Abstract
To understand the relevance of p53 missense mutations in vivo, we generated a mouse containing an arg-to-his substitution at p53 amino acid 172, which corresponds to the R175H hot-spot mutation in human tumors by homologous recombination. Inadvertently, this mouse contains the additional deletion of a G nucleotide at a splice junction that attenuates levels of mutant p53 to near wild-type levels. Mice heterozygous for the mutant allele differed from p53+/− mice in tumor spectrum, with a significant increase in the number of carcinomas and a slight decrease in the number of lymphomas. More importantly, the osteosarcomas and carcinomas that developed in these mutant mice frequently metastasized (69% and 40%, respectively). In contrast, metastasis is rare in osteosarcomas of p53+/− mice. Loss of heterozygosity studies of tumors indicated loss of heterozygosity in only 1 of 11 tumors. These data indicate clear differences between a p53 missense mutation and a null allele in tumorigenesis in vivo and suggest that the p53R172HΔg mutant represents a gain-of-function allele.
Mutations in the p53 tumor suppressor are common in the etiology of many tumors and disrupt the ability of p53 to induce growth arrest or apoptosis. In particular, the R175H mutation corresponds to one of six hot spot mutations in p53 and accounts for ≈6% of the missense mutations identified in human cancers (1).
Mutant p53 proteins containing missense mutations have been studied in various assays, leading to suggestions that some may yield gain-of-function or dominant negative phenotypes. For example, expression of mutant p53 in p53 null cells yields tumors in syngeneic mice (2) and makes established cells highly tumorigenic (3). When assayed in comparison to other mutants, the p53R175H mutant was always the most detrimental (4–7). The p53R175H mutant cooperates better than any other mutant in ras-mediated transformation (4, 5). Cells expressing the p53R175H mutant are the most tumorigenic, as measured by growth in soft agar (6). In another experiment, genomic instability was measured in fibroblasts derived from Li-Fraumeni syndrome patients heterozygous for the p53R175H mutant (8). These cells exhibited a disrupted spindle checkpoint control and accumulated a polyploid DNA content as compared with cells with other p53 missense mutations, which arrest with a 4n DNA content. These data, coupled with the fact that most p53 mutations (>80%) occurring in human tumors are missense mutations, suggest that cells with p53 missense mutations have a growth advantage.
These in vitro experiments, however, cannot assay the effect of mutant p53 on tumor growth rate, survival, and tumor spectrum in vivo. The generation of mice null for p53 (9, 10) has yielded a plethora of data as to the importance of p53 in vivo. Mice heterozygous or homozygous for a p53 null allele are predisposed to multiple tumor types such as lymphomas, sarcomas, and other tumors resembling to some extent the types of tumors seen in the Li-Fraumeni syndrome. Thymocytes from mice lacking p53 are resistant to cell death by DNA-damaging agents compared with cells from normal mice (11). However, the null mouse model may be inadequate in that the most common alteration in p53 both in somatic cells (1) and in the germline of patients with Li-Fraumeni syndrome is a missense mutation. Thus, it is possible that the existing model has shed light only on the tumorigenic effect of those p53 mutations that are loss of function. A mouse that contains a missense mutation in one of the endogenous p53 genes would be invaluable to understanding p53 missense mutations in vivo.
To examine the in vivo significance of p53 missense mutations, we generated a mouse containing an arg-to-his substitution at amino acid 172 (equivalent to the 175 arg-to-his in human) in one of the endogenous p53 alleles by using homologous recombination. Mice with this mutation express low levels of mutant p53. Mice heterozygous for the R172H mutation exhibit differences in tumor spectrum and an increased rate of metastasis as compared with p53+/− mice. Importantly, most tumors from heterozygous mice do not show loss of heterozygosity (LOH). This allele thus exhibits a gain-of-function phenotype in vivo.
Materials and Methods
Gene Targeting and Generating the p53R172HΔg Mice.
