Abstract
We have found that a major target for effectors secreted by Pseudomonas syringae is the abscisic acid (ABA) signalling pathway. Microarray data identified a prominent group of effector-induced genes that were associated with ABA biosynthesis and also responses to this plant hormone. Genes upregulated by effector delivery share a 42% overlap with ABA-responsive genes and are also components of networks induced by osmotic stress and drought. Strongly induced were NCED3, encoding a key enzyme of ABA biosynthesis, and the abscisic acid insensitive 1 (ABI1) clade of genes encoding protein phosphatases type 2C (PP2Cs) involved in the regulation of ABA signalling. Modification of PP2C expression resulting in ABA insensitivity or hypersensitivity led to restriction or enhanced multiplication of bacteria, respectively. Levels of ABA increased rapidly during bacterial colonisation. Exogenous ABA application enhanced susceptibility, whereas colonisation was reduced in an ABA biosynthetic mutant. Expression of the bacterial effector AvrPtoB in planta modified host ABA signalling. Our data suggest that a major virulence strategy is effector-mediated manipulation of plant hormone homeostasis, which leads to the suppression of defence responses.
Keywords: ABA, Arabidopsis thaliana, Pseudomonas syringae, transcriptomics, type III effectors
Introduction
Plant–pathogen interactions involve a dynamic process of cut, thrust and counterthrust as the host attempts to contain the insurgent within the complex intracellular and extracellular landscape of the battlefield. Plant basal defences provide several strategic layers of constitutive and induced defences, the latter based upon the ability to detect and respond to conserved molecular patterns associated with the invading microbe. The various repertoires of predominately surface-expressed ligands, commonly referred to as PAMPs (pathogen-associated molecular patterns), engage different combinations of plant-surface receptors whose output is customised to reflect the degree of ‘danger' posed by the particular pathogen. The basal defense response manifests itself in the form of biochemical and structural defences designed to restrict microbial multiplication and nutrition, including rapid ionic changes and phosphorylation cascades, ultimately leading to transcription of defence-related genes and the formation of defensive cell wall depositions, termed papillae (Nurnberger et al, 2004; Zipfel and Felix, 2005).
The frontline weapons of bacterial virulence comprise a collection of chemical virulence factors and ∼40 proteinaceous ‘effectors'. The latter are delivered into the plant cell via a type III protein secretion system (T3SS; Mudgett, 2005). Evidence is emerging that the type III effectors (T3Es) interfere with host signalling and metabolism to promote suppression of basal defence and pathogen nutrition. However, specific function has been assigned to very few T3Es, and our knowledge of the synergistic collaborations with other effectors that may be required for successful parasitism is rudimentary. Three phytohormones, salicylic acid (SA), jasmonic acid (JA) and ethylene (ET), are known to participate in regulating defence responses in plants. SA is predominately associated with resistance against biotrophic and hemi-biotrophic pathogens and the establishment of systemic acquired resistance (Grant and Lamb, 2006). By contrast, JA- and ET-dependent defence mechanisms generally contribute to resistance against necrotrophic pathogens, suggesting that the signalling network engaged by the host is dependent upon the nature of the pathogen and its mode of pathogenicity (Glazebrook, 2005). The plant hormone abscisic acid (ABA) is involved in plant responses to several abiotic stresses (drought, salt and cold) as well as seed germination and plant growth (Seo and Koshiba, 2002; Nambara and Marion-Poll, 2005). Additionally, exogeneous ABA treatment increases the susceptibility of various plant species to bacterial and fungal pathogens (Henfling et al, 1980; Ward et al, 1989; McDonald and Cahill, 1999; Mohr and Cahill, 2003; Thaler and Bostock, 2004). ABA-deficient mutants showed a reduction in susceptibility to the necrotroph Botrytis cinerea (Audenaert et al, 2002) and virulent isolates of Pseudomonas syringae pv tomato DC3000 in tomato (Thaler and Bostock, 2004), and the oomycete Hyaloperonospora parasitica in Arabidopsis (Mohr and Cahill, 2003). Collectively, these data suggest that ABA behaves as a negative regulator of defence responses.
We are interested in how virulence factors (including T3Es) produced by DC3000 in Arabidopsis suppress plant basal defences. Building upon analysis of genome-wide transcriptome changes during early stages of the A. thaliana/DC3000 interaction (Truman et al, 2006) we now present results supporting effector-mediated manipulation of ABA biosynthetic and signalling pathways as a core virulence mechanism. The changes in plant hormones observed during infection by bacteria and fungi have traditionally been considered as ‘side effects of successful parasitism'. By contrast, we now show that the successful manipulation of the ABA hormonal network has probably been a fundamental step in the evolution of a plant pathogenic bacterium.
Results
Microarrays reveal a role for ABA in virulence
We have analysed previously the temporal dynamics of host gene expression during bacterial infection of Arabidopsis leaves by global transcriptional profiling (Truman et al, 2006). Three challenges designed to report background inoculation effects (mock; 10 mM MgCl2), PAMP responses (DC3000hrpA−, a mutant compromised in T3SS) or disease (wild-type DC3000), and carefully chosen sampling times enabled the identification of differentially expressed genes associated with the activation of basal defence and establishment of pathogenesis. By removing the basal defence signature attributable to challenge with the hrpA− mutant, we captured the specific effects of bacterial virulence factors on host transcription. Hierarchical clustering identified transcripts coregulated by T3E delivery 12 h post-inoculation (12 hpi). These transcripts represent genes upregulated by T3Es, either due to the action of the effectors themselves or, alternatively, as a host response to T3E activities. We have now examined in detail one coregulated cluster consisting of 47 T3E-induced genes (Table I). The group of genes is characterised by an over-representation of transcripts encoding protein phosphatases of the 2C class (PP2C), which are implicated in responses to ABA, including ABI1 (abscisic acid insensitive 1; Schweighofer et al, 2004). Remarkably, NCED3 (At3g14440), which encodes the enzyme that catalyses the early limiting step in water stress-induced ABA biosynthesis (Qin and Zeevaart, 1999; Tan et al, 2003), was induced more than 15-fold by T3Es. Other ABA response-associated genes resident in this cluster include AFP (abscisic acid insensitive-5 binding protein), an ABA signalling regulator (At1g69260) (Lopez-Molina et al, 2003), and four NAC (for NAM, ATAF1, 2 and CUC2) transcription factors (At1g52890, At3g15500, At4g27410 [RD26] and At5g39610) previously shown to be ABA, drought and NaCl-inducible (Tran et al, 2004).
Table 1.
A coregulated set of T3E-induced genes responsive to ABA identified by hierarchial clustering
| AGI number | Function (name) | ABA/stress reference | ARE | FC |
|---|---|---|---|---|
| ABA biosynthetic genes | ||||
| At3g14440 | ABA synthesis (NCED3) | Qin and Zeevaart (1999) | B | 17.0 |
| Protein phosphatase 2C clade A | ||||
| At5g59220 | PP2C | Xin et al (2005) | 2D | 10.0 |
| At1g72770 | PP2C (HAB1) | Saez et al (2004) | 2A2D | 2.1 |
| At1g07430 | PP2C | AB4D | 8.7 | |
| At3g11410 | PP2C (AtPP2CA) | Kuhn et al (2006) | AC2D | 4.2 |
| At5g57050 | PP2C (ABI2) | Merlot et al (2001) | B | 3.2 |
| At4g26080 | PP2C (ABI1) | Merlot et al (2001) | C | 2.9 |
| NAC domain transcription factors | ||||
| At5g39610 | NAC TF (AtNAC6) | He et al (2005) | 3.6 | |
| At1g52890 | NAC TF (ANACO19) | Tran et al (2004) | 3C2D | 2.9 |
| At3g15500 | NAC TF (AtNAC3) | Tran et al (2004) | C3D | 7.9 |
| At4g27410 | NAC TF (RD26) | Fujita et al (2004) | A2CD | 4.5 |
| Transcription factors of various families | ||||
| At1g66550 | WRKY TF (WRKY67) | D | 10.2 | |
| At5g49450 | bZIP family TF | 2D | 4.3 | |
| At1g24440 | C3HC4 zinc finger? | 3.1 | ||
| Other signalling pathways | ||||
| At1g69260 | ABI5 binding protein | Lopez et al (2003) | 2C2D | 3.1 |
| At1g05100 | MAP3K (MAPKKK18) | BC3D | 10.9 | |
| At2g02710 | PAC motif containing | BD | 2.1 | |
| At5g62540 | Ubiquitin conjugating | D | 2.2 | |
| Assorted physiological processes | ||||
| At2g03760 | Steroid sulphotransferase | 7.0 | ||
| At5g13750 | Transporter (ZIFL1) | C2D | 2.7 | |
| At3g11340 | UDP glucosyl transferase | 2.4 | ||
| At3g46660 | UDP glucosyl transferase | D | 5.3 | |
| At3g06500 | β-Fructofuranosidase | B3D | 4.3 | |
| At4g21680 | Oligopeptide transporter | 2.5 | ||
| At1g05560 | UDP-glucose transferase | Tran et al (2004) | D | 3.3 |
| At1g26770 | Expansin (AtEXPA10) | 4.4 | ||
| At5g13820 | Telomeric DNA-binding-1 | Nagaoka and Takano (2003) | C3D | 3.2 |
| At3g14660 | Cytochrome P450 | C | 2.1 | |
| Genes of unknown function | ||||
| At3g01650 | Copine related | 3.3 | ||
| At1g24600 | Expressed protein | A2D | 3.2 | |
| At3g48350 | Cysteine proteinase like | 3D | 8.1 | |
| At3g28007 | Nodulin MtN3 family | 4D | 2.7 | |
| At3g29575 | Expressed protein | 4C3D | 3.3 | |
| At1g69480 | ERD1/XPR1/SYG1 | BC | 3.4 | |
| At2g28400 | Expressed protein | 3.2 | ||
| At4g33980 | Expressed protein | 3C | 2.4 | |
| At5g04250 | OTU cysteine protease? | C3D | 5.1 | |
| At5g54730 | WD40 repeat protein | 3D | 3.9 | |
| At5g42900 | Expressed protein | C2D | 11.5 | |
| At1g33110 | MATE efflux protein | 3.1 | ||
| At5g64230 | Expressed protein | D | 6.2 | |
| At5g65660 | HPRG protein | 2D | 2.8 | |
| At1g58270 | MATH domain protein | 2A4D | 3.0 | |
| At3g07350 | Expressed protein | ACD | 6.5 | |
| At3g28210 | Zinc-finger protein | Xin et al (2005) | D | 2.1 |
| At5g13360 | Auxin responsive GH3 | D | 2.1 | |
| At5g64250 | 2-Nitropropane diox. | 2.7 | ||
| Each of the 880 genes, significantly differentially expressed in response to T3E at 12 hpi, were median-centred and logged before clustering. This table presents a cluster of 47 genes containing a significant over-representation of ABA response elements in their promoters (1 kb upstream). Most genes of known function had previously been associated with abiotic stress. ARE—presence or absence of an ABA response element as defined in the PLACE database (Higo et al, 1999). Scoring is non-redundant and the number of occurrences precedes the motif. (A) ABREMOTIFAOSOSEM—TACGTGTC; (B) ABREATCONSENSUS—[CT]ACGTGGC; (C) ACGTABREMOTIFA2OSEM—ACGTG[GT]C; (D) ABRELATEDD1—ACGTG. FC—fold change. | ||||
Analysis of the proximal 1 kb promoter regions of the coregulated genes using the PLACE Signal Scan programme (Higo et al, 1999) identified that more than 50% contain one or more ABA response elements (ABRE; ACGTG[GT]C), which could be further elaborated into three overlapping regions of increasing complexity. All three contained the core ACGTG motif, which itself was present in more than 75% of the promoters (Table I). The significant over-representation of these motifs was confirmed using POBO (Kankainen and Holm, 2004).
In summary, our analysis suggested that one mechanism of T3E action is to elevate components of the ABA signalling and biosynthesis pathways.
ABA levels in challenged tissues
In a compatible interaction leading to disease, the induction of NCED3 between 4 and 12 hpi follows T3E delivery. As NCED is a key regulatory enzyme in ABA biosynthesis, we examined whether T3Es were directly manipulating levels of ABA. We measured ABA at 8, 12 and 18 h following mock, DC3000 or DC3000 hrpA− challenges. Increases in ABA occurred rapidly, and by 18 hpi ABA levels were ∼10-fold higher in DC3000 than in hrpA− or mock-challenged leaves (Figure 1A) and were directly correlated to the level of DC3000 inoculum (Figure 1B). Although DC3000 multiplies in planta, there was no significant difference in bacterial numbers (hrpA− versus DC3000) at 10 hpi despite increasing ABA levels (data not shown), and 15-fold less virulent bacteria produced more than six-fold more ABA than the non-pathogenic hrpA− mutant (Figure 1B). As the hormones SA and JA have already been demonstrated to play a role in plant defences, we compared SA, JA and ABA levels 18 hpi in uninoculated and pathogen-challenged leaves (Figure 1C). Foliar ABA and JA were significantly induced by DC3000, suggesting DC3000 induces both ABA and JA biosynthesis, whereas free SA did not differ significantly between hrpA− and DC3000.
Figure 1.
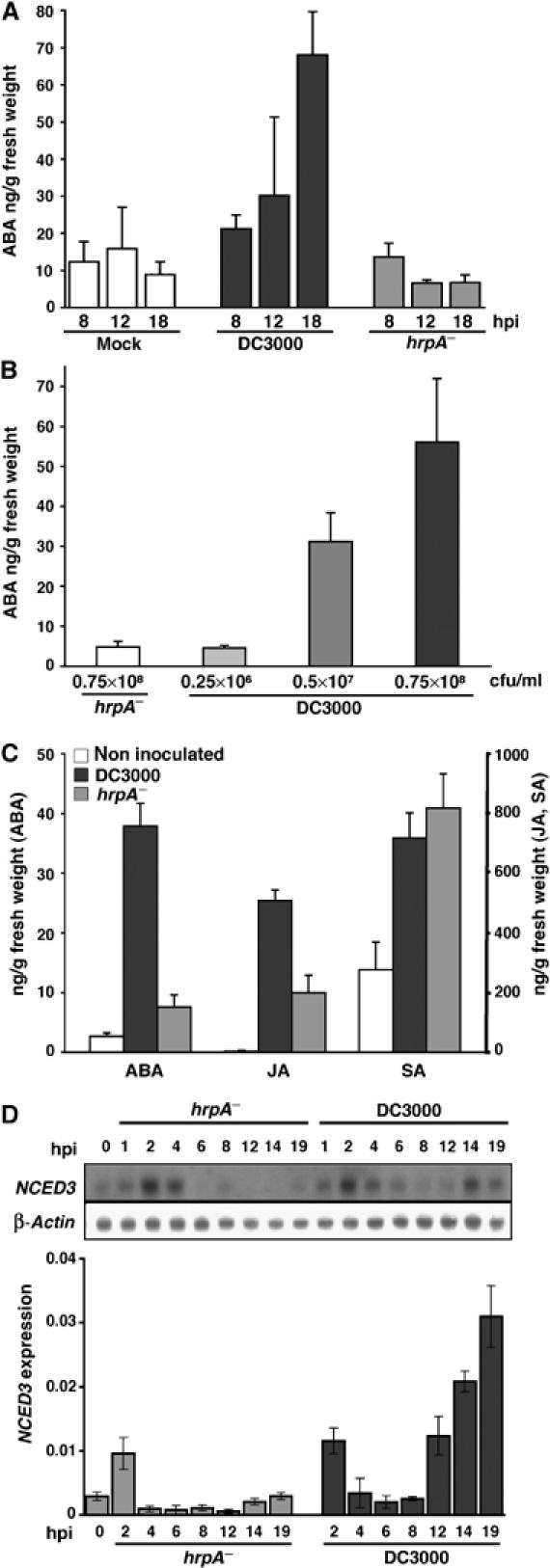
ABA levels increase during a compatible interaction in an inoculum concentration-dependent manner. (A) Time course of changes in ABA levels in Col-5 leaves after mock infiltration (MgCl2, 10 mM) or challenge with DC3000 or DC3000 hrpA−. (B) ABA levels are dependent on DC3000 concentration. ABA levels in leaves challenged with DC3000 hrpA− or increasing concentrations of DC3000 were measured 18 hpi. (C) Relative in planta levels of ABA, JA and SA during compatible (DC3000) and basal defence (DC3000hrpA−) responses. Hormones were measured in both non-inoculated and challenged (0.5 × 108 CFU/ml) Col-5 leaves at 18 hpi. (D) 9-cis-Epoxycarotenoid-dioxygenase3 (NCED3) is induced by T3Es concomitant with elevated foliar ABA. Time course of NCED3 steady-state mRNA levels following DC3000 and DC3000hrpA− challenge determined, in independent experiments, by RNA blot (upper panel) or RT–PCR. Bacterial titres and RT–PCR values (copies of target transcript/copy actin 2) are means of triplicate samples and error bars represent 1 s.d. All experiments were repeated at least twice with similar results.
NCED3, strongly induced by T3Es at 12 hpi, encodes the major stress-induced ABA regulatory enzyme, and is the most likely candidate to contribute to elevated ABA levels in the compatible interaction. In agreement with our microarray results NCED3, steady-state levels increased from 8 hpi only after challenge with DC3000 (Figure 1D). The transient increase in NCED3 transcript at 2 hpi evident in both DC3000 and DC3000hrpA−-challenged leaves resulted from the inoculation procedure, as it was also induced by mock infiltration (data not shown).
ABA application and pathogenicity responses share significant overlap
To identify common ABA signalling components induced by T3Es we interrogated experiments in the NASC arrays (http://affymetrix.arabidopsis.info) database indicative of host responses to hormone application or abiotic and biotic stresses. Hierarchical clustering of the 880 genes significantly differentially regulated between DC3000 and DC3000hrpA− challenges at 12 hpi closely associated abiotic stress experiments with compatibility. In addition to ABA treatments at 30 min, 1 and 3 h, genes modified by T3Es were also found to respond to salt and osmotic stress treatments (Figure 2A). A major inducible cluster (i) was shared between compatible responses (DC3000 challenge at 6, 12 and 24 hpi) and osmotic stress. This cluster contains a subcluster (ii) that comprises the majority of the genes represented in Table I, including seven PP2C-s, NCED3 and three NAM-s (annotated in Supplementary Table I). Two other notable clusters represent genes commonly suppressed both by T3Es and osmotic stress (iii) and genes differing in response between osmotic stress and T3Es activity (iv).
Figure 2.
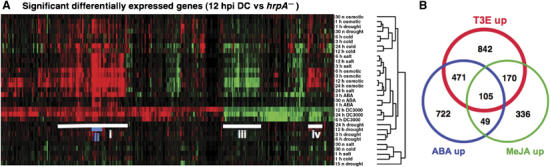
Hierarchal clustering of T3E-responsive transcripts shows a strong overlap with abiotic stresses. (A) The 880 probesets identified previously as significantly differentially expressed in response to T3Es at 12 hpi were clustered using GeneChip expression data from the AtGenExpress consortium. Experiments reporting the effects of cold, drought, salt stress, osmotic stress (mannitol) and ABA application were included as well as additional time points reporting T3E activity. All data sets were normalised and interpreted using the GCRMA function of the Bioconductor microarray analysis package (http://www.bioconductor.org/). Hierarchical clustering was applied using an uncentred correlation and complete linkage clustering. Genes induced relative to their control are coloured red, those suppressed are coloured green, whereas genes unchanged in their expression levels are coloured black. Cluster i, genes sharing strong similarity to regulatory networks induced by abiotic stresses, in particular, osmotic stress. Cluster ii, all the Clade A PP2Cs originally identified in Table I are highlighted in blue. Cluster iii, genes suppressed by both T3E and abiotic stresses. Cluster iv, genes suppressed by T3E but induced by abiotic stresses. (B) Venn diagram showing the commonality between transcripts differentially induced by T3Es and those strongly induced by ABA or MeJA application. SAM (Tusher et al, 2001) was used to identify genes induced by DC3000 relative to DC3000hrpA− or DC3000hrcC at 6, 12 and 24 hpi from two independent but similar experiments, with a minimum fold change of 2 and a false discovery rate of less than 5%. ABA- and MeJA-induced genes were identified solely on fold change taking the average of two replicates.
Given that T3Es increase foliar ABA and JA levels by 18 hpi (Figure 1) and the strong similarity between transcripts induced by ABA and T3Es (Figure 2A), we examined the overlap between responses to each hormone treatment and T3Es using experiments from the NASC database repository. The ABA response has a substantial overlap with T3E-upregulated transcripts (42%). Interestingly, 41% of the transcripts responsive to JA are also T3E induced (Figure 2B, Supplementary Table II). Collectively, these data suggest transcriptional reprogramming induced by T3E activity is highly similar to that induced through the ABA response pathway, and that ABA as well as JA signalling pathways contribute to pathogenicity.
Exogenous application of ABA enhances multiplication of wild-type DC3000 and a hrpA− mutant
The effect of exogenous application of ABA on the multiplication of DC3000 and DC3000hrpA− in challenged leaves was examined. If de novo ABA biosynthesis contributed to DC3000 virulence, we hypothesised that ABA application would allow the hrpA− mutant to overcome host basal defences. ABA pretreatment 24 h before bacterial inoculation increased DC3000 virulence by an order of magnitude (Figure 3C). Furthermore, ABA application also enabled hrpA− bacteria partially to overcome basal defences, consistently permitting a ∼five-fold increase in growth (Figure 3A, statistical significance confirmed by ‘t'-tests). Thus, exogenously applied ABA promotes colonisation by both virulent and normally non-pathogenic P. syringae.
Figure 3.
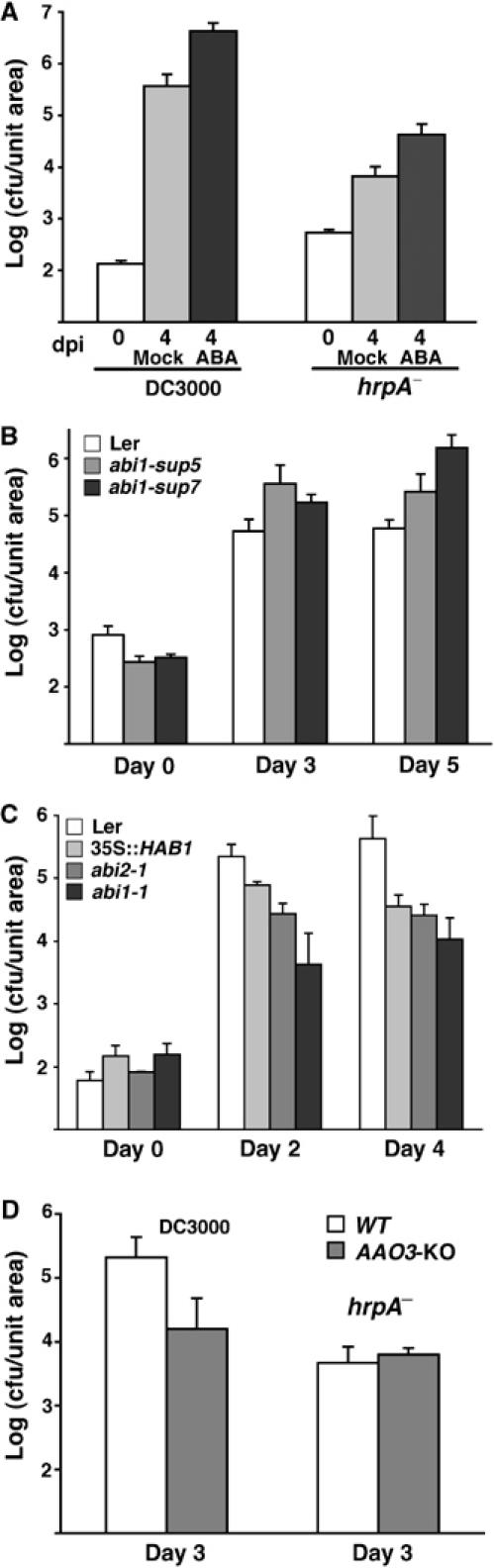
Effect of exogenous application of ABA and ABA signalling mutants on the growth of compatible bacteria. (A) Growth of DC3000 and DC3000 hrpA− on Col-0 plants sprayed 24 h before inoculation with ABA (100 μM in 0.2% ethanol) or mock (0.2% ethanol). (B) Growth of DC3000 in the leaves of wild-type Ler and the ABA hypersensitive mutants abi1-sup7 and abi1-sup5. (C) Growth of DC3000 in wild-type Ler and the ABA-insensitive mutants abi1-1, abi2-1 or transgenic 35S∷HAB1 leaves. (D) Growth of DC3000 and DC3000 hrpA− in leaves of wild-type Col-0 and the ABA biosynthetic mutant aao3. Figures are representative of at least three replicate experiments. Error bars represent 1 s.d.
ABA signalling mutants in defence
To examine the potential role of ABA signalling in pathogenicity, we tested the T3E-induced Clade A PP2Cs ABI1, ABI2 and HAB1 to determine their role in mediating the suppression of basal defense. The dominant mutations abi1-1, abi2-1 as well as 35S∷HAB1 plants show reduced sensitivity to ABA compared with wild type (Kornneef et al, 1984; Saez et al, 2004). Conversely, intragenic revertants of the originally dominant abi1-1 mutant, abi1-1sup5 and abi1-1sup7 behave as recessive alleles of ABI1 (Gosti et al, 1999). These mutants were tested for their ability to support bacterial multiplication. Figure 3B shows that the ABA-hypersensitive lines, abi1-1sup5 and abi1-1sup7, supported up to 30-fold more DC3000 multiplication. Conversely, the ABA-insensitive abi1-1 and abi2-1 mutants and the 35S∷HAB1 transgenic line were 20–80-fold more resistant than wild type (Figure 3C). In contrast to ABA application, no significant growth differences were obtained for DC3000hrpA− (not shown). Arabidopsis ABA aldehyde oxidase 3 (At2g27150; AAO3), a cytosolically localised enzyme, catalyses the last step of ABA biosynthesis in response to drought stress (Seo and Koshiba, 2002). Multiplication of virulent DC3000 in an AAO3 T-DNA knockout line (SALK_072361) was significantly restricted compared with wild type. We interpret these data to suggest that both de novo ABA biosynthesis induced by T3E delivery and PP2C activities collaborate to regulate pathogenicity.
ABA attenuates callose deposition associated with basal defence
The susceptible phenotype of the ABA hypersensitive abi1suppressor mutants correlated with enhanced chlorosis and formation of water-soaked lesions on leaves 3–4 dpi (days post-infection) (Figure 4A). A hallmark of basal defence to attempted bacterial and fungal penetration is the deposition of callose in paramural deposits. We used aniline blue staining (Keshavarzi et al, 2004) to monitor callose deposition at the cellular level (Figure 4B). Compared with wild-type leaves challenged with DC3000 at 12 hpi (Figure 4Ba), both abi1-sup5 (data not shown) and abi1-sup7 (Figure 4Bb) were completely devoid of callose-associated fluorescence. By contrast, the ABA-insensitive mutants abi1-1 (Figure 4Bc), abi2-1 and the 35S∷HAB1 line (Figure 4Bd) showed augmented callose deposition, although no significant differences in the response were detected in mutant leaves following challenge with DC3000hrpA− (data not shown). Next, mock- (Figure 4Be) and ABA-treated (Figure 4Bf) leaves were compared following virulent DC3000 challenge. As predicted, less callose-associated fluorescence was detected in ABA-treated plants 12 hpi after DC3000 challenge than in control inoculated leaves (compare Figure 4Be with 4Bf).
Figure 4.
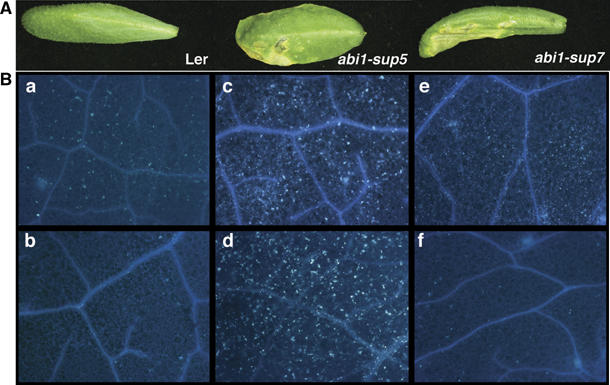
ABA signalling mutants have altered macroscopic and microscopic disease symptoms. (A) ABA-hypersensitive plants display enhanced chlorosis and necrosis 3 dpi following challenge with DC3000 (2.5 × 105 CFU/ml) compared with wild-type Ler. (B) Callose deposition in wild-type Ler (a) contrasts to reduced callose deposition in abi1-sup7 (b) and enhanced callose deposition in abi1-1 mutant and the transgenic 35S∷HAB1 line (c and d, respectively), 12 hpi with DC3000. Callose deposition in mock-treated (e) or ABA-treated (f) wild-type Col-0, 12 hpi with DC3000.
ABA suppression of genes involved in basal defense
We have shown that effectors delivered intracellularly are able to downregulate certain PAMP-induced genes (PIGs; de Torres et al, 2006; Truman et al, 2006). To examine whether ABA itself contributes to suppression of these basal defence components, we used RNA blot and RT–PCR to examine the expression level of two PIGs suppressed by T3Es, FRK1 (flagellin-induced receptor kinase 1) and the glycerol kinase encoding NHO1 (Kang et al, 2003; Truman et al, 2006) in ABA-insensitive mutants challenged with DC3000 or DC3000hrpA−. Figure 5 shows that suppression of both FRK1 (Figure 5A and B) and NHO1 (Figure 5A and C) is delayed in both abi1-1 and abi2-1 (ABA-insensitive) backgrounds. The effects of the abi1-1 and abi1-2 mutations appear to be to stabilise the relative amounts of defence transcripts. We are therefore unable to determine whether the delayed suppression of defence transcripts is due to enhanced innate immunity or an inability to activate suppression mechanisms. Whichever mechanism is operative, the elevated basal defence transcripts in the compatible interaction are consistent with the enhanced resistance to virulent DC3000 as shown in Figure 2.
Figure 5.
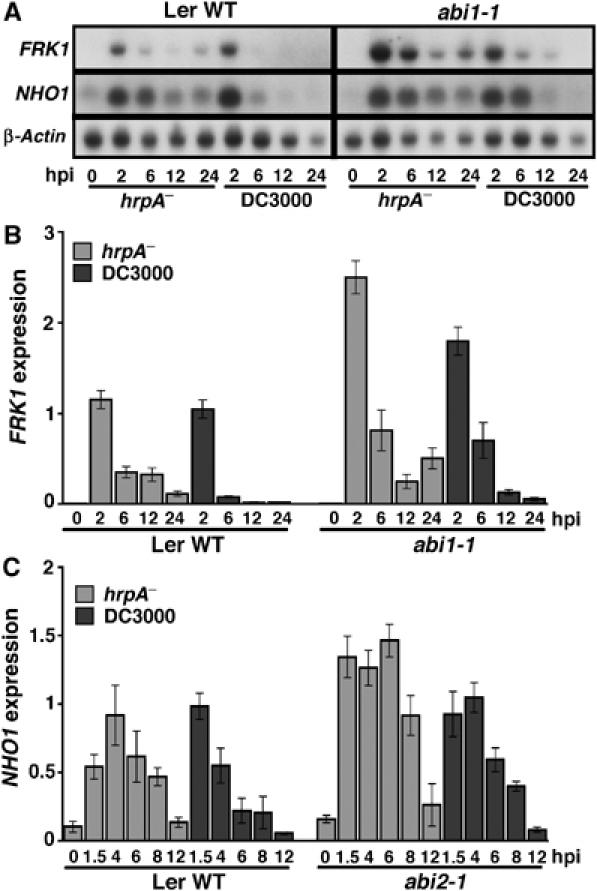
Suppression of defence gene transcripts is attenuated in ABA-insensitive mutants. (A) RNA blot of FRK1 and NHO1 transcript accumulation over time following DC3000hrpA− or DC3000 challenges of wild-type Ler (left hand panel) or the ABA-insensitive mutant, abi1-1. (B) Independent RT–PCR experiment showing suppression of FRK1 accumulation in the abi1-1 mutant is restricted after challenge with DC3000 compared with hrpA−. (C) RT–PCR reveals that suppression of NHO1 accumulation by DC3000 is also compromised in the abi2-1 mutant. RT–PCR experiments measured copies of target transcript/copy actin 2, and were in triplicate. Error bars represent 1 s.d.
Expression of the bacterial effector AvrPtoB in planta increases ABA levels
Conditional expression of the conserved P. syringae effector AvrPtoB increases susceptibility to DC3000hrpA−, suppresses callose deposition and dramatically suppresses PIG transcripts (de Torres et al, 2006). Using transgenic plants carrying avrPtoB under the control of a dexamethasone (Dex)-activated promoter, we examined NCED3, NHO1 and FRK1 transcripts in response to MgCl2 or DC3000hrpA− inoculations following Dex induction by RNA blot (Figure 6A) or RT–PCR (Figure 6B). As expected, Dex-treated leaves suppressed induction of NHO1 and FRK1 by DC3000hrpA−. However, avrPtoB expression induced NCED3 irrespective of the inoculation, suggesting that AvrPtoB alone can modify ABA signalling responses (Figure 6A and B). Notably, the levels of NCED3 mRNA induced by Dex are ∼5–6 times the levels obtained in a compatible interaction.
Figure 6.
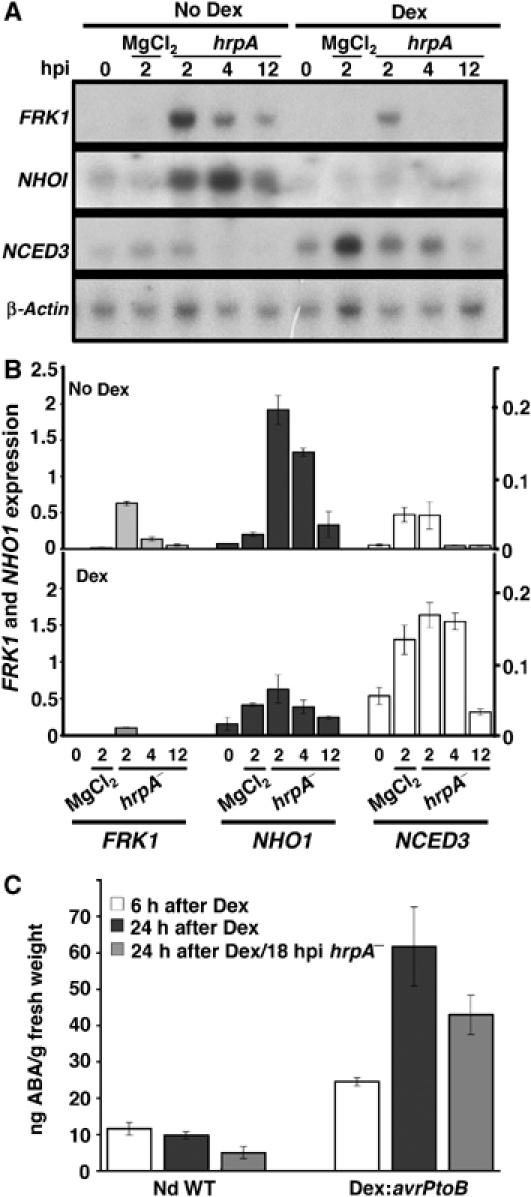
The bacteria effector, AvrPtoB, modifies basal defence transcripts and triggers elevated foliar ABA levels. (A, B) Six hours after Dex or mock treatment, Dex∷avrPtoB transgenic plants were inoculated (t=0) with MgCl2 or DC3000hrpA− and harvested for RNA extraction at the times indicated. (C) Six hours after Dex treatment leaves from Nd-0 wild type or Dex∷avrPtoB transgenic plants were inoculated with and without DC3000hrpA−. ABA levels were determined at the times indicated after Dex treatment. (A) RNA blot showing time course of defence-related (FRK1, NHO1) or ABA biosynthesis (NCED3)-related transcript accumulation in control or avrPtoB-expressing plants after challenge with DC3000hrpA−. The actin 2 transcript was used as a loading control. (B) Independent experiment of (A), measuring transcript accumulation by RT–PCR with expression levels determined relative to the equivalent actin 2 internal standard (copies of transcript/copy actin 2). Error bars represent 1 s.d. (C) Measurement of foliar ABA levels in hrpA−-challenged and unchallenged Nd-0 (control) or Dex∷avrPtoB plants 24 h following Dex induction. Error bars represent 1 s.d.
Consistent with these data, Dex treatment of Dex∷avrPtoB transgenic plants resulted in a significant increase in ABA levels within 6 h of application (Figure 6C), confirming that AvrPtoB activity alone can induce ABA synthesis. In conjunction with analyses of the PP2C mutants, these data support the hypothesis that elevated ABA contributes to the modified basal defence phenotypes previously described by conditional induction of AvrPtoB (de Torres et al, 2006).
Discussion
Examples of signalling crosstalk between biotic and abiotic stress are emerging (Xiong and Yang, 2003; Anderson et al, 2004), but results are conflicting. Antagonism between biotic and abiotic stress responses has been reported for rice (Xiong and Yang, 2003). By contrast, other studies (Park et al, 2001; Mengiste et al, 2003; Chini et al, 2004) suggest that biotic and abiotic stress responses share common components, the eventual outcome being determined by the nature of the stress and the host genotype. Here we show that bacterial virulence factors specifically manipulate components of the ABA biosynthetic and response machinery, leading to an increase in ABA levels and signal responses in leaves of Arabidopsis plants. Application of ABA at physiologically relevant concentrations enhances the susceptibility of A. thaliana to two already virulent strains of P. syringae, DC3000 and M4 (data not shown), and also nonpathogenic DC3000hrpA−. Collectively, these results suggest that ABA modifies general cellular metabolic homeostasis to facilitate bacterial growth, and can itself partially substitute for T3E activity. Moreover, conditional expression of a single bacterial effector, AvrPtoB, can enhance bacterial growth, elevate ABA levels and suppress PAMP-responsive genes. Our data suggest that de novo ABA biosynthesis is regulated by T3Es such as AvrPtoB through the induction of NCED3. These findings provide a mechanistic explanation for previous observations that ABA and/or drought stress increased susceptibility to the fungus B. cinerea, the oomycete H. parasitica and the avirulent P. syringae strain 1065 (Audenaert et al, 2002; Mohr and Cahill, 2003; Thaler and Bostock, 2004). Interestingly, apparently contradictory results demonstrate that ABA primes for callose accumulation and thereby enhances basal resistance in response to Blumeria graminis f sp hordei and activates induced resistance in response to the necrotrophic fungi Alternaria brassicicola and Plectosphaerella cucumerina (Ton and Mauch-Mani, 2004; Wiese et al, 2004). The apparently conflicting reports illustrate that ABA, like the other defence hormones JA/ET and SA, has effects which are dependent upon the mode of pathogenicity of the respective pathogen and possibly, also the nature of the PAMPs carried by each microbe.
Foliar ABA levels increased significantly within 12 hpi in a T3SS-dependent manner. DC3000 multiplied more rapidly and to greater titre in ABA hypersensitive plants compared with wild type. Restriction of virulent DC3000 growth was detected in the ABA-insensitive mutants abi1-1 and abi2-1, in lines constitutively overexpressing HAB1 and in the ABA biosynthetic mutant aao3. In abi1-1 mutants, DC3000 multiplication only slightly outperformed the non-pathogenic hrpA− mutant, corroborating our observation that hrpA− multiplication appears unaffected in abi1-1 and abi2-1 genetic backgrounds but is increased by ABA treatment in an ABI-1/ABI-2-dependent manner. ABI1 and ABI2 are PP2Cs that negatively regulate many aspects of ABA signalling. The reduced sensitivity to ABA observed in the abi1-1 and abi2-1 dominant mutants, and replicated in the aao3 biosynthetic mutant, is probably owing to the formation of inactive complexes between the corresponding mutant proteins and master positive regulators of ABA signalling (Leung et al, 1997).
What is triggering ABA accumulation? Abiotic stress-induced ABA accumulation is a key factor in controlling downstream adaptive responses, including stomatal aperture, osmolyte accumulation and gene expression. However, the mechanism by which plants sense and signal the stress-induced changes in water status and initiate adaptive responses such as ABA accumulation and osmoregulation are unknown. The increased ABA levels observed under drought and salt stress are attained by the induction of genes that catalyse hormone synthesis. NCED3 is the best candidate for direct regulation by upstream signalling and shows the strongest induction after water stress among all nine members of the gene family (Tan et al, 2003). NECD3 was induced between 4 and 12 hpi following DC3000 challenge, consistent with the elevated foliar ABA levels within ∼9 hpi (or ∼6 h after avrPtoB Dex-induced expression). NCED3 induction has therefore emerged as a possible target for effectors such as AvrPtoB.
What is the mechanism of ABA action? Manipulation of ABA biosynthesis and response pathways by T3Es represents a very powerful virulence strategy, as hormone homeostasis has a global impact on multiple cellular processes. One obvious physiological benefit for the pathogen would be ABA-mediated stomatal closure and consequent reduction in water loss. We examined the possibility that increases in ABA might lead to stomatal closure and, thereby, maintain potentially beneficial high water availability within the apoplast. Stomatal apertures were measured during the light periods 8 and 12 hpi with the hrpA− mutant or DC3000, but no significant difference in stomatal width/length ratios were found. Similarly, expression of AvrPtoB in the plant did not lead to stomatal closure 8, 12 or 27 h after Dex-mediated induction (see Supplementary data). We hypothesise that the effect of ABA accumulation on virulence is not simply through change in the physical conditions in the leaf, but through alteration of the signalling networks that coordinate plant defences. Suppression of the defensive transcriptional response requires a functional ABA signalling pathways and correlates positively with ABA levels.
It is fascinating that T3Es appear to have evolved to overcome basal defences, at least in part, through manipulating levels of a plant hormone. AvrPtoB, which is a member of the large HopAB family of effectors, singly appears to be able to suppress many components of basal defence, including alterations to the cell wall such as callose deposition (de Torres et al, 2006). Here we show that AvrPtoB induces NCED3 and elevates foliar ABA levels when conditionally expressed in planta. Moreover, transcripts associated with key regulators of basal defence, FRK1 and NHO1, are suppressed. Whether other individual effectors have evolved the ability to confer virulence through modifying ABA homeostasis remains to be determined.
ABA is normally associated with responses to abiotic stress and T3E-mediated increases in ABA may therefore exploit endogenous stress pathways to compensate for an enhanced metabolic load associated with pathogen nutrition in the apoplast. In addition to abiotic stress, ABA signal transduction is interlinked to the regulatory circuits of primary metabolism, cell growth and cell division. For instance, ABA plays a crucial role in the regulation of vegetative development by glucose (Cheng et al, 2002). The restriction of Arabidopsis growth, frequently associated with compatible Pseudomonas/Arabidopsis interactions (M de Torres, unpublished observation), may therefore represent a negative effect associated with elevated ABA levels. However, at the seed stage, ABA action is required for proper development, desiccation tolerance and long-term seed viability (Ooms et al, 1993), and therefore priming of ABA biosynthesis could confer enhanced survival to seed-borne Pseudomonads such as P. syringae pv. tomato.
Antagonism between ABA and the JA/ET signalling pathways was recently proposed as a strategy to regulate abiotic stress-related and biotic response pathways (Anderson et al, 2004). Given the large, independent overlap of ABA and JA responses with T3E-specific transcriptional reprogramming (Figure 3), our data suggest that both JA and ABA contribute to pathogenicity by the hemi-biotroph DC3000. These data are particularly interesting in light of recent results showing that ABA and SA-dependent PAMP-induced stomatal closure can be overcome by the P. syringae virulence factor coronatine, a JA mimic (Melotto et al, 2006). Thus, ABA appears to have a role in both pre-invasion innate immunity and post-invasion virulence. Analysis of relative ratios of these three hormones following challenge with coronatine-deficient strains of P. syringae pv. tomato (Brooks et al, 2004) should help resolve this issue.
Shen et al (2006) recently identified the H-subunit of Mg-chelatase as an ABA receptor (ABAR), which mediates seed germination, stomatal movement and post-germination growth. RNAi ABAR plants have corresponding ABA-insensitive phenotypes for these processes. The ABAR transcript is suppressed in plants challenged by DC3000 (Truman et al, 2006), suggesting that in addition to inducing negative regulators of ABA signalling (PP2Cs), T3Es also attenuate levels of the ABA receptor.
In summary, we have found that T3Es specifically target hormone homeostasis to promote virulence, and highlight a role for ABA alongside JA, SA and ET as phytohormone mediators of defence responses. How modification of these signalling processes contributes to pathogen multiplication remains to be elucidated. Whether or not ABA's role is simply in abrogating defensive cell wall alterations, or if it has additional roles in providing pathogen nutrition and the post-translational regulation of PIGs remains to be determined.
Materials and methods
Plants
Plants were grown under short days and 70% humidity, as previously described (de Torres et al, 2003). Source reference for the other Arabidopsis genotypes used are as follows: abi1-1 and abi2-1 (Kornneef et al, 1984), background La-er; 35S∷HAB1 (Saez et al, 2004), background Col-0; abi1-1sup5 and abi1-1sup7 (Gosti et al, 1999), background La-er; Dex∷avrPtoB (de Torres et al, 2006), background Nd-0.
In planta bacterial population counts and time courses
Bacterial cultures were maintained, prepared and used for inoculation as described (de Torres et al, 2003, 2006). Final cell density was adjusted to OD600 0.0002 (∼1 × 105 CFU ml−1) in 10 mM MgCl2 for bacterial populations counts. For hormone determinations, RNA expression studies or phenotpye assays, bacterial cell densities were typically adjusted to OD600 0.2 (∼1 × 108 cfu ml−1), unless otherwise indicated.
ABA treatment
ABA (Sigma, Dorset, UK) was solubilised in ethanol diluted in water. This ABA solution (100 μM, 0.2% ethanol) was sprayed on Arabidopsis plants. Control plants were treated identically with a solution of 0.2% ethanol. Assays were performed 24 h after ABA application.
Dex treatment
Conditional expression of avrPtoB was achieved by brushing leaves of Dex∷avrPtoB plants with a Dex (Sigma, Dorset, UK) solution (6 μM in 0.02% Silwet). Control treatment was 0.02% Silwet alone (de Torres et al, 2006).
Hormone measurements
A detailed protocol for hormone measurements is included in Supplementary data.
Callose deposition
Epifluorescence microscopy was used to detect callose after staining with aqueous aniline blue as described in Keshavarzi et al (2004). Images were recorded using a Zeiss Axiophot camera and captured using Zeiss Axiovision software.
RNA extraction and RNA blots
Total RNA was isolated as described (de Torres et al, 2003). Probe templates were amplified by PCR from cDNAs or genomic DNA and labelled with [32P]dCTP using the Prime-It®II labelling kit (Stratagene, California, USA). The genes, their AGI numbers and primers used and resultant amplicon size are detailed in Supplementary data.
RT–PCR
cDNA was generated from 1 μg of total RNA with SuperscriptIII (Invitrogen Corporation, California, USA) following the manufacturer's instructions. Quantitative PCR was performed on the cDNA using the QuantiTect SYBR Green PCR kit (Qiagen, West Sussex, UK) on a Rotor-Gene 3000 (Corbett Research, Cambridge, UK). Absolute quantification was determined by plotting standard curves using serial dilutions of the appropriate cDNA PCR products containing the target sequence. Actin 2 was used as an internal standard to normalise cDNA content in the samples. Expression levels were calculated as number of copies of each particular mRNA per number of Actin 2 copies. The primers used for RT–PCR and amplicon size are described in Supplementary data.
AAO3 T-DNA knockout
Seeds from SALK-072361 line were screened using the following gene-specific primers: F:5′-TTCTATTGGAAATGCATTGCC-3′; R:5′-CCATGTCTGCATGTTTCTGTG-3′ and the insertion-specific primer LBb1. Progeny from a homozygous plant were used for insertion subsequent experiments.
Supplementary Material
Supplementary Information
Supplementary Table I
Supplementary Table II
Supplementary Legends
Acknowledgments
We thank Wendy Byrne for expert technical assistance. This work was supported by a Wain Fellowship to MT and BBSRC grants BB/C514115/1, P18600 and P14635 to MG.
References
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K (2004) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audenaert K, De Meyer GB, Hofte MM (2002) Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol 128: 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DM, Hernandez-Guzman G, Kloek AP, Alarcon-Chaidez F, Sreedharan A, Rangaswamy V, Penaloza-Vazquez A, Bender CL, Kunkel BN (2004) Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact 17: 162–174 [DOI] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, Koshiba T, Sheen J (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14: 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Grant JJ, Seki M, Shinozaki K, Loake GJ (2004) Drought tolerance established by enhanced expression of the CC-NBS-LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. Plant J 38: 810–822 [DOI] [PubMed] [Google Scholar]
- de Torres M, Mansfield JW, Grabov N, Brown IR, Ammouneh H, Tsiamis G, Forsyth A, Robatzek S, Grant M, Boch J (2006) Pseudomonas syringae effector AvrPtoB suppresses basal defence in Arabidopsis. Plant J 47: 368–382 [DOI] [PubMed] [Google Scholar]
- de Torres M, Sanchez P, Fernandez-Delmond I, Grant M (2003) Expression profiling of the host response to bacterial infection: the transition from basal to induced defence responses in RPM1-mediated resistance. Plant J 33: 665–676 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39: 863–876 [DOI] [PubMed] [Google Scholar]
- Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M, Lamb C (2006) Systemic immunity. Curr Opin Plant Biol 9: 414–420 [DOI] [PubMed] [Google Scholar]
- Henfling J, Bostock R, Kuc J (1980) Effect of abscisic acid on rishitin and lubimin accumulation and resistance to Phytophthora infestans and Cladosporium cucumerinum in potato tuber tissue-slices. Phytopathology 70: 1074–1078 [Google Scholar]
- He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44: 903–916 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L, Li J, Zhao T, Xiao F, Tang X, Thilmony R, He S, Zhou JM (2003) Interplay of the Arabidopsis nonhost resistance gene NHO1 with bacterial virulence. Proc Natl Acad Sci USA 100: 3519–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankainen M, Holm L (2004) POBO, transcription factor binding site verification with bootstrapping. Nucleic Acids Res 32: W222–W229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavarzi M, Soylu S, Brown I, Bonas U, Nicole M, Rossiter J, Mansfield J (2004) Basal defenses induced in pepper by lipopolysaccharides are suppressed by Xanthomonas campestris pv. vesicatoria. Mol Plant Microbe Interact 17: 805–815 [DOI] [PubMed] [Google Scholar]
- Kornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic-acid insensitive mutants of Arabidopsis thaliana. Plant Physiol 61: 377–383 [Google Scholar]
- Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI (2006) The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol 140: 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 17: 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald K, Cahill D (1999) Influence of abscisic acid and the abscisic acid biosynthesis inhibitor, norflurazon, on interactions between Phytophthora sojae and soybean. Eur J Plant Pathol 105: 651–658 [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Mengiste T, Chen X, Salmeron J, Dietrich R (2003) The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15: 2551–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25: 295–303 [DOI] [PubMed] [Google Scholar]
- Mohr P, Cahill D (2003) Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora parasitica. Funct Plant Biol 30: 461–469 [DOI] [PubMed] [Google Scholar]
- Mudgett MB (2005) New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu Rev Plant Biol 56: 509–531 [DOI] [PubMed] [Google Scholar]
- Nagaoka S, Takano T (2003) Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J Exp Bot 54: 2231–2237 [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Nurnberger T, Brunner F, Kemmerling B, Piater L (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev 198: 249–266 [DOI] [PubMed] [Google Scholar]
- Ooms J, Leon-Kloosterziel KM, Bartels D, Koornneef M, Karssen CM (1993) Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana (a comparative study using abscisic acid-insensitive abi3 mutants). Plant Physiol 102: 1185–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Park CJ, Lee SB, Ham BK, Shin R, Paek KH (2001) Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13: 1035–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Zeevaart JA (1999) The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc Natl Acad Sci USA 96: 15354–15361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL (2004) Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J 37: 354–369 [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9: 236–243 [DOI] [PubMed] [Google Scholar]
- Seo M, Koshiba T (2002) Complex regulation of ABA biosynthesis in plants. Trends Plant Sci 7: 41–48 [DOI] [PubMed] [Google Scholar]
- Shen YY, Wang XF, Wu FQ, Du SY, Cao Z, Shang Y, Wang XL, Peng CC, Yu XC, Zhu SY, Fan RC, Xu YH, Zhang DP (2006) The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443: 823–826 [DOI] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Thaler JS, Bostock RM (2004) Interactions between abscisic-acid-mediated responses and plant resistance to pathogens and insects. Ecology 85: 48–58 [Google Scholar]
- Ton J, Mauch-Mani B (2004) Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J 38: 119–130 [DOI] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W, de Zabala MT, Grant M (2006) Type III effectors orchestrate a complex interplay between transcriptional networks to modify basal defence responses during pathogenesis and resistance. Plant J 46: 14–33 [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward EWB, Cahill DM, MK B (1989) Abscisic acid suppression of phenylalanine ammonia lyase activity and mNA, and resistance of soybeans to Phytophthora megasperma f.sp. glycinea. Plant Physiol 91: 23–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese J, Kranz T, Schubert S (2004) Induction of pathogen resistance in barley by abiotic stress. Plant Biol (Stuttgart) 6: 529–536 [DOI] [PubMed] [Google Scholar]
- Xin Z, Zhao Y, Zheng ZL (2005) Transcriptome analysis reveals specific modulation of abscisic acid signaling by ROP10 small GTPase in Arabidopsis. Plant Physiol 139: 1350–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Yang Y (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15: 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Felix G (2005) Plants and animals: a different taste for microbes? Curr Opin Plant Biol 8: 353–360 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Table I
Supplementary Table II
Supplementary Legends


