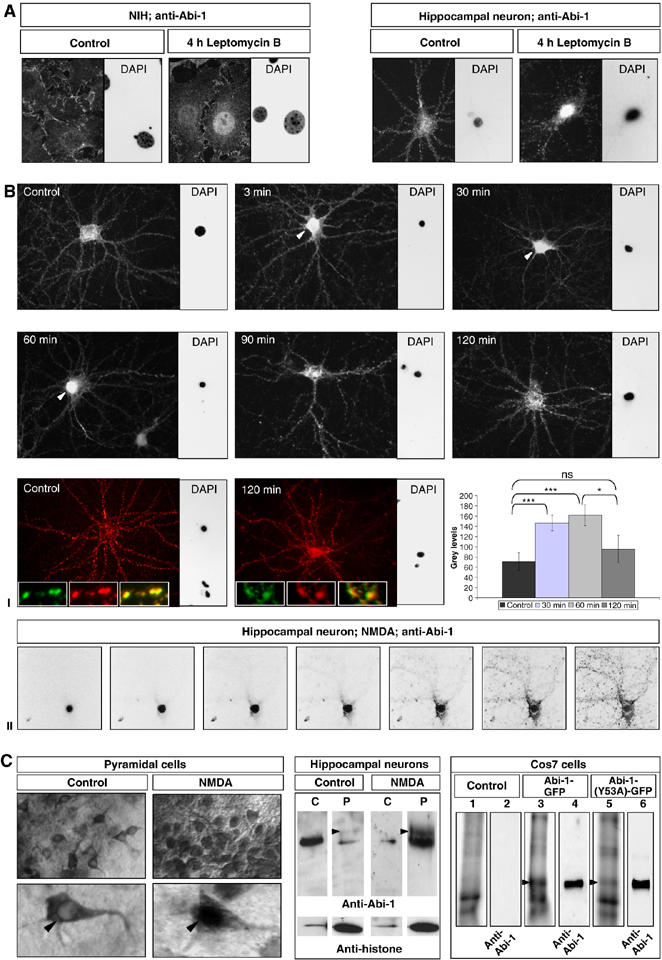
Abi-1 translocates from the PSD to the nucleus after NMDA treatment. (A) Treatment of NIH cells as well as hippocampal neurons with the nuclear export inhibitor leptomycinB results in the nuclear accumulation of Abi-1. (B) Application of NMDA (100 μM) to DIV21 neurons for 3 min leads to nuclear translocation of Abi-1. The most intense and significant nuclear staining of Abi-1 was observed after 60 min; in parallel, the dendritic compartment becomes more and more devoid of Abi-1 protein. Redistribution of Abi-1 into the dendrites can be observed from 90 min up to 2 h. The synaptic depletion of Abi-1 does not influence the gross morphology and number of synaptic contacts as shown by Bassoon staining (red) and Abi-1 staining (green) at time points 0 and 120 min. For quantification of nuclear Abi-1 staining, the total fluorescence as nuclear gray levels (linear scale of 0–256) was measured (I). Confocal analysis showed nuclear localization of Abi-1 in an NMDA-treated hippocampal neuron (II). (C) Acute rat brain slices were incubated with NMDA for 30 min, followed by Abi-1 DAB immunostaining. More than 90% of the pyramidal neurons in the stimulated slices exhibited nuclear staining (arrowheads). For Western blot analysis of Abi-1 protein, lysates of these neurons were separated into a nuclear/pellet (P) and a cytoplasmic fraction (C). Histone-H2B antibodies were used as a nuclear marker. Note the slight shift of Abi-1 protein from the cytoplasm to the nuclear pellet fraction. Arrowheads indicate the appearance of slightly larger Abi-1 bands that have already been characterized as phosphorylated Abi-1 protein. For an in vitro phosphorylation assay, Abi-1-GFP (lane 3) and the mutated Abi-1(Y53A)-GFP protein (lane 5) were transfected into Cos7 cells (lane 1 as untransfected control), immunoprecipitated on a column and incubated with a recombinant abelson tyrosine kinase using radioactive 32P-labeled ATP. After gel separation of the proteins, blotting and exposure to X-ray film, it could be shown in independent experiments that the WT-Abi-1-GFP was heavily phosphorylated by the abelson kinase, whereas the mutated form (Abi-1(Y53A)-GFP) showed nearly no signal on the X-ray film (loaded Abi-1 protein amounts are shown by immunodetection, lanes 4 and 6). Significances: *0.01, **0.001, ***0.0001.
Targeting of Abi-1 to the PSD is SH3 domain dependent, whereas the nuclear accumulation is regulated by the phosphorylation of tyrosine 53
To investigate the targeting mechanisms for the synaptic and nuclear compartment, we employed Abi-1-GFP deletion constructs. The full-length protein as well as the SH3 domain alone was perfectly targeted to synaptic contacts as revealed by Bassoon staining. In contrast, constructs missing the SH3 domain were evenly distributed in the cytoplasm and neurites (Figure 4A).
Figure 4.