A 15.3-kb NotI-NotI p53 fragment was isolated from a murine 129/Sv genomic library and was subcloned. The double replacement procedure was used essentially as described (12). The targeting vector for the first targeting step contained the 2.4-kb HindIII-BamHI fragment of p53 intron 1, the PGKneoNTRtkpA cassette (12) (a gift from R. Jaenisch, Massachusetts Institute of Technology), and the 0.9-kb partial PstI-PstI fragment containing p53 intron 6 through intron 9. The resulting plasmid Pgl6 was linearized with SalI and was electroporated into the AB-1 embryonic stem (ES) cells. DNA was harvested, was digested by EcoRI, and was subjected to mini-Southern blot analysis (13).
One correctly targeted ES cell, 3C12, was expanded and used for the second step of targeting. The targeting plasmid contained the 15.3-kb p53 genomic fragment with the R172H mutation in exon 5. Site-directed PCR mutagenesis protocol (14) was utilized to generate the p53 arg-to-his mutation (CGC to CAC) at codon 172. The PCR product containing the mutation was cloned and sequenced to confirm the desired mutation. The resulting targeting plasmid (36N6) was linearized and electroporated into ES cell clone 3C12. Single strand conformation polymorphism (SSCP) analysis was performed on DNA from ES clones by using a pair of primers flanking the p53 exon 5-intron 5 region: A3 (5′-TAC TCT CCT CCC CTC AAT AAG C-3′) and A4 (5′-AAT TAC AGA CCT CGG GTG GC-3′) as described (15). The PCR to detect the TK gene was performed by using the TK primer (5′-ACT GAA GGC TTT ACT ATT GC-3′) and A6 (5′-CCT CTG TAG CAT GGG CAT CC-3′). The DNA from targeted ES cell clones were digested with EcoRI and were subjected to the same Southern blot analysis performed in the first step targeting to check the integrity of the locus.
Mutant ES cell lines were used for injection into C57/BL6 blastocysts to generate chimeric mice. High percentage chimeras from clone 3C10 were crossed with C57/BL6 female mice and contributed to the germline of mice. The resulting heterozygous mice were either intercrossed to generate heterozygous mice and homozygous mice or were crossed back with C57/BL6 to generate heterozygous mice. Mice in the tumorigenic study came from both crosses.
Genotyping of p53R172HΔg mice was performed by using an allele-specific primer reaction. Primers 172W (5′-CAC ATG ACG GAG GTC GTG ACA CG-3′) and exon 7 (5′-ATG GTG GTA TAC TCA GAG CC-3′) amplified the wild-type p53 allele and primers 172M (5′-CAC ATG ACG GAG GTC GTG ACA CA-3′), and exon 7 amplified the R172H mutant allele in separate reactions.
Immunoprecipitation and Reverse Transcription–PCR.
Mouse embryonic fibroblasts were obtained from dissecting and mincing 12.5-day embryos individually and were maintained in DMEM containing 15% fetal bovine serum. Mouse embryonic fibroblasts were labeled and proteins immunoprecipitated with p53 antibody PAb421 as described (16).
Total RNA was isolated from mouse embryonic fibroblast cells homozygous for a p53R172HΔg allele by using a RNAzol B solution (Tel-Test, Friendswood, TX). Forty micrograms of RNA were used to purify mRNA by using Dynabeads (Dynal, Great Neck, NY) according to the manufacturer's instructions. First-strand cDNA was synthesized by using either a polydT primer or p53 primer E11. Using the first-strand cDNA as substrate, primers E1a (5′-GAG TTC ATT GGG ACC ATC CTG-3′) and E11 (5′-AGG ATT GTG TCT CAG CCC TGA-3′) were used to generate full-length p53 cDNA for sequencing analysis. Primers E2 (5′-AGCCAGGAGACATTTTCACC-3′) and E4 (5′-GCAGGAGCTCCTGACACT CGG-3′) were also used to clone different splicing variants in the mouse embryonic fibroblasts that were homozygous for the p53R172HΔg allele.
Statistical Analysis.
The binomial distribution test for comparing two proportions was used to analyze the statistical significance of the tumor spectrum difference between p53R172HΔg/+ mice and two groups of p53+/− mice.
LOH Analysis.
DNA was isolated from paraffin-embedded tumor samples identified by a pathologist using laser-capture technology. PCR amplification using primers A3 and A4 resulted in a product 384 base pairs long. Although it can be difficult to generate PCR products of this length from paraffin-extracted DNA, we were able to analyze eight of the samples. All samples were also analyzed by using primers A3 and miE5R (5′-CCA TCA CCA TCG GAG CAG CG-3′), which generated a PCR product of 184 base pairs. Both primer sets gave identical results. PCR products were generated by using touchdown thermocycling conditions: initial denaturation at 95°C for 5 min followed by 10 cycles of denaturation at 94°C for 1 min, annealing at 65°C for 1 min (decreasing 1°C per min until 56°C), and extension at 72°C for 1 min. This was followed by 35 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min and final extension at 72°C for 10 min. The PCR reaction was supplemented with [α-32P]dCTP, and 5 μl of the labeled products were denatured and separated on a 0.5× MDE (FMC) polyacrylamide gel. Electrophoresis was performed for 16 hours at 4.5 W. Gels were dried and bands were visualized by autoradiography. The quantitative in vitro reconstitutional test was performed by SSCP analysis with mutant DNA (representing an LOH tumor sample) mixed with increasing proportions of heterozygous genomic DNA (representing a potential contaminating source) and quantifying the intensities of the mutant and wild-type bands.
Results
Generation of p53R172HΔg Mutant Mice.
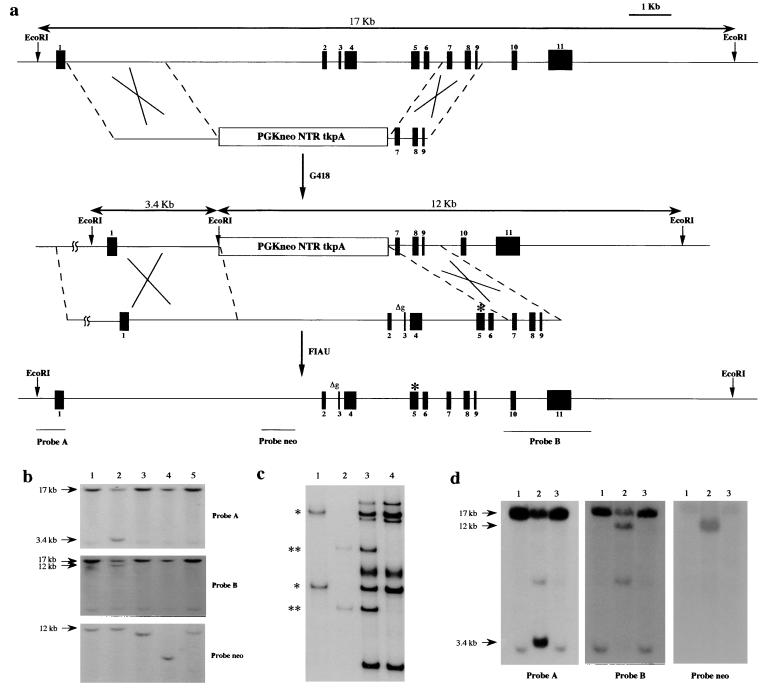
To examine the in vivo significance of p53 missense mutations, we generated a mouse containing an arg-to-his substitution at amino acid 172 (equivalent to the 175 arg-to-his in human) in one of the endogenous p53 alleles by using homologous recombination. The double-replacement method (12) was used to introduce the mutation into ES cells (Fig. 1a). In brief, the first electroporation with a targeting vector containing the neo and TK genes between p53 homologous sequences marked the locus of interest. The first round of selection occurred in G418, and the G418-resistant ES cell colonies were screened by Southern blotting for the correct targeting event (Fig. 1b). The targeted clone 3C12 was expanded and used for electroporation with a second plasmid containing the p53 gene with the R172H mutation (Fig. 1a). To generate a missense mutation at amino acid 172, a PCR-based site-directed mutagenesis protocol was used. A CGC to CAC substitution at codon 172 was confirmed by sequencing; no other changes were observed. Selection against TK was performed in the presence of 1-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-s-iodouracil (FIAU). In the case of a homologous recombination event, the neo and TK genes are lost and the p53 gene is reconstituted. To identify the targeted missense mutation in the ES cells, we performed SSCP (Fig. 1c) and sequencing analyses to confirm the mutation in the p53 locus. In addition, the restriction digests of the locus confirmed that the p53 gene was reconstituted (Fig. 1d). Two ES cell clones were obtained, and both were used in blastocyst injections. One gave rise to chimeric mice, which resulted in germline transmission of the allele with the p53R172H mutation.
Figure 1.
The generation of a mouse containing a p53R172H substitution. (a) The genomic organization of the p53 gene and the targeting vectors are shown. The first recombination step results in incorporation of the neo and tk genes (PGKneoNTRtkpA) at the p53 locus. In the second step, the neo and tk genes are replaced with the genomic p53 containing a point mutation (asterisk), resulting in an arg-to-his substitution at amino acid 172. Δg represents the deletion of a G nucleotide at the intron 2 splice acceptor site. (b) Southern hybridization of EcoRI-digested DNA from several ES cell clones electroporated with the PGKneoNTRtkpA targeting vector. The 17-kb EcoRI fragment represents the wild-type p53 allele whereas the 3.4- and 12-kb EcoRI fragments are the expected sizes for the mutant. The ES cell clone (3C12) in lane 2 is correctly targeted. (c) SSCP analysis using primers spanning the mutation. Lanes: 1, plasmid containing wild-type p53 genomic sequences; 2, plasmid containing mutant p53 genomic sequences; 3, genomic DNA from targeted ES cell clone; 4, genomic DNA from normal ES cells. *, wild-type bands; **, mutant-specific bands. (d) Southern blot hybridization of EcoRI-digested DNA from derivatives of 3C12 cells targeted with a construct containing a missense mutation in p53. Lanes: 1, normal ES cell DNA; 2, 3C12 DNA; 3, DNA from a correctly targeted ES cell clone.
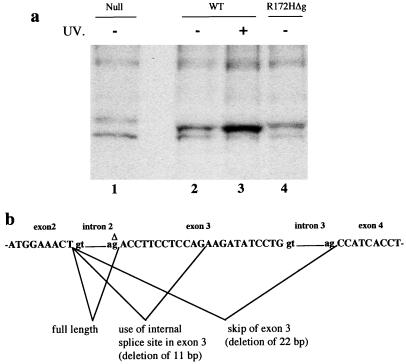
Mice homozygous and heterozygous for the p53 mutant allele were established. Fibroblasts from embryos with these genotypes were also established in culture. Immunoprecipitation analysis with a p53 antibody, PAb421, indicated that homozygous mutant cells expressed levels of the p53R172H mutant protein comparable to wild-type p53 (Fig. 2a). This was not expected because the corresponding mutation in human p53 results in an increased half-life and thus a higher level of protein (5). Detailed analysis by reverse transcription–PCR revealed the presence of several p53 mRNAs from cells homozygous for the mutant allele, and sequencing indicated three mRNA products: full length, deletion of exon 3, and deletion of a part of exon 3 with the aberrant splicing products being the major species (Fig. 2b). Sequencing of genomic DNA in this region revealed deletion of a G nucleotide at the splice acceptor site of intron 2 (Fig. 2b). The incorrectly spliced mRNA species result in a frameshift of the p53 coding sequence. This deletion explains both the presence of multiple mRNA transcripts and the low levels of mutant protein being produced. Additional experiments performed to determine whether smaller proteins were made by this allele showed the absence of smaller p53 protein products. The deletion occurred in the last cloning step generated by the process of cloning in Escherichia coli because only the targeting construct had this mutation. We named this p53 mutant allele p53R172HΔg.
Figure 2.
(a) Analysis of p53 protein levels in fibroblasts from embryos null for p53 (lane 1), wild-type (WT; lanes 2 and 3), or homozygous for the p53R172HΔg mutation (lane 4). Wild-type fibroblasts were treated with UV to show induction of p53 and confirm position on the gel (lane 3). (b) The structure of the p53 gene around exons 2–4 and the spliced products identified in fibroblasts from homozygous p53R172HΔg mutant mice. Δ denotes the G nucleotide in intron 2 that is deleted.
Analysis of Survival, Tumor Spectrum, and Metastasis in p53R172HΔg Mice.
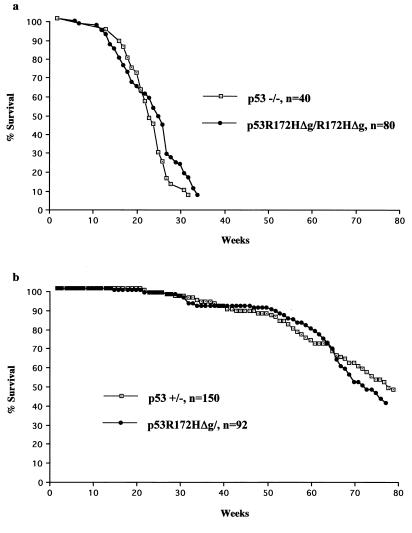
Because mice were making mutant p53 protein, albeit at a reduced level, we examined a cohort of mice homozygous and heterozygous for the p53R172HΔg mutation. There were no differences between the survival curves when homozygous and heterozygous p53R172HΔg mice were compared with p53−/− and p53+/− mice of the same mixed C57/BL6 and 129/Sv background (10, 17) (Fig. 3). Like p53−/− mice, the p53R172HΔg homozygous mice were highly susceptible to early onset spontaneous tumor formation. The predominant tumor type that developed in these mice (over 75%) was malignant thymic lymphoma.
Figure 3.
Comparisons of survival between p53 null mice and p53R172HΔg mice. The p53−/− mice survival data set is from our lab, and the p53+/− mice survival data set is from Harvey et al. (17). (a) Survival curves of 80 p53R172HΔg/R172HΔg mice and 40 p53−/− mice. (b) Survival curves of 92 p53R172HΔg/+ mice and 150 p53+/− mice.
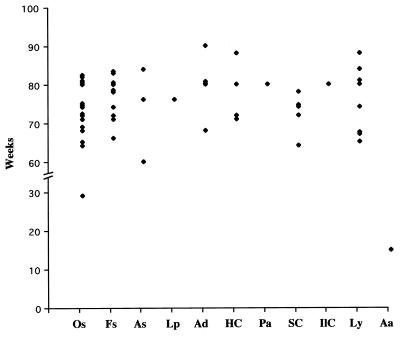
In contrast, the tumor spectrum in the p53R172HΔg heterozygous mice differed significantly from that of p53+/− mice (Table 1). Even though the p53R172HΔg/+ heterozygous mice developed sarcomas at a frequency similar to p53+/− mice [54 versus 57% (10, 17)], carcinomas were significantly more frequent in the p53R172HΔg/+ mice than in the p53+/− mice [29% vs. 10% (17) and 11% (10); P < 0.005 and P < 0.025, respectively]. The lymphoma incidence was slightly lower in the p53R172HΔg/+ mice than in two independent studies of p53 null mice [16% vs. 25% (10) and 32% (17); P < 0.25 and P < 0.025, respectively]. It is also interesting to note that the latencies of the major tumor types that developed in p53R172HΔg/+ mice were very similar (Fig. 4).
Table 1.
Tumor spectrum of p53R172HΔg/+ mice
| Tumor types | No. | Metastasis |
|---|---|---|
| Sarcomas | ||
| Osteosarcomas | 16 | 11 |
| Fibrosarcomas | 10 | 0 |
| Angiosarcomas | 3 | 0 |
| Lyposarcomas | 1 | 0 |
| Carcinomas | ||
| Adenocarcinomas | 5 | 2 |
| Hepatocellular carcinomas | 4 | 0 |
| Poorly differentiated carcinomas | 1 | 1 |
| Squamous cell carcinomas | 5 | 2 |
| Islet cell carcinomas | 1 | 1 |
| Lymphoma | 9 | 0 |
| Adenoma | 1 | 0 |
In a group of 51 p53R172HΔg/+ mice, one or more tumors had developed in 43. A total of 56 tumors were detected and used to analyze the tumor distribution.
Figure 4.
Tumor latency of different tumor types from 56 tumors that developed in 51 p53R172HΔg/+ mice. Os, osteosarcomas; Fs, fibrosarcomas; As, angiosarcomas; Lp, lyposarcomas; Ad, adenocarcinomas; HC, hepatocellular carcinomas; Pa, poorly differentiated carcinomas; SC, squamous cell carcinomas; IlC, islet cell carcinomas; Ly, lymphomas; Aa, adenoma.
In addition to differences in tumor spectrum, tumors in the p53R172HΔg heterozygous animals demonstrated a high metastatic potential, especially the osteosarcomas and carcinomas (Fig. 5). In 43 of 51 mice that had tumors, 17 had obvious metastasis nodules in the lung, liver, or both. Eleven of sixteen (69%) osteosarcomas had metastasized. Adenocarcinomas and squamous cell carcinomas also metastasized at high frequency, both at 40%. One unusual tumor, an islet cell carcinoma, also metastasized. Interestingly, none of 10 fibrosarcomas exhibited evidence of metastatic lesions. In contrast, several studies have reported no metastasis (17) or a very low rate of metastasis (10%) (18) from the osteosarcomas in the p53+/− mice of the same genetic background. There have been no reports of metastasis from carcinomas in p53+/− mice. Additionally, in our laboratory, we have not seen metastasis in p53+/− mice nor in p53 null mice with varying copies of the mdm2 null allele (19), suggesting that laboratory conditions per se do not alter this phenotype.
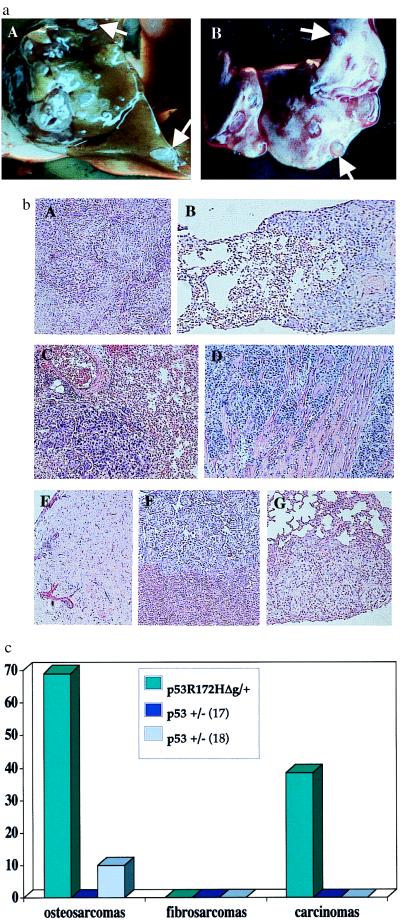
Figure 5.
Frequency of metastasis from tumors of p53R172HΔg/+ mice. (a) Metastatic lesions from an osteosarcoma that developed in a p53R172HΔg/+ mouse. (A) Liver. (B) Lung. (b) Representative histologies of tumors and their metastatic spread. A, C, and E represent primary tumors; B, D, F, and G represent their corresponding metastases. (A and B) Hepatocelluar carcinomas with metastasis in lung. (C and D) Lung adenocarcinomas with metastasis in myocardium (E, F, and G) Osteosarcoma with metastasis in liver (F) and lung (G). (c) Frequency of metastasis from p53R172HΔg/+ mice vs. p53+/− mice. p53+/− metastasis data are from Harvey et al. (17) and Tervana et al. (18), respectively.
Loss of Heterozygosity.
Loss of heterozygosity is a hallmark for loss of function of tumor suppressor genes in tumorigenesis. Because the data presented above suggested a gain-of-function activity for the p53R172HΔg allele, we checked the status of the wild-type p53 allele in different types of tumors from the p53R172HΔg/+ mice. SSCP analysis can readily distinguish the wild-type and mutant alleles (see Fig. 1c). The in situ laser-capture procedure was used to isolate tumor sample from adjacent normal tissue. In addition, quantitative in vitro reconstitution experiments demonstrated that even up to 15% of normal cell contamination would not obscure the LOH results (data not shown). The LOH results from 11 different tumor samples are shown in Fig. 6 and are summarized in Table 2. Ten of eleven tumors retained the wild-type allele and thus demonstrