Abstract
Mycobacterium tuberculosis is responsible for nearly 3 million human deaths worldwide every year. Understanding the mechanisms and bacterial factors responsible for the ability of M. tuberculosis to cause disease in humans is critical for the development of improved treatment strategies. Many bacterial pathogens use pili as adherence factors to colonize the host. We discovered that M. tuberculosis produces fine (2- to 3-nm-wide), aggregative, flexible pili that are recognized by IgG antibodies contained in sera obtained from patients with active tuberculosis, indicating that the bacilli produce pili or pili-associated antigen during human infection. Purified M. tuberculosis pili (MTP) are composed of low-molecular-weight protein subunits encoded by the predicted M. tuberculosis H37Rv ORF, designated Rv3312A. MTP bind to the extracellular matrix protein laminin in vitro, suggesting that MTP possess adhesive properties. Isogenic mtp mutants lost the ability to produce Mtp in vitro and demonstrated decreased laminin-binding capabilities. MTP shares morphological, biochemical, and functional properties attributed to bacterial pili, especially with curli amyloid fibers. Thus, we propose that MTP are previously unidentified host-colonization factors of M. tuberculosis.
Keywords: adherence, antigen, laminin, amyloid
Tuberculosis remains the most devastating bacterial cause of human mortality (1). Despite improved diagnosis, surveillance, and treatment regimens, the incidence of TB increases annually (2). The ability to combat this deadly pathogen hinges on the dissection and understanding of the mechanisms of pathogenesis for Mycobacterium tuberculosis. Central to the ability of the microbe to cause disease is the capability to survive and replicate within macrophages by avoiding lysosomal fusion with the mycobacteria-containing phagosome (3). M. tuberculosis interacts with and invades various human and animal epithelial cells in culture and appears to possess multiple mechanisms of entry into macrophages (4–6). Despite these long-standing and recent observations, very little is known regarding host–microbe interactions and events between M. tuberculosis and host cells before survival and replication within the macrophage. Furthermore, the specific bacterial adhesins involved in the complex interplay between M. tuberculosis and the human host are largely unknown. Nevertheless, a few potential adherence factors are considered, including a heparin-binding hemagglutinin (HBHA), a fibronectin-binding protein family or antigen 85 complex, and the subfamily of polymorphic acidic, glycine-rich proteins, called PE_PGRS. HBHA is a surface-exposed protein that is involved in binding of the bacillus to epithelial cells but not to phagocytes (7–9), and the experimental data available suggest that HBHA is important in extrapulmonary spread after the initial long-term colonization of the host. Fibronectin-binding proteins, first identified as the 30-kDa or α-antigen (10, 11), are mycolyltransferase enzymes (12) that can bind to the extracellular matrix (ECM) protein fibronectin in vitro (11). This property may represent a mechanism of tissue colonization. The surface-exposed PE_PGRS proteins are found in M. tuberculosis and Mycobacterium bovis (13–16). M. bovis bacillus Calmette–Guérin with a mutation in the PE_PGRS gene Rv1818 was found to be less aggregative in liquid culture and showed reduced ability to infect J774 macrophages (14). The M. tuberculosis PE_PGRS protein encoded by the ORF Rv1759c also shows fibronectin-binding properties (17).
Many bacteria pathogenic to plants and animals produce polymeric adhesive organelles termed pili, or fimbriae, to facilitate the initial attachment and subsequent successful colonization of eukaryotic cells (18). Pili are polymeric, hydrophobic, proteinaceous structures generally composed of a major repeating subunit called pilin and, in some cases, a minor tip-associated adhesin subunit. Pili are involved in many virulence-associated functions, such as agglutination of human and animal erythrocytes, bacterial aggregation, biofilm formation, adherence, and colonization of mucosal surfaces (18, 19). Because of their key role in bacterial pathogenesis, pili are viewed as virulence factors and, therefore, as important targets for vaccine development (18). It is widely held that mycobacteria do not produce pili. Here, we provide compelling ultrastructural, biochemical, and genetic data that show that M. tuberculosis produces pili, whose pilin subunit is encoded by the Rv3312A gene. Sera from convalescent TB patients contain antibodies that specifically react to the previously unidentified fibrous organelle. Furthermore, the role of MTP as adhesive structures is supported by our findings that purified MTP bind to the extracellular matrix protein laminin and that mtp mutants are unable to bind to laminin. In all, our data suggest that MTP could be used by the mycobacteria as a mechanism to colonize the human host.
Results
M. tuberculosis Cells Produce Pili.
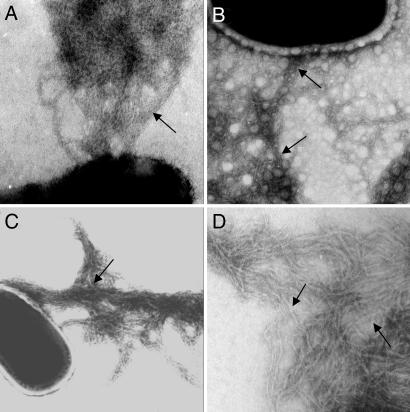
Pili are present on many bacteria that cause disease in the human respiratory tract including the Gram-positive pathogens Group B Streptococcus and Corynebacterium diptheriae (20, 21); this latter being a close relative of Mycobacterium in terms of phylogeny and complex cell-wall architecture (22). Previous ultrastructural analyses have suggested that pathogenic mycobacteria produce fibers reminiscent of pili (23, 24). We investigated the production of pili in M. tuberculosis cells propagated under various standard culture conditions by negative staining and transmission electron microscopy (TEM). We found that M. tuberculosis H37Rv, when grown for 2–3 weeks on Middlebrook 7H10 or 7H11 solid mycobacteriological medium, produced a dense fibrillar meshwork composed of thin (2- to 3-nm-wide), coiled-coil, aggregated fibers, resembling pili that extended many microns away from the bacterial surface (Fig. 1A). These organelles were named Mycobacterium tuberculosis pili or MTP. At least 10% of individual mycobacteria (n = 100) examined under these culture conditions produced these MTP structures. Similar structures were seen in avirulent strain H37Ra (Fig. 1B) and the clinical isolate CDC1551 (Fig. 1C), suggesting that MTP production is a common property among M. tuberculosis strains. The morphology of MTP is strikingly similar to that of the well characterized pili called curli produced by Escherichia coli and Salmonella enterica (25, 26).
Fig. 1.
Demonstration of pili production by M. tuberculosis strains. Transmission electron micrographs of negatively stained M. tuberculosis strains H37Rv (A), avirulent H37Ra (B), clinical isolate CDC1551 (C), and purified MTP (D). Black arrows point to MTP pili fibers. (Magnification: ×45,000.)
M. tuberculosis ORF Rv3312A Encodes the MTP Structural Subunit.
MTP was isolated from M. tuberculosis by using mechanical shearing and differential centrifugation techniques commonly used to prepare pili from other microorganisms (27). Because of the lipid-rich nature of the M. tuberculosis cell wall, an additional extraction step involving chloroform and methanol was performed to remove unwanted lipids from the pili preparation. Copious amount of intact pili were observable by TEM in the chloroform/methanol-insoluble MTP fraction (Fig. 1D). However, after analysis by SDS/PAGE and Coomassie brilliant blue and silver staining procedures, no obvious MTP monomer in the range typical for pilins, 10–25 kDa, was obtained [see supporting information (SI) Fig. 5]. Thus, it appears that MTP are not readily dissociated into subunits by standard reducing and denaturing conditions, a characteristic consistent with the biochemical properties known for curli and type I pili (26, 28).
Because of the difficulty in dissociating MTP into monomeric subunits, we sought to identify the nature of the pilin subunit by direct analysis using liquid chromatography-tandem mass spectrometry of acid hydrolysates from purified MTP (SI Fig. 6). Three independently prepared pili-enriched fractions produced a mass spectrum (SI Fig. 6A) associated with a peptide fragment having a monoisotopic mass (Mr) of 1,086.55 Da and a sequence of PGAAPPPPAAGGGA (SI Fig. 6B). The MTP-associated sequence corresponded to the carboxyl terminus of a predicted protein of 10.5 kDa encoded by M. tuberculosis H37Rv ORF Rv3312A (herein called mtp) (SI Fig. 6C) (13). The MTP-associated protein is predicted to have a transmembrane domain located between residues 10 and 30 (SI Fig. 7). These are traits consistent with a pilin protein (29). Affinity-purified antibodies produced against a peptide containing amino acids 60–79 (SI Fig. 6C) of the identified mtp-encoded protein, specifically detected a 5-kDa product in Western blots of purified MTP (SI Fig. 8). Preimmune serum did not react with protein (SI Fig. 8A, lane 2). This result is in agreement with the MS/MS analysis of MTP and suggests the presence of a cleaved signal sequence or other proteolytic modification of the prepilin, Mtp. These observations regarding the molecular mass of the mature mtp-encoded protein are also consistent with the internal amino terminus sequence noted in the reannotation of this particular M. tuberculosis ORF (30). ImmunoGold electron microscopy experiments provided further evidence that the mtp gene product is the MTP structural subunit, because Mtp peptide-specific antibodies bound to purified fibers (SI Fig. 8B). Nearly all of the gold particles are seen decorating individual filaments. In contrast, no labeling was obtained with preimmune serum (data not shown). To further confirm these findings, M. tuberculosis H37Rv mtp was amplified from chromosomal DNA, cloned, and expressed as a recombinant His6-tagged protein in E. coli BL21. The recombinant His-Mtp migrated with an apparent molecular mass of 14.5 kDa (SI Fig. 8C, lane 2) and reacted specifically with anti-Mtp peptide-specific antibodies (anti-Mtp) by Western blotting (SI Fig. 8C, lane 3). The fact that recombinant Mtp ran as a full-length product suggests that E. coli lacks the mechanism for the putative cleavage of the Mtp prepilin because the mature molecule from purified MTP migrates at 5 kDa. Taken together, these data show that MTP are composed of the mtp (Rv3312A) gene product in M. tuberculosis.
Rv3312A Is Required for Production of MTP Fibers in M. tuberculosis.
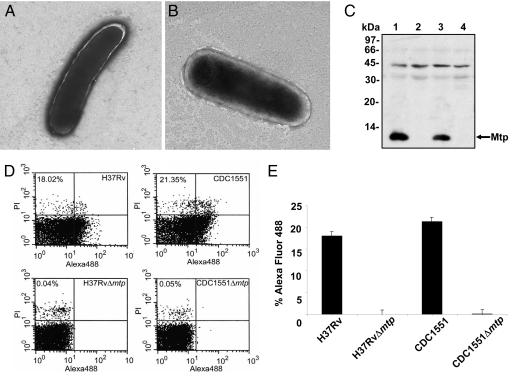
To provide genetic evidence of the role of mtp in production of the fibers, mtp of H37Rv and the homologous ORF MT3413 of CDC1551 were targeted for mutation by means of specialized transduction (31). The mutations were confirmed by PCR using internal gene-specific primers and flanking primers (SI Fig. 9), and the effect of the deletion on the ability of the mutants to produce pili was determined by TEM and Western blotting. Neither H37RvΔ mtp nor CDC1551Δ mtp were able to produce MTP (Fig. 2 A and B), and no synthesis of the subunit occurred in the mutants (Fig. 2C). Lastly, surface production of Mtp in the wild-type strains and lack thereof in the derivative mtp mutants was confirmed by flow cytometry using anti-Mtp antibody (Fig. 2D). Eighteen percent of H37Rv and 20% of CDC1551 cells analyzed (n = 50,000), expressed MTP on their surface, whereas the mtp mutants lacked detectable MTP (Fig. 2E).
Fig. 2.
Isogenic M. tuberculosis mtp mutants lack production of pili fibers. (A and B) Electron micrographs of H37RvΔ mtp (A) and CDC1551Δ mtp (B) showing no filamentous structures. (Magnification: ×25,000.) (C) Anti-Mtp Western blot of whole bacteria extracts showing production of the pilus protein in H37Rv (lane 1) and CDC1551 (lane 3) and absence of the pilin in both H37RvΔ mtp (lane 2) and CDC1551Δ mtp (lane 4). (D) Surface detection of MTP in wild-type parental and derivative mtp deletion strains by flow cytometry (n = 50,000) using anti-Mtp antisera. MTP–antibody complexes were detected by using Alexa Fluor conjugate. (E) Plot of the flow cytometry values obtained in D.
M. tuberculosis Pili Bind to Laminin.
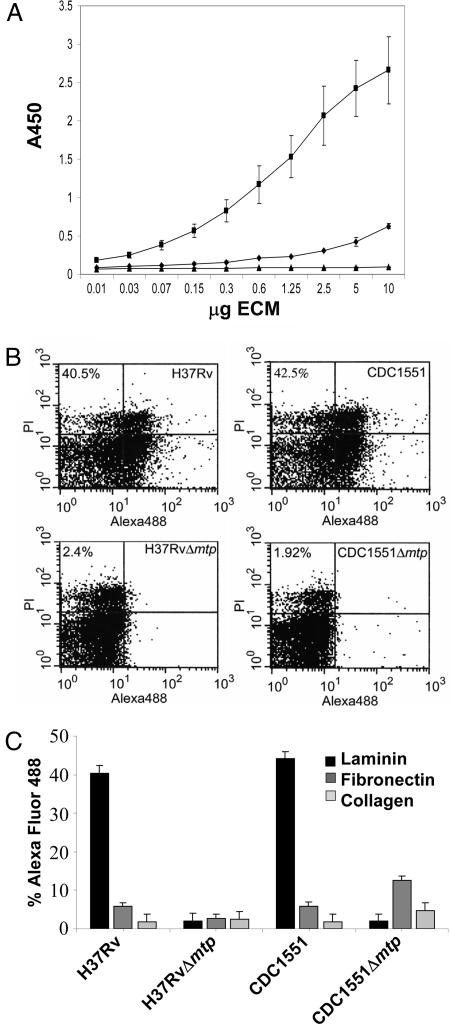
Pili mediate specific recognition of host-cell receptors facilitating close contact and tissue colonization (18, 32). Previous studies have shown that M. tuberculosis preferentially attaches to and invades damaged areas of the human respiratory mucosa in an organ culture system (33). In these instances of tissue damage, ECM proteins may be more highly exposed than in healthy tissue. Purified MTP was tested for its ability to interact with ECM proteins by using an ELISA-based assay. We found that MTP has a strong dose-dependent affinity for laminin (Fig. 3A) but lacked significant binding affinity for fibronectin or type IV collagen. To determine whether the laminin-binding property observed for the native MTP fibers was attributable to the mtp gene product, the ability of M. tuberculosis H37RvΔ mtp and CDC1551Δ mtp to bind laminin were compared with the wild-type parental strains by using flow cytometry. It was found that both mtp mutants demonstrated a 40-fold reduction in laminin-binding capacity versus the wild-type strains (Fig. 3 B and C). These data suggest that MTP mediates adhesion of M. tuberculosis to the extracellular matrix, an event that would facilitate direct interaction between the bacilli and the host epithelium during TB infection in the lung or other tissues.
Fig. 3.
MTP binds to laminin. (A) ELISA-based assay to measure the binding of increasing concentrations of fibronectin (♦), laminin (■), and collagen type IV (▴) to MTP-coated plates. Binding was detected by using either rabbit anti-fibronectin, anti-laminin, or mouse anti-collagen type IV. All ELISA values given are averaged from triplicate experiments. Error bars represent the standard deviation from the mean. (B) Flow cytometry demonstration of binding of laminin to MTP-producing M. tuberculosis but not to mtp mutants. Binding to fibronectin and collagen was negligible (n = 50,000). Binding was determined by using anti-ECM antibodies described above followed by detection using Alexa Fluor conjugate. (C) Bars indicate relative Alexa Fluor signal from flow cytometry. Data are presented from triplicate experiments.
Sera from TB Patients Contain MTP-Reactive IgG Antibodies.
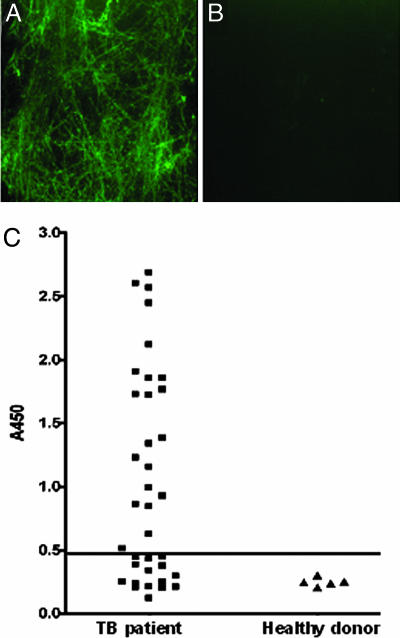
During TB infection, M. tuberculosis-specific molecules are recognized as antigens by the host immune system and induce the production of antibodies. The presence of these antibodies represents a powerful indication that the antigen is produced during natural infection. If M. tuberculosis produces MTP during infection of the human host, it would be possible to detect anti-Mtp antibodies in sera collected from TB patients. Thus, MTP was used in immunofluorescence (IF) microscopy experiments using sera from TB patients (n = 36) in various stages of treatment to detect antibody-MTP complexes. Sera from healthy donors (n = 5) were used as negative controls. Among the 36 individual TB patient sera tested, 60% produced a positive reaction defined by the presence of a characteristic fibrous meshwork of intense fluorescence (Fig. 4A). In contrast, none of the control sera gave a positive reaction with MTP because no fluorescent fibers were observed (Fig. 4B). No reactivity was obtained between purified MTP and goat anti-human IgG Alexa Fluor conjugate (in the absence of human sera), confirming the specificity of the IF reaction (data not shown).
Fig. 4.
MTP are produced during human TB infection. (A) IF showing that MTP incubated with TB patient sera produced fluorescent MTP fibers. (B) No reaction was observed when MTP was incubated with sera from healthy donors. (Original magnification: ×1,000.) (C) Sera from TB patients (n = 36) and from healthy donors (n = 5) were tested for anti-MTP IgG antibodies by ELISA. Most of the patients' sera (60%) showed a significant IgG titer against immobilized MTP fibers. Results obtained at sera diluted 1:3,200 are shown. The horizontal line represents the cut-off value of two times the average ELISA reading of healthy control sera. All of the sera positive by ELISA produced a positive IF reaction as shown in A.
ELISA was used to determine the titer of anti-MTP IgG in the panel of TB patient sera. It was observed that 60% had anti-MTP IgG titers >3,200, whereas healthy control sera did not react significantly with MTP (Fig. 4C). As predicted, nearly all of the human sera that displayed positive reactivity to MTP by IF had ELISA anti-MTP IgG titers of 3,200 or greater (Fig. 4C). The finding that MTP-specific antibodies were found in human sera obtained from TB patients provides compelling evidence that MTP are immunogenic and are produced by M. tuberculosis during infection of the human host. Showing that MTP are antigenic is supported by the reannotation of Rv3312A as a secreted antigen (30).
Discussion
Pili-mediated attachment to phagocytic cells is important for host-defense mechanisms either through direct pilus–macrophage interaction or by the presence of anti-pilin opsonizing antibody (19). It is well documented that M. tuberculosis adheres to and invades macrophages and epithelial cells; however, the bacterial factors involved in the colonization of the host are largely unknown. Here, we provide compelling ultrastructural, biochemical, and genetic data that support the notion that M. tuberculosis produces pili, herein called MTP, and this observation has important implications regarding our understanding of both pathogenesis and basic physiology of the tubercle bacillus.
Although much study has been devoted to the identification of cell surface components involved in virulence of and protective immunity against M. tuberculosis, the majority of studies have been conducted on the complex lipids associated with the cell wall (34–36). The lipid-rich cell wall of mycobacteria has been described as wax-like, and many ultrastructural analyses attempted to relate visible structures with known cell-wall chemical entities (23, 37, 38). These glycopeptidolipids or mycosidic surface teguments of mycobacteria often appeared fibrillar and, unlike MTP, are transparent by TEM (23, 37–39).
Recently, pili have been identified in Gram-positive Group B Streptococcus and C. diphtheriae (20, 21, 40). Assembly of pili in these pathogens is associated with sortase genes, whose products are involved in covalent linkage of the pilins to the cell wall peptidoglycan (20, 21, 40). Because M. tuberculosis lacks any identifiable sortase-encoding genes (41), it is unlikely that MTP are assembled in a sortase-dependent manner. Because M. tuberculosis has a unique cell-wall architecture, it remains possible that the Mtp assembly factors are not similar to known pili assembly genes. Further studies screening M. tuberculosis mutants for the loss of MTP expression could reveal the factors required to assemble the MTP structures.
The ORF that encodes Mtp in the M. tuberculosis H37Rv was annotated as Rv3312A (herein called mtp) and deletion of this ORF in two virulent strains abrogated production of MTP. mtp is not organized in an operon or clustered with genes that are generally associated with pili biogenesis. Thus, it is possible that the biogenesis genes are scattered away from mtp on the M. tuberculosis chromosome, as is the case of the biogenesis genes of type IV pili of Pseudomonas aeruginosa (19). Among the available mycobacterial genome databases, the mtp gene is present only in M. tuberculosis, M. bovis, and Mycobacterium avium ssp paratuberculosis but was not found in Mycobacterium smegmatis by using BLAST analysis. This suggests that MTP production may be limited to pathogenic mycobacteria.
It is not surprising that Mtp lacks homology to known pilins because of the fact that few Gram-positive pili have been molecularly characterized. The Mtp protein does, however, possess many characteristics of pilin molecules. Namely, it is a low-molecular-weight protein that undergoes processing, 60% of the amino acid sequence is hydrophobic, and it contains two cysteine residues at the carboxy terminus, which may be important for disulfide formation and protein stability.
In several respects, MTP resembles a unique pili family of bacterial amyloids called curli (42). Properties in common are (i) MTP and curli are morphologically indistinguishable; they are both highly sticky, aggregative, and insoluble fibers; (ii) both pili types bind laminin (25, 43); the interaction between MTP and laminin could be attributed to the presence of a collagen-like repeat beginning at residue 30 of the Mtp protein. Additionally, binding to laminin may be an important feature of M. tuberculosis, and evidence exists to suggest that another protein, malate synthase, is also a laminin-binding adhesin (44) that is recognized by antibodies contained within TB patient sera (45); (iii) both pili types bind Congo red dye (data not shown), a property associated with bacterial aggregation and human amyloidogenic proteins; and (iv) biochemically, although no obvious primary sequence homology between MTP and curli exists, both proteins contain large numbers of glycine and proline residues, which account for the hydrophobicity that is a common property of pilins.
Several bacterial pili have been shown to bind ECMs. During TB, an orchestrated inflammatory cell-mediated immune response is generated by the host in an attempt to contain the infection within a granuloma. Exposure of ECMs during tissue damage due to inflammatory immune responses may present targets for binding of invasive pathogens, like M. tuberculosis, to host tissues. We demonstrated that purified MTP and piliated M. tuberculosis strains bound to laminin. Thus, the sticky nature of MTP and its ability to bind laminin are adhesive properties.
If MTP-mediated events are critical for M. tuberculosis to establish TB infection, as in other microbes, then Mtp could be considered an attractive vaccine candidate. Because MTP are produced in vivo, and the M. tuberculosis habitat is the human body and is transmitted directly from person to person, it is likely that pili play an important role in some aspect of human TB infection. Furthermore, TB patients contain antibodies to the native MTP antigen, providing evidence that the pili are produced in vivo. In support of their possible role in M. tuberculosis pathogenesis, studies have shown that M. tuberculosis mutants in the two component signal-transduction system PhoPR are attenuated in vivo (46, 47) and that mtp (Rv3312A) is among 44 genes positively regulated by that system (46). From the knowledge of how pili function for other bacterial pathogens and the studies presented here, it is reasonable to surmise that M. tuberculosis, like many bacterial pathogens, could use structural organelles known as pili to facilitate colonization of the human host.
Materials and Methods
Strains and Culture Conditions.
M. tuberculosis H37Rv (virulent) and H37Ra (attenuated) are laboratory strains and M. tuberculosis strain CDC1551 is a recent clinical isolate, whereas M. smegmatis mc2155 is a saprophyte. Culture and manipulation of virulent strains were performed under standard biosafety level 3 (BSL-3) laboratory procedures. M. tuberculosis was propagated on Middlebrook 7H10 or 7H11 complete agar media (Difco, Sparks, MD) containing 10% oleic acid, albumin, dextrose, catalase (OADC) and Tween 80 (Tw) for 2–3 weeks at 37°C in a humidified incubator containing a 5% CO2 atmosphere. M. smegmatis was grown on 7H10 agar. All cultures were Gram and Ziehl–Neelson stained to confirm their purity.
Electron Microscopy and ImmunoGold Labeling.
For ultrastructural analyses by TEM, virulent M. tuberculosis strains were first fixed in 4% paraformaldehyde (Ted Pella, Redding, CA). Ten microliters of a bacterial suspension or pili preparation were negatively stained with 1% phosphotungstic acid (pH 7.4) on 300-mesh carbon formvar copper grid (Ted Pella) and viewed by using a CM12 electron microscope (Philips, Eindhoven, The Netherlands) at 80 kV at the University of Arizona Electron Microscopy Core Facility. For ImmunoGold labeling, samples were incubated for 1 h with a 1:10 dilution of primary antibody, followed by 1-h incubation with the anti- rabbit IgG 10-nm gold conjugate (BBL, Cockeysville, MD) (48).
Preparation of Purified MTP.
MTP were purified by a procedure previously described, with modifications (27). Briefly, M. tuberculosis H37Ra was cultured in 7H9-OADC-Tw at 37°C with shaking for 48–72 h. The starter culture was used to inoculate 100 7H11 agar plates, as lawns, modified by the exclusion of OADC and incubated for 2 weeks as described above. The bacteria were harvested by using a sterile glass spreader and suspended in 200 ml of PBS. The suspension was divided into 25-ml aliquots, and the pili were mechanically sheared from the bacterial surface by vigorous vortexing for 5 min in 50-ml conical tubes containing 1 cc of sterile 3-mm glass beads. After shearing, the bacterial suspensions were centrifuged at 3,000 × g for 1 h, and the supernatant was collected. Bacterial pellets were washed with PBS to recover more pili, and the supernatants were pooled. Remaining bacterial cells and debris were eliminated by two centrifugations at 3,000 × g, followed by a centrifugation at 18,000 × g for 10 min. Contaminating lipids were removed after vigorous mixing for 1 h with an equal volume of chloroform/methanol (2:1). After centrifugation at 18,000 × g for 30 min to partition phases in 35-ml centrifuge tubes (Fisher, Rockville, MD), the aqueous and interphase fractions were carefully collected and extracted twice more, and the soluble material (lipids) in the organic solvents was discarded. The final aqueous and interphase fractions were ultracentrifuged at 120,000 × g for 3 h at 4°C in a Ti-56 fixed angle rotor (Beckman, Fullerton, CA). The resulting MTP-enriched pellets (as determined by TEM) were resuspended in PBS and analyzed by 16% SDS/PAGE (49) or Tricine-PAGE. Protein concentrations were determined by the BCA protein assay (Pierce, Rockford, IL).
Antibody Production and Western Blotting.
An immunogenic portion of the pilin was selected from the deduced protein sequence of ORF Rv3312A. The peptide CHDDFHRDSDGPDHSRDYPG (residues 60–79 of ORF Rv3312A) was synthesized and conjugated to keyhole limpet hemocyanin (KLH) to obtain rabbit polyclonal peptide-specific antisera (Zymed, San Francisco, CA) by using TiterMax adjuvant (CytRx). For Western blotting, 100 μg of total protein or normalized bacterial suspensions were electrophoresed in 16% polyacrylamide gel, blotted onto a PVDF PSQ membrane (Millipore, Bedford, MA), and reacted for 1 h with prebleed or immune sera (anti-Mtp peptide) diluted 1:2,500. Next, goat anti-rabbit IgG HRP conjugate (Pierce) diluted 1:20,000 was added for 1 h and the reaction detected with a chemiluminescent substrate (Pierce).
IF.
Fifty micrograms of enriched MTP fibers were dried onto sterile 12-mm glass coverslips (Fisher) before fixation in 3–4% paraformaldehyde. TB patient sera (n = 36) or healthy (PPD-negative) donor sera (n = 5) were used as primary antibodies at 1:1,000 dilutions in PBS containing 10% FBS. After 1-h incubations and washes with PBS, goat anti-human IgG Alexa Fluor 488 conjugate (Molecular Probes, Eugene, OR) was added for 1 h. The samples mounted for IF visualization with UV light on a TE 2000S microscope (Nikon, East Rutherford, NJ) (48). Images were obtained as TIFF files by using Metacam software and adjusted for consistent contrast by using Photoshop 7.0 (Adobe Systems, Mountain View, CA).
ELISA.
Flat-bottom ELISA plates (Greiner, Monroe, NC) were coated overnight at 4°C with 1.5 μg per well of MTP in 150 mM carbonate buffer, pH 9.5. After washing and blocking, the wells were incubated with serial dilutions of human sera, followed by incubation with the secondary antibody (DAKO, Glostrup, Denmark). The reaction was developed with TMB single solution substrate (Zymed) and stopped with 1 M HCl before reading absorbance at 450 nm by using a microtiter plate reader (Bio-Rad, Hercules, CA). Background absorbance from empty control wells was subtracted from test samples.
Flow Cytometry.
Flow cytometry was used to detect the production of Mtp and to measure binding of the mycobacteria to ECMs. M. tuberculosis cells were harvested from 7H11 plates into sterile PBS, vortexed, and allowed to settle. To avoid clumping bacteria, the upper portion of the suspension was removed and diluted to an optical density of 1.1, and 45-μl aliquots were incubated with 25 μl of anti-Mtp peptide antibodies by using dilutions of 1:1,000 for 1 h on ice. After three gentle washes with PBS, the bacteria were resuspended in 25 μl of a dilution of goat anti-rabbit IgG (H+L) Alexa Fluor conjugate (Invitrogen, Carlsbad, CA). After 1-h incubation at 4°C, the bacteria were gently washed three times with PBS and resuspended in 800-μl final volume of PBS. For the analysis, the bacteria were labeled with 5 μl of a propidium iodide solution (Sigma, St. Louis, MO). Propidium iodide was visualized through a 42-nm band pass centered at 585. These experiments were repeated in triplicate. The FITC fluorescence emission was collected through a 30-nm band-pass filter centered at 530 in which 50,000 events were measured. The samples were analyzed at the ARL Biotechnology/ACCC Cytometry Core Facility at the University of Arizona, by using a FACScan (Becton Dickinson, Franklin Lakes, NJ). For measuring the binding of bacteria to ECMs, mycobacteria were incubated for 1 h at 4°C with 0.125 μg of laminin, fibronectin, or type IV collagen. After incubation, the cells were washed with PBS and incubated with anti-ECM antibodies. Before analysis, the ECM-antibody complexes were detected by using anti-rabbit or anti-mouse Alexa Fluor conjugate (Invitrogen).
ECM Binding Assay.
A sandwich ELISA was used to detect and quantitate the binding affinity of MTP for ECM proteins by using 1.5 μg of purified MTP immobilized onto ELISA plates as described above. Serial dilutions of fibronectin, laminin, or type IV collagen were added for 1 h, followed by incubation with rabbit anti-fibronectin, rabbit anti-laminin, or mouse anti-type IV collagen (Sigma). The sandwiched complex was detected by using anti-rabbit or anti-mouse IgG peroxidase conjugate (Sigma), developed and read as described above.
Construction of mtp Mutants by Specialized Transduction.
Propagation and titering of mycobacteriophages, construction of allelic exchange substrates, and specialized transduction of M. tuberculosis were performed essentially as described (31). Additional details are provided as SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Esteban A. Roberts and James W. Moulder for critical review of the manuscript and Frederick J. Cassels, Lee W. Riley, and Bentley A. Fane for helpful discussions. This work was supported by National Institutes of Health Grant AI45537-01 (to R.L.F.) and a University of Arizona Small Grant (to R.L.F. and J.A.G.).
Abbreviations
- ECM
extracellular matrix
- IF
immunofluorescence
- MTP
M. tuberculosis pili
- TEM
transmission electron microscopy.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0602304104/DC1.
References
- 1.Dye C, Scheele S, Dolin P, Pathania V, Raviglione MC. J Am Med Assoc. 1999;282:677–686. doi: 10.1001/jama.282.7.677. [DOI] [PubMed] [Google Scholar]
- 2.Frieden TR, Sterling TR, Munsiff SS, Watt CJ, Dye C. Lancet. 2003;362:887–899. doi: 10.1016/S0140-6736(03)14333-4. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong JA. J Exp Med. 1971;134:713–740. doi: 10.1084/jem.134.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shepard CC. Proc Soc Exp Biol Med. 1955;90:392–396. doi: 10.3181/00379727-90-22043. [DOI] [PubMed] [Google Scholar]
- 5.Schlesinger LS, Bellinger-Kawahara CG, Payne NR, Horwitz MA. J Immunol. 1990;144:2771–2780. [PubMed] [Google Scholar]
- 6.Bermudez LE, Goodman J. Infect Immun. 1996;64:1400–1406. doi: 10.1128/iai.64.4.1400-1406.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pethe K, Alonso S, Biet F, Delogu G, Brennan MJ, Locht C, Menozzi FD. Nature. 2001;412:190–194. doi: 10.1038/35084083. [DOI] [PubMed] [Google Scholar]
- 8.Menozzi FD, Rouse JH, Alavi M, Laude-Sharp M, Muller J, Bischoff R, Brennan MJ, Locht C. J Exp Med. 1996;184:993–1001. doi: 10.1084/jem.184.3.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menozzi FD, Bischoff R, Fort E, Brennan MJ, Locht C. Proc Natl Acad Sci USA. 1998;95:12625–12630. doi: 10.1073/pnas.95.21.12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abou-Zeid C, Ratliff TL, Wiker HG, Harboe M, Bennedsen J, Rook GA. Infect Immun. 1988;56:3046–3051. doi: 10.1128/iai.56.12.3046-3051.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ratliff TL, McGarr JA, Abou-Zeid C, Rook GA, Stanford JL, Aslanzadeh J, Brown EJ. J Gen Microbiol. 1988;134:1307–1313. doi: 10.1099/00221287-134-5-1307. [DOI] [PubMed] [Google Scholar]
- 12.Belisle JT, Vissa VD, Sievert T, Takayama K, Brennan PJ, Besra GS. Science. 1997;276:1420–1422. doi: 10.1126/science.276.5317.1420. [DOI] [PubMed] [Google Scholar]
- 13.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, et al. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 14.Brennan MJ, Delogu G, Chen Y, Bardarov S, Kriakov J, Alavi M, Jacobs WR., Jr Infect Immun. 2001;69:7326–7333. doi: 10.1128/IAI.69.12.7326-7333.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banu S, Honore N, Saint-Joanis B, Philpott D, Prevost MC, Cole ST. Mol Microbiol. 2002;44:9–19. doi: 10.1046/j.1365-2958.2002.02813.x. [DOI] [PubMed] [Google Scholar]
- 16.Delogu G, Pusceddu C, Bua A, Fadda G, Brennan MJ, Zanetti S. Mol Microbiol. 2004;52:725–733. doi: 10.1111/j.1365-2958.2004.04007.x. [DOI] [PubMed] [Google Scholar]
- 17.Espitia C, Laclette JP, Mondragon-Palomino M, Amador A, Campuzano J, Martens A, Singh M, Cicero R, Zhang Y, Moreno C. Microbiology. 1999;145(Pt 12):3487–3495. doi: 10.1099/00221287-145-12-3487. [DOI] [PubMed] [Google Scholar]
- 18.Finlay BB, Falkow S. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strom MS, Lory S. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 20.Ton-That H, Schneewind O. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 21.Lauer P, Rinaudo CD, Soriani M, Margarit I, Maione D, Rosini R, Taddei AR, Mora M, Rappuoli R, Grandi G, Telford JL. Science. 2005;309:105. doi: 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- 22.De Sousa-D'Auria C, Kacem R, Puech V, Tropis M, Leblon G, Houssin C, Daffe M. FEMS Microbiol Lett. 2003;224:35–44. doi: 10.1016/S0378-1097(03)00396-3. [DOI] [PubMed] [Google Scholar]
- 23.Barksdale L, Kim KS. Bacteriol Rev. 1977;41:217–372. doi: 10.1128/br.41.1.217-372.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xalabarder C. Mikroskopie. 1957;11:406–410. [PubMed] [Google Scholar]
- 25.Olsen A, Jonsson A, Normark S. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 26.Collinson SK, Emody L, Muller KH, Trust TJ, Kay WW. J Bacteriol. 1991;173:4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giron JA, Ho AS, Schoolnik GK. Science. 1991;254:710–713. doi: 10.1126/science.1683004. [DOI] [PubMed] [Google Scholar]
- 28.McMichael JC, Ou JT. J Bacteriol. 1979;138:969–975. doi: 10.1128/jb.138.3.969-975.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 30.Camus JC, Pryor MJ, Medigue C, Cole ST. Microbiology. 2002;148:2967–2973. doi: 10.1099/00221287-148-10-2967. [DOI] [PubMed] [Google Scholar]
- 31.Bardarov S, Bardarov S, Jr, Pavelka MS, Jr, Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, Jacobs WR., Jr Microbiology. 2002;148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- 32.Klemm P, Schembri MA. Int J Med Microbiol. 2000;290:27–35. doi: 10.1016/S1438-4221(00)80102-2. [DOI] [PubMed] [Google Scholar]
- 33.Middleton AM, Chadwick MV, Nicholson AG, Dewar A, Groger RK, Brown EJ, Ratliff TL, Wilson R. Tuberculosis (Edinburgh) 2002;82:69–78. doi: 10.1054/tube.2002.0324. [DOI] [PubMed] [Google Scholar]
- 34.Andersen RJ. Fortschr Chem Org Naturst. 1939;3:145–202. [Google Scholar]
- 35.Andersen RJ. Yale J Biol Med. 1943;15:311–345. [PMC free article] [PubMed] [Google Scholar]
- 36.Draper P. Front Biosci. 1998;3:D1253–D1261. doi: 10.2741/a360. [DOI] [PubMed] [Google Scholar]
- 37.Barrow WW, Ullom BP, Brennan PJ. J Bacteriol. 1980;144:814–822. doi: 10.1128/jb.144.2.814-822.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Draper P, Rees RJ. Nature. 1970;228:860–861. doi: 10.1038/228860a0. [DOI] [PubMed] [Google Scholar]
- 39.Kim KS, Salton MR, Barksdale L. J Bacteriol. 1976;125:739–743. doi: 10.1128/jb.125.2.739-743.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ton-That H, Marraffini LA, Schneewind O. Mol Microbiol. 2004;53:251–261. doi: 10.1111/j.1365-2958.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- 41.Paterson GK, Mitchell TJ. Trends Microbiol. 2004;12:89–95. doi: 10.1016/j.tim.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 42.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olsen A, Herwald H, Wikstrom M, Persson K, Mattsson E, Bjorck L. J Biol Chem. 2002;277:34568–34572. doi: 10.1074/jbc.M206353200. [DOI] [PubMed] [Google Scholar]
- 44.Kinhikar AG, Vargas D, Li H, Mahaffey SB, Hinds L, Belisle JT, Laal S. Mol Microbiol. 2006;60:999–1013. doi: 10.1111/j.1365-2958.2006.05151.x. [DOI] [PubMed] [Google Scholar]
- 45.Samanich K, Belisle JT, Laal S. Infect Immun. 2001;69:4600–4609. doi: 10.1128/IAI.69.7.4600-4609.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walters SB, Dubnau E, Kolesnikova I, Laval F, Daffe M, Smith I. Mol Microbiol. 2006;60:312–330. doi: 10.1111/j.1365-2958.2006.05102.x. [DOI] [PubMed] [Google Scholar]
- 47.Perez E, Samper S, Bordas Y, Guilhot C, Gicquel B, Martin C. Mol Microbiol. 2001;41:179–187. doi: 10.1046/j.1365-2958.2001.02500.x. [DOI] [PubMed] [Google Scholar]
- 48.Giron JA, Torres AG, Freer E, Kaper JB. Mol Microbiol. 2002;44:361–379. doi: 10.1046/j.1365-2958.2002.02899.x. [DOI] [PubMed] [Google Scholar]
- 49.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.