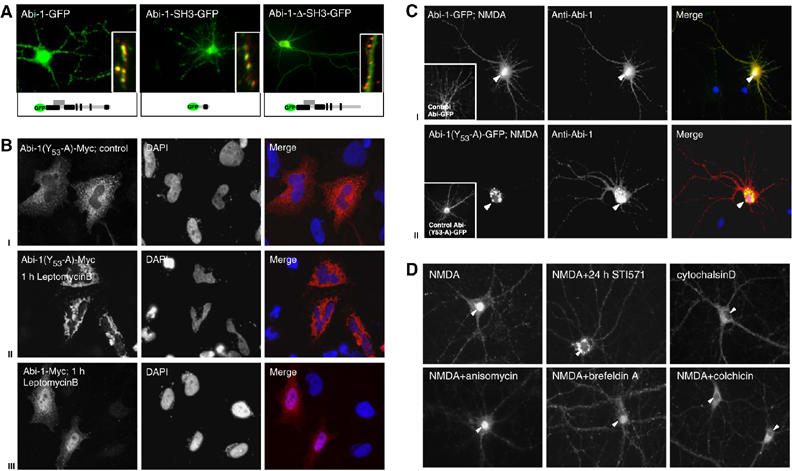
Determinants of Abi-1 targeting to the nuclear and synaptic compartments. (A) Full-length Abi-1-GFP protein and the Abi-1 SH3 domain colocalize with the presynaptic marker protein Bassoon (red, insets). Deletion of the SH3 domain results in the loss of synaptic targeting. (B) Phosphorylation of Abi-1 tyrosine 53 is a prerequisite for nuclear import leptomycinB treatment of Abi-1-(53Y-A)-Myc-transfected HeLA cells (localized in the cytoplasm under control conditions, I) leads to accumulation of the protein in the perinuclear area (II). Wild-type Abi-1-Myc (III) is enriched in the nucleus after leptomycinB treatment. (C) After NMDA application to hippocampal neurons, full-length Abi-1-GFP as well as the endogenous Abi-1 (red) readily translocates into the nucleus (yellow staining, I). Transfected Abi-1 protein, mutated at tyrosine 53 (Abi-1-(53Y-A)-GFP), accumulates after NMDA application in the perinuclear area. Endogenous Abi-1 (red) is enriched in the nucleus (II). (D) Inhibition of c-Abl by the specific compound STI571 for 24 h before NMDA treatment results in a perinuclear accumulation and no nuclear enrichment of Abi-1 (compare B and C). Nuclear translocation can also be prevented by the disturbance of cytoskeletal components with colchicin or cytochalasinD. The application of anisomycin or brefeldinA does not influence nuclear accumulation.
In order to analyze whether the phosphorylation of tyrosine 53 influences synaptic or nuclear targeting, we designed Abi-1-GFP and Myc-tagged fusion proteins with a mutated tyrosine at position 53(Y-A). After application of leptomycinB for 4 h, the mutated Abi-1(53Y-A) fusion protein did not accumulate in the nucleus but accumulated in the perinuclear area (Figure 4B(I and II)). Abi-1-Myc, however, readily entered the nucleus (Figure 4B(III)). Next, we transfected hippocampal neurons with the described Abi-1-GFP chimeras, applied NMDA, fixed and additionally counterstained with the Abi-1 antibody to detect the wild-type protein simultaneously as an internal control. Abi-1-GFP as well as Abi-1(53Y-A)-GFP was targeted to synaptic sites. After NMDA treatment, Abi-1-GFP accumulated in the nucleus, whereas the mutated Abi-1(53Y-A)-GFP was enriched in the perinuclear area (Figure 4C(I and II)), indicating that the tyrosine 53 phosphorylation is not important for synaptic targeting but a prerequisite for nuclear entry.
To substantiate these findings, we applied the small-molecule inhibitor of abl family tyrosine kinases, STI571 (Buchdunger et al, 1996), for 24 h before NMDA application. Specific inhibition of c-Abl prevented the translocation of wild-type Abi-1 into the nucleus but led to an accumulation of the protein in the perinuclear area, similar to the mutated Abi-1(53Y-A)-GFP chimera (Figure 4D). To differentiate between the proposed synapto-nuclear transport of Abi-1 and novel protein synthesis, we inhibited the translation machinery and the Golgi apparatus with anisomycin and brefeldinA. Both treatments did not affect nuclear accumulation of Abi-1. The treatment of hippocampal neurons with colchicin or cytochalasinD, however, prevented nuclear translocation, suggesting that Abi-1 nuclear transport is directly or indirectly dependent on a functional microtubular and microfilament system (Figure 4D).
Abi-1 protein expression in developing neurons regulates dendritic outgrowth and branching and determines the shape and number of synaptic contacts
According to the discrete distribution of Abi-1 during developmental maturation (Figure 2C), we analyzed the functional importance of Abi-1 for neuronal cell morphology and synapse formation by altered Abi-1 protein concentrations. To that end, we elevated Abi-1 protein levels by Abi-1 (over)expression, or applied small RNAi technology to selectively knock down Abi-1. Rescue experiments were performed with mutated RNAi-resistant Abi-1-Myc fusion proteins that carry seven conservative nucleotide exchanges localized at the RNAi targeting area. In contrast to the control vector, Abi-1 protein levels and staining intensity were markedly reduced in RNAi-transfected HeLa and neuronal cells (Figure 5A and B). Hippocampal neurons displayed a highly branched dendritic tree when transfected with the RNAi construct. The increased branching of MAP2-positive dendrites, as determined by the total number of dendrites or branching points (Quitsch et al, 2005), was highly significant at both time points investigated (Figure 5C–G). Interestingly, early reduction of Abi-1 (day 3) had a strong effect on primary dendrites, and transfection at day 7 resulted in a significant increase of secondary and tertiary dendrites (Figure 5F). The analysis of the total number of branching points (TNBPs) in neurons transfected at day 7 accordingly showed a significant increase (Figure 5G). Overexpression of Abi-1 had an opposite effect and reduced the TNBPs (Figure 5E and G); the Abi-1(53Y-A) fusion protein was less effective in reducing TNBPs compared with the wild-type protein. The RNAi-resistant Mut-Abi-1(53Y-A) was unable to rescue the RNAi-induced phenotype in neurons depleted of wild-type Abi-1 protein. When the RNAi construct was transfected at later stages of neuronal development, no obvious dendritic phenotype could be observed (data not shown).
Figure 5.
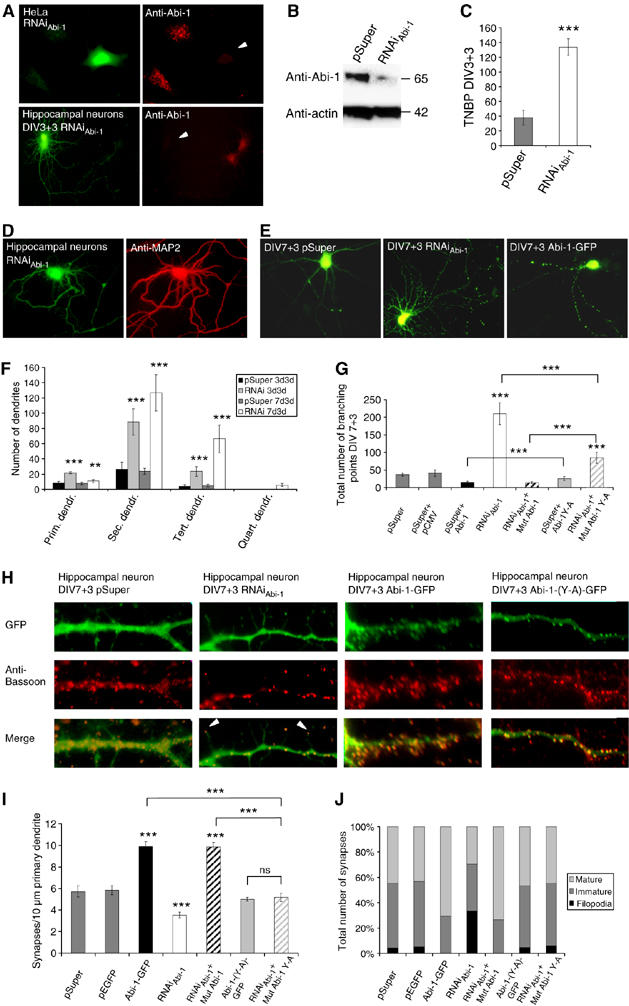
Abi-1 protein levels influence dendritogenesis and synaptic maturation. (A, B) The transfection of HeLa cells and hippocampal neurons with the Abi-1-RNAi construct reduces the level of endogenous Abi-1 protein to barely detectable levels. (C, D) In neurons transfected at day 3, the downregulation of Abi-1 leads to a highly branched MAP2-positive dendritic tree. The TNBPs is significantly elevated in RNAi-transfected neurons (E–G). This phenotype is also observed after transfection on day 7, showing especially high numbers of secondary and tertiary dendrites. Overexpression of Abi-1 protein results in a simplified dendritic tree, with a reduction of the TNBPs compared with controls. Significances compared with the empty vector controls are indicated above the bars. Note that RNAi-resistant wild-type Abi-1 cotransfected with RNAi (RNAiAbi-1+Mut Abi-1) reduces TNBPs significantly, whereas the RNAi-resistant Abi-1-(53Y-A) construct is not able to compensate (or overcompensate) the loss of endogenous Abi-1 (RNAiAbi-1+Mut-Abi-1-(53Y-A)) (H–J). Analysis of synaptic contacts reveals a significant reduction of Bassoon-positive boutons in Abi-1-RNAi-transfected neurons. Moreover, RNAi-transfected neurons show a high percentage of immature synaptic contacts on thin filopodia-like protrusions with apical Bassoon-positive presynaptic terminals (arrow heads). In contrast, the overexpression of Abi-1-GFP results in a significant increase of mature synaptic contacts at the dendritic shaft. Note that RNAi-resistant Abi-1 construct cotransfected with RNAi (RNAiAbi-1+Mut Abi-1) also significantly increases the synapse number as well as maturity. In contrast, the RNAi-resistant Abi-1-(53Y-A) construct alone or cotransfected with RNAiAbi-1 is not able to increase synapse number or change synapse morphology. Significances compared with the empty vector controls pSuper and pSuper+pEGFP are indicated above the bars. Significances: **0.001%; ***0.0001; ns, not significant.
Subsequently, we analyzed the number and morphology of synapses. The overexpression of Abi-1 increased the total number of synaptic contacts per 10 μm dendrite and the percentage of mature contact sites (Figure 5H–J). In RNAi-transfected neurons, the total number of synaptic contacts was significantly reduced and the morphology significantly differed from neurons transfected with the vector control. Abi-1-RNAi-transfected hippocampal neurons were characterized by a high percentage of thin filopodia-like protrusions, with apical Bassoon-positive presynaptic terminals (Figure 5H–J, arrow heads). Synapses with a mature morphology were reduced. RNAi-resistant Abi-1 protein could rescue the RNAi synaptic phenotype and led to synapse number and morphology comparable with Abi-1 overexpression. In contrast, the Abi-1(53Y-A)-mutated expression construct could not change synapse number and morphology, and cotransfection of the RNAi-resistant Abi-1-(53Y-A) fusion protein with RNAi rescued the phenotype, but could neither increase nor change the morphology of synaptic contacts comparable to the overexpression of wild-type Abi-1 protein (Figure 5I and J).
Abi-1 protein can bind to the Myc/Max complex and enhance E-box-regulated gene transcription
In a broad screening approach, we tested for the direct interaction of Abi-1 with known DNA recognition sequences. To that end, we determined the binding of 24 important transcription factors (only three of them are displayed here) from HeLa nuclear extracts to their corresponding DNA cis elements in the presence or absence of Abi-1-GFP purified protein. We used a commercially available enzyme-linked immunosorbent assay (ELISA)-based kits (Clontech) that measures protein amounts bound to oligonucleotides (recognition sequences) affixed to the bottom of a 24-well plate. A direct interaction of Abi-1 with the tested DNA recognition sequences could not be observed. However, the addition of increasing amounts of purified Abi-1-GFP protein (Figure 6A) to the HeLa nuclear extract showed significantly elevated levels for the binding of the transcription factor Max to its corresponding recognition sequence (Figure 6A). Staining of hippocampal neurons with Max antibodies revealed localization of the protein in the nucleus of neurons, as well as within the dendritic compartment in minor concentrations, as shown by Bassoon and Abi-1 staining (Figure 6B). Preabsorption of the Max antibody with the immunogenic peptide abolished the immunohistochemical stainings and Western blot signals. Co-immunoprecipitations from nuclear extracts after stimulation showed that Abi-1 can be detected in the Max precipitate. Myc that is known to be tightly associated with the Max protein was used as a positive control. On the other hand, precipitations with the Abi-1 antibody resulted in the detection of the Myc/Max heterodimer (Figure 6C). Next, we analyzed whether Abi-1 can influence transcriptional activity at the E-box-regulated promoters that regulate secreted alkaline phosphatase (SEAP) expression (Figure 6D). A significant increase (Figure 6D) in the amount of the secreted phosphatase could be detected by Abi-1-GFP cotransfection, similar to the pSEAP transfection used as a positive control. These data point toward an enhancement of the promotor activity by the Abi-1/Myc/Max complex after EGF stimulation of the cells.
Figure 6.
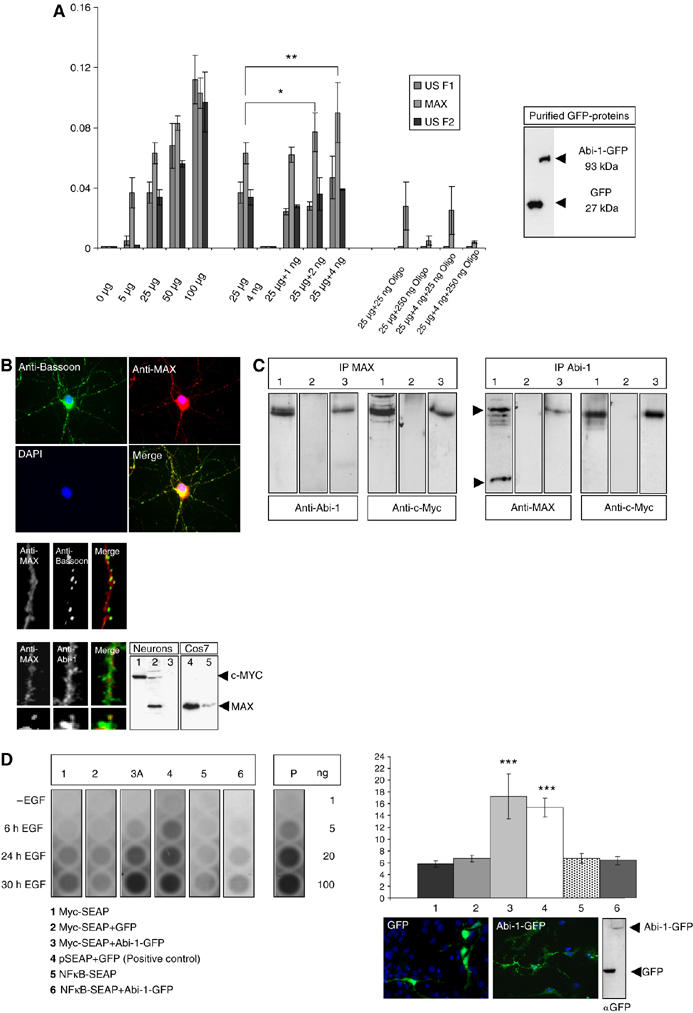
Abi-1 binds to the Myc/Max complex and enhances E-box-regulated gene transcription. (A) Concentrations of bound transcription factors from HeLa cell nuclear extracts were determined after binding to oligonucleotides with the respective recognition sequence (Transfactor kit, ELISA-based). A total of 24 different transcription factors were tested. Here, only the results for the E-box binding proteins USF1, USF2 and Max are depicted. The first cluster of bars indicates the increasing amounts of transcription factors relative to the total amount of nuclear extract measured (5–100 μg). The next cluster shows the values for 25 μg HeLa nuclear extract and 4 ng of purified Abi-1-GFP protein (also seen on Western blot). Then the values for the transcription factors after coincubation of 25 μg nuclear extract with increasing amounts of Abi-1-GFP protein are given. The relative amount of bound Max protein significantly increased by the coincubation of Abi-1-GFP. The last cluster of bars shows the internal control values after coincubation with unbound competing oligonucleotides. (B) Myc (lane 1) and Max (lanes 2 and 4, Cos7 cells as a positive control) are expressed in neurons and are mainly localized within the nucleus. Some Max-positive signals were also detected in the cytoplasm and within the dendritic compartment partly overlapping with Abi-1 and Bassoon stainings. Preabsorption of the Max antibody with the immunizing peptide results in the loss of signal (lanes 3 and 5). (C) Co-IP from nuclear extracts after stimulation (lane 1, input; lane 2, IgG control; lane 3, IP, 5 μg of protein loaded). Complexes that are precipitated with a Max antibody contain the Abi-1 protein. As a control, we stained also against Myc showing the Myc/Max highly stable complex at a size of about 63 kDa. On the other hand, experiments using Abi-1 antibodies result in the detection of the Myc/Max complex as consecutively shown by Myc staining. (D) E-box promoter assay. Neuronal cells were transfected/cotransfected with Myc-SEAP (secreted alkaline phosphatase vector carrying the E-box element), GFP-Abi-1 (lane 3) and pEGFP (lane 2). In addition, we tested NFκB-SEAP (coding for a NFκB-responsive promotor) (lane 5), NFκB-SEAP/GFP-Abi-1 (lane 6) as well as Myc-SEAP alone (lane 1) as negative control and the vector pSEAP with pEGFP (lane 4) as positive control (as recommended and provided by the manufacturer). Chemiluminescence reaction was controlled by the measurement of different amounts of purified alkaline phosphatase (AP, given in ng). Transfection rates were tested by counting cotransfected cells and Western blotting as shown. Cells were stimulated by EGF (20 ng/μl) as recommended by the supplier of the vector kit. Compared with controls (lanes 1, 2, 5 and 6), secreted SEAP was significantly elevated to levels of the positive control vector (lane 4) after 24 and 30 h EGF treatment when Abi-1-GFP was cotransfected with the Myc SEAP vector (lane 3).
Discussion
Abi-1, initially localized in growth cones, is part of the PSD matrix and interacts with ProSAP/Shank
We have shown that in neurons, Abi-1, which was first discovered as a c-Abl interacting protein that can suppress v-Abl-mediated transformation of cultured cells (Shi et al, 1995), is tightly bound to the PSD matrix via interaction with the PSD protein ProSAP2/Shank3. This interaction is mediated by a proline-rich stretch of eight amino acids that binds the Abi-1 SH3 domain. In hippocampal neurons, Abi-1, one of the key molecules for the rearrangement of the actin cytoskeleton, is initially found in neurites and is especially enriched in growth cones that are characterized by a high actin turnover. When synaptic formation starts, Abi-1 is relocated into filopodia and spiny dendritic protrusions, and becomes part of the PSD matrix of synaptic contacts. From the data presented it is likely that during development Abi-1 is recruited to the PSD via the ProSAP2/Shank3 interaction, as the deletion of Abi-SH3 domain results in a loss of synaptic targeting. Moreover, the expression pattern is quite similar to the expression profile of ProSAP/Shank proteins (Du et al, 1998; Boeckers et al, 2002). Interestingly, all other components of the Abi-1 trimeric complex (Innocenti et al, 2003; Eps8, Sos1) as well as WAVE1 are also specifically localized in dendritic spines and/or PSDs. From previous data, one can assume that actin reorganization in spiny protrusions is essentially organized by the cooperative activity of this protein complex (Innocenti et al, 2002, 2003, 2004). Eps8 is a prominent substrate of the EGF and PDGF receptors (Scita et al, 1999, 2001), and recent data underline its functional role in the growth of actin filaments by its barbed-end capping activity (Disanza et al, 2004). It might, therefore, well be that at synaptic sites, Eps8 as well as the Rac GTPase Sos-1 links growth factor receptor signaling directly to the reorganization of the local actin-based cytoskeleton in a Rac-dependent manner (Fan and Goff, 2000; Scita et al, 2001; Di Fiore and Scita, 2002).
Abi-1 shuttles to the nuclear compartment upon synaptic activation in a c-abl phosphorylation-dependent manner
One of the peculiarities of Abi-1 is the localization of the protein in two distinct cellular compartments (Echarri et al, 2004). As already shown for c-Abl (Taagepera et al, 1998), Abi-1 could be detected in nuclei after treatment with the nuclear export inhibitor leptomycinB, pointing to a dual role of this molecule in distinct cellular compartments. Noteworthy in this regard is the fact that several actin-binding proteins are also found in the nucleus. For example, the actin-regulatory protein WASP enters the nucleus to regulate transcription of HSP90 (Suetsugu and Takenawa, 2003). Similarly, WAVE1, which binds Abi-1, has also been identified in the nucleus but its nuclear role is hitherto unknown (Westphal et al, 2000). Recently Innocenti et al (2005) reported that Abi-1 can interact with both WAVE and N-WASP, leading to either Rac-dependent membrane protrusions/macropinocytosis (WAVE) or actin-based vesicular transport and the regulation of cell surface distribution of epidermal growth factor receptors and transferrin receptors (N-WASP). Therefore, actin reorganization and gene transcription might be regulated by similar protein complexes. In our study, we could show that nuclear Abi-1 shuttling is present in neurons, and that nuclear localization of Abi-1 can be induced by NMDA application. In turn, NMDA treatment clearly reduced Abi-1 staining intensity at synaptic sites. This effect was abolished by the destruction of the microtubular and microfilament systems, but independent from protein translation or processing, strongly supporting the concept of an active synapse to nucleus transport. We further analyzed this intriguing phenomenon of a synaptic molecule that quickly changes the intracellular compartment upon extracellular stimuli. To this end, we sought to identify the synaptic and nuclear targeting signals. Using Abi-1-GFP deletion constructs, we could show that the ProSAP2/Shank3 interacting Abi-1 SH3 domain alone is necessary and sufficient for synaptic targeting. Therefore, we assume that Abi-1 is localized and attached to PSDs via interaction with ProSAP2/Shank3. Next we tested whether c-Abl, a protein that also shuttles between cytoplasm and nucleus (Taagepera et al, 1998; Moresco and Koleske, 2003) and was shown to be also highly enriched with the PSD fraction, is involved in these intracellular transport mechanisms. Experiments with the mutated Abi-1-(53Y-A) construct that was nearly resistant to c-Abl phosphorylation as well as STI571 treatment indicated that c-Abl phosphorylation of Abi-1 seems to be irrelevant for dendritic transport but a prerequisite for nuclear import. The relationship between non-phosphorylated, cytoplasmic and phosphorylated Abi-1 proteins (Juang and Hoffmann, 1999; Courtney et al, 2000), being metabolized/activated (i.e. at synaptic sites), could be a molecular switch that is responsible for changes in gene transcription depending upon synaptic activation.
Abi-1 levels are essential for regulated dendrite and synapse formation
According to the spatial localization of Abi-1 during neuronal development, we were especially interested to analyze the functional role of Abi-1 in dendrite formation and during early stages of synaptogenesis. Most strikingly, developing neurons expressing the Abi-RNAi construct displayed an obvious altered morphology, with a significant exuberant increase of total branching points. Experiments showed that especially the initial formation of dendritic structures is dependent upon physiological Abi-1 levels. In contrast, overexpression of Abi-1 resulted in the simplification of the dendritic tree. Interestingly, we could show that the dendritic phenotype induced by Abi-1 depletion cannot be rescued by the Abi-1-(53Y-A) construct, pointing toward the importance of tyrosine phosphorylation (see also below). The significant role of the c-Abl pathways in a correct neuronal development is already well documented (Koleske et al, 1998; van Etten, 1999). In a recent paper, Jones et al (2004) showed that the inhibition or overexpression of a constitutively active c-Abl in hippocampal neurons results in similar structural changes as described here. From the published data, one has to assume that c-Abl and Abi-1 are counteracting with respect to actin stability. Abi-1 stabilizes actin filaments, whereas its absence leads to the ongoing outgrowth of immature dendrites and filopodia.
Moreover, we found that transfection of the RNAi construct significantly reduced the number of synaptic contacts and resulted in the formation of immature appearing synapses (about 30%) at the very tip of thin filopodia-like dendritic structures (Hering and Sheng, 2001). In contrast, overexpression of Abi-1 resulted in an increase in the relative amounts of mature, mushroom-like spines at synapses. Again, the overexpression of the Abi-1-(53Y-A) protein did not lead to dramatic changes in synapse number and morphology. In line with these results, an RNAi-resistant Abi-1-(53Y-A) construct could rescue the synaptic phenotype, with respect to number and morphology; however, no overcompensation with a shift toward mature synapses could be observed. One striking difference between the filopodia protrusions and mature spines is the presence of high concentrations of actin in dendritic spines (Matus, 2000). In light of our results, Abi-1 has to be regarded as a regulator of organized actin polymerization at the leading edge of neuronal filopodia controlling the maturation of synaptic contacts. As the remodeling of spines via reorganization of the actin cytoskeleton is essential for the maintenance of LTP (Fukazawa et al, 2003), one has to assume that Abi-1 can potentially influence learning and memory formation. Previously, it has been reported that overexpression of ProSAP/Shank is involved in the functional and morphological maturation of spines (Sala et al, 2001), and expression of ProSAP2/Shank3 is sufficient to induce spines in cerebellar aspiny neurons (Roussignol et al, 2005). From our results, one can speculate that this phenomenon might be explained, in part, by the recruitment of Abi-1 complexes via ProSAP2/Shank3 molecules to postsynaptic contact sites.
Abi-1 is part of the Myc/Max complex of transcription factors
We could provide evidence that Abi-1 binds to the Myc/Max complex of transcription factors and influences the efficiency of an E-box-regulated transcription of a reporter gene in vitro. At this moment, we can only speculate on the functional role of a Myc/Max/Abi-1 complex in neuronal nuclei after NMDA treatment, but a mechanism comparable to that of synaptic CASK/Lin-2, which binds DNA as a transcriptional coactivator, is quite conceivable (Hsueh et al, 2000). In this respect, Abi-1 is a good candidate for the proposed synaptically activated second messenger and transcription factor to the nucleus that is required for long-lasting forms of learning-related synaptic plasticity (Alberini, 1999). Assuming that phosphorylation of Abi-1 at tyrosine 53 is not essential for the local synaptic function of Abi-1, the rescue experiments with the Abi-1-(53Y-A) proteins are of special importance. The experiments show that whenever Abi-1 proteins are expressed in neurons that are unable to enter the nucleus, synapse number as well as synapse morphology cannot be altered toward a more mature phenotype. This in turn would indicate that nuclear entry and consecutive transcriptional regulation of target genes is also an important functional role of Abi-1. What are the neuronal target genes that could be regulated by this complex and how is the expression regulated? Max is strongly bound to its partner protein Myc (Adhikary and Eilers, 2005). Myc/Max heterodimers are known to activate transcription by binding to a DNA sequence (CACA/GTG), called the E-box element. Max can, however, also bind to several c-Myc-related proteins like N-Myc, Mad or Mnt, thereby inhibiting transcription of E-box-regulated promoters. Several thousand genes are potentially regulated by Myc/Max and, to date, only the in vivo regulation of some specific genes involved in cell cycle regulation, transformation, tumorigenesis and development is characterized (Baudino and Cleveland, 2001). Analysis of the functional role of Myc and Myc-related proteins in postmitotic neurons is just beginning. Recently, it could be shown that c-Myc represses the expression of the GABA-A receptors (Vaknin and Hann, 2006). In search of target genes that are regulated by N-Myc, Perini et al (2005) found that the EGF receptor is one of very few genes that are directly regulated by the N-Myc/Max complex. The EGF receptor is certainly a very attractive candidate gene as members of the EGFR family are localized at the postsynaptic sites (Wong and Guillaud, 2004), and the role of EGF receptors and their ligands in neuronal migration, neurite formation and synapse physiology is well documented (Burden and Yarden, 1997; Huang et al, 2000). Interestingly, the cell surface distribution is regulated by an Abi-1/N-WASP complex (Innocenti et al, 2005). Future studies should especially concentrate on EGF receptors as potential target genes that might be specifically regulated by Abi-1 upon synaptic activation.
Materials and methods
The Materials and methods are only described very briefly. For further information, see the Supplementary data section.
Yeast two-hybrid screen
The YTH screen was carried out with a proline-rich region next to the PDZ domain of ProSAP2/Shank3 as bait. A pretransformed cDNA library from rat brain was screened and six independent clones coding for Abi-1 were isolated.
GST fusion proteins, antibodies, GST pull-down and immunoprecipitation experiments
The different protocols, materials and plasmids used are described in detail in the Supplementary data section.
Cell culture, expression vectors and transfection experiments
The protocols are described in detail in the Supplementary data section.
Isolation of subcellular protein fractions and Western blot analysis
The protocols are described in detail in the Supplementary data section.
Immunohistochemistry (in situ hybridization) of hippocampal culture and rat brain sections, and image acquisition and quantification
Cell culture experiments of rat hippocampal primary neurons and transfections, immunohistochemistry and in situ hybridization were performed as described previously (Boeckers et al, 1999, 2005).
Small interference RNA experiments
Knockdown of Abi-1 was achieved by RNAi following published methods using the pSUPER vector (OligoEngine, Seattle, WA, USA).
Pharmacological treatment
Neurons or freshly prepared brain slices (50 μm) were treated with NMDA, 100 μM final concentration, for the indicated time. The other agents (purchased from Sigma-Aldrich (Taufkirchen, Germany) if not stated differently) were used at the following concentrations (in μM): 3.8 Anisomycin (Serva, Heidelberg, Germany), 7.1 BrefeldinA, 1.9 cytochalasinD, 10 colchicin, 5 STI571 (kindly provided by Novartis, Basel, Switzerland), 0.02 leptomycinB and EGF 20 ng/μl.
In vitro phosphorylation of Abi-1 fusion proteins
Cos7 cells were transfected with Abi-1-GFP and Abi-1-(Y53A)-GFP. Proteins were immunoprecipitated, loaded on μMACS-microcolumns and incubated with 100 μCi [γ-32P]ATP (>5000 Ci/mmol) and 500 U of recombinant Abl protein tyrosine kinase (c-Abl, New England Biolabs). After blotting, the phosphorylated proteins were detected by exposition to an X-ray film. Loaded Abi-1 fusion proteins were visualized by an Abi-1-specific antibody.
Transcription factor and promoter activity assay
To test a direct or indirect association of Abi-1 with consensus DNA binding sequences, we used a commercially available ELISA-based kit (TransFactor Kit, Clontech, SanDiego, CA, Oncogenesis1-3, Inflammation1). Immunopurified GFP-tagged Abi-1 was either tested for direct binding to the different consensus binding sequences or added to different amounts of HeLa cell nuclear lysate (MBL, Woburn, MA, USA).
Supplementary Material
Supplementary data
Supplementary Figures
Acknowledgments
This study was supported by grants from the Deutsche Forschungsgemeinschaft (DFG, Bo1718/2-2 to TMB; SFB497/B8 to TMB and JB; and SFB426 to EDG) and the Land Baden Württemberg to TMB, StL and CP (grant 1423/74). We gratefully acknowledge the professional technical assistance of U Pika-Hartlaub and N Damm. STI571 was kindly provided by Novartis (Basel, Switzerland) and Shank1a cDNA by C Sala (Milano, Italy).
References
- Adhikary S, Eilers M (2005) Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 6: 635–645 [DOI] [PubMed] [Google Scholar]
- Alberini CM (1999) Genes to remember. J Exp Biol 202: 2887–2891 [DOI] [PubMed] [Google Scholar]
- Baron MK, Boeckers TM, Vaida B, Faham S, Gingery M, Sawaya MR, Salyer D, Gundelfinger ED, Bowie JU (2006) An architectural framework that may lie at the core of the postsynaptic density. Science 311: 531–555 [DOI] [PubMed] [Google Scholar]
- Baudino TA, Cleveland JL (2001) The Max network gone mad. Mol Cell Biol 21: 691–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckers TM, Bockmann J, Kreutz MR, Gundelfinger ED (2002) ProSAP/Shank proteins—a family of higher order organizing molecules of the postsynaptic density with an emerging role in human neurological disease. J Neurochem 81: 903–910 [DOI] [PubMed] [Google Scholar]
- Boeckers TM, Kreutz MR, Winter C, Zuschratter W, Smalla K-H, Sanmarti-Vila L, Wex H, Langnaese K, Bockmann J, Garner CC, Gundelfinger ED (1999) Proline-rich synapse-associated protein-1/cortactin binding protein 1 (ProSAP1/CortBP1) is a PDZ-domain protein highly enriched in the postsynaptic density. J Neurosci 19: 6506–6518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckers TM, Liedtke Th, Dresbach Th, Bockmann J, Kreutz MR, Gundelfinger ED (2005) C-terminal synaptic targeting elements for postsynaptic density proteins ProSAP1/Shank2 and ProSAP2/Shank3. J Neurochem 92: 519–524 [DOI] [PubMed] [Google Scholar]
- Buchdunger E, Zimmermann J, Mett H, Meyer T, Muller M, Druker BJ, Lydon NB (1996) Inhibition of the Abl protein-tyrosine kinase in vitro and in vivo by a 2-phenylaminopyrimidine derivate. Cancer Res 56: 100–104 [PubMed] [Google Scholar]
- Burden S, Yarden Y (1997) Neuregulins and their receptors: a versatile signaling module in organogenesis and oncogenesis. Neuron 18: 847–855 [DOI] [PubMed] [Google Scholar]
- Courtney KD, Grove M, Vandongen H, Vandongen A, LaMantia AS, Pendergast AM (2000) Localization and phosphorylation of Abl-interactor proteins, Abi-1 and Abi-2, in the developing nervous system. Mol Cell Neurosci 16: 244–257 [DOI] [PubMed] [Google Scholar]
- Di Fiore PP, Scita G (2002) Eps8 in the midst of GTPases. Int J Biochem Cell Biol 34: 1178–1183 [DOI] [PubMed] [Google Scholar]
- Disanza A, Carlier MF, Stradal TE, Didry D, Frittoli E, Confalonieri S, Croce A, Wehland J, Di Fiore PP, Scita G (2004) Eps8 controls actin-based motility by capping the barbed ends of actin filaments. Nat Cell Biol 6: 1180–1188 [DOI] [PubMed] [Google Scholar]
- Du Y, Weed SA, Xiong WC, Marshall TD, Parsons JT (1998) Identification of a novel cortactin SH3 domain-binding protein and its localization to growth cones of cultured neurons. Mol Cell Biol 18: 5838–5851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echarri A, Lai MJ, Robinson MR, Pendergast AM (2004) Abl interactor 1 (Abi-1) wave-binding and SNARE domains regulate its nucleocytoplasmic shuttling, lamellipodium localization, and wave-1 levels. Mol Cell Biol 24: 4979–4993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan PD, Goff SP (2000) Abl interactor 1 binds to Sos and inhibits epidermal growth factor- and v-Abl-induced activation of extracellular signal-regulated kinases. Mol Cell Biol 20: 7591–7601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa Y, Saitoh Y, Ozawa F, Ohta Y, Mizuno K, Inokuchi K (2003) Hippocampal LTP is accompanied by enhanced F-actin content within the dendritic spine that is essential for late LTP maintenance in vivo. Neuron 38: 447–460 [DOI] [PubMed] [Google Scholar]
- Hering H, Sheng M (2001) Dendritic spines: structure, dynamics and regulation. Nat Rev Neurosci 2: 880–888 [DOI] [PubMed] [Google Scholar]
- Hsueh YP, Wang TF, Yang FC, Sheng M (2000) Nuclear translocation and transcription regulation by the membrane-associated guanylate kinase CASK/LIN-2. Nature 16: 298–302 [DOI] [PubMed] [Google Scholar]
- Huang YZ, Won S, Ali DW, Wang Q, Tanowitz M, Du QS, Pelkey KA, Yang DJ, Xiong WC, Salter MW, Mei L (2000) Regulation of neuregulin signaling by PSD-95 interacting with ErbB4 at CNS synapses. Neuron 26: 443–455 [DOI] [PubMed] [Google Scholar]
- Ichigotani Y, Fujii K, Hamaguchi M, Matsuda S (2002) In search of a function for the E3B1/Abi2/Argbp1/NESH family. Int J Mol Med 9: 591–595 [PubMed] [Google Scholar]
- Innocenti M, Frittoli E, Ponzanelli I, Falck JR, Brachmann SM, Di Fiore PP, Scita G (2003) Phosphoinositide 3-kinase activates Rac by entering in a complex with Eps8, Abi1, and Sos-1. J Cell Biol 160: 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti M, Gerboth S, Rottner K, Lai FP, Hertzog M, Stradal TE, Frittoli E, Didry D, Polo S, Disanza A, Benesch S, Fiore PP, Carlier MF, Scita G (2005) Abi1 regulates the activity of N-WASP and WAVE in distinct actin-based processes. Nat Cell Biol 7: 969–976 [DOI] [PubMed] [Google Scholar]
- Innocenti M, Tenca P, Frittoli E, Faretta M., Tocchetti A, Di Fiore PP, Scita G (2002) Mechanisms through which Sos-1 coordinates the activation of Ras and Rac. J Cell Biol 156: 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti M, Zucconi A, Disanza A, Frittoli E, Areces LB, Steffen A, Stradal TE, Di Fiore PP, Carlier MF, Scita G (2004) Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat Cell Biol 6: 319–327 [DOI] [PubMed] [Google Scholar]
- Jones SB, Lu HY, Lu Q (2004) Abl tyrosine kinase promotes dendrogenesis by inducing actin cytoskeletal rearrangements in cooperation with Rho family small GTPases in hippocampal neurons. J Neurosci 29: 8510–8521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang JL, Hoffmann FM (1999) Drosophila abelson interacting protein (dAbi) is a positive regulator of abelson tyrosine kinase activity. Oncogene 16: 5138–5147 [DOI] [PubMed] [Google Scholar]
- Koleske AJ, Gifford AM, Scott ML, Nee M, Bronson RT, Miczek KA, Baltimore D (1998) Essential roles for the Abl and Arg tyrosine kinases in neurulation. Neuron 21: 1259–1272 [DOI] [PubMed] [Google Scholar]
- Leng Y, Zhang J, Badour K, Arpaia E, Freeman S, Cheung P, Siu M, Siminovitch K (2005) Abelson-interactor-1 promotes WAVE2 membrane translocation and Abelson-mediated tyrosine phosphorylation required for WAVE2 activation. Proc Natl Acad Sci USA 102: 1098–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Huang J, Wang Y, Eipper BA, Mains RE (2003) Kalirin, a multifunctional Rho guanine nucleotide exchange factor, is necessary for maintenance of hippocampal pyramidal neuron dendrites and dendritic spines. J Neurosci 19: 10593–10603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus A (2000) Actin-based plasticity in dendritic spines. Science 290: 754–758 [DOI] [PubMed] [Google Scholar]
- Moresco EM, Koleske AJ (2003) Regulation of neuronal morphogenesis and synaptic function by Abl family kinases. Curr Opin Neurobiol 13: 535–544 [DOI] [PubMed] [Google Scholar]
- Pak DT, Yang S, Rudolph-Correia S, Kim E, Sheng M (2001) Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron 2: 289–303 [DOI] [PubMed] [Google Scholar]
- Pendergast AM (2002) The Abl family kinases: mechanisms of regulation and signaling. Adv Cancer Res 85: 51–100 [DOI] [PubMed] [Google Scholar]
- Perini G, Diolaiti D, Porro A, Della Valle G (2005) In vivo transcriptional regulation of N-Myc target genes is controlled by E-box methylation. Proc Natl Acad Sci USA 102: 12117–12122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quitsch A, Berhörster K, Liew CW, Richter D, Kreienkamp HJ (2005) Postsynaptic Shank antagonizes dendrite branching induced by leucine-rich repeat protein Densin 180. J Neurosci 25: 479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussignol G, Ango F, Romorini S, Tu JC, Sala C, Worley PF, Bockaert J, Fagni L (2005) Shank expression is sufficient to induce functional dendritic spine synapses in aspiny neurons. J Neurosci 25: 3560–3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala C, Piech V, Wilson NR, Passafaro M, Liu G, Sheng M (2001) Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron 31: 115–130 [DOI] [PubMed] [Google Scholar]
- Scita G, Nordstrom C, Carbone R, Tenca P, Giardina G, Gutkind S, Bjarnegard M, Betsholtz C, Di Fiore PP (1999) EPS8 and E3B1 transduce signals from Ras to Rac. Nature 401: 290–293 [DOI] [PubMed] [Google Scholar]
- Scita G, Tenca P, Areces LB, Tocchetti A, Frittoli E, Giardina G, Ponzanelli I, Sini P, Innocenti M, Di Fiore PP (2001) An effector region in Eps8 is responsible for the activation of the Rac-specific GEF activity of Sos-1 and for the proper localization of the Rac-based actin-polymerizing machine. J Cell Biol 154: 1031–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Kim E (2000) The Shank family of scaffold proteins. J Cell Sci 113: 1851–1856 [DOI] [PubMed] [Google Scholar]
- Shi Y, Alin K, Goff SP (1995) Abl-interactor-1, a novel SH3 protein binding to the carboxy-terminal portion of the Abl protein, suppresses v-abl transforming activity. Genes Dev 9: 2583–2597 [DOI] [PubMed] [Google Scholar]
- Stradal T, Courtney KD, Rottner K, Hahne P, Small JV, Pendergast AM (2001) The Abl interactor proteins localize to sites of actin polymerization at the tips of lamellipodia and filopodia. Curr Biol 11: 891–895 [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Takenawa T (2003) Translocation of N-WASP by nuclear localization and export signals into the nucleus modulates expression of HSP90. J Biol Chem 278: 42515–42523 [DOI] [PubMed] [Google Scholar]
- Taagepera S, McDonald D, Loeb JE, Whitaker LL, McElroy AK, Wang JY, Hope TJ (1998) Nuclear–cytoplasmic shuttling of C-ABL tyrosine kinase. Proc Natl Acad Sci USA 95: 7457–7462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani K, Sato S, Sukezane T, Kojima H, Hirose H, Hanafusa H, Shishido T (2003) Abl interactor 1 promotes tyrosine 296 phosphorylation of mammalian enabled (Mena) by c-Abl kinase. J Biol Chem 13: 21685–21692 [DOI] [PubMed] [Google Scholar]
- Vaknin UA, Hann SR (2006) The alpha1 subunit of GABAA receptor is repressed by c-myc and is pro-apoptotic. J Cell Biochem 97: 1094–1103 [DOI] [PubMed] [Google Scholar]
- Van Etten RA (1999) Cycling, stressed-out and nervous: cellular functions of c-Abl. Trends Cell Biol 9: 179–186 [DOI] [PubMed] [Google Scholar]
- Westphal RS, Soderling SH, Alto NM, Langeberg LK, Scott JD (2000) Scar/WAVE-1, a Wiskott–Aldrich syndrome protein, assembles an actin-associated multi-kinase scaffold. EMBO J 19: 4589–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RW, Guillaud L (2004) The role of epidermal growth factor and its receptors in mammalian CNS. Cytokine Growth Factor Rev 15: 147–156 [DOI] [PubMed] [Google Scholar]
- Wong WT, Faulkner-Jones BE, Sanes JR, Wong RO (2000) Rapid dendritic remodeling in the developing retina: dependence on neurotransmission and reciprocal regulation by Rac and Rho. J Neurosci 1: 5024–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T (2004) Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat Rev Neurosci 5: 24–34 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary Figures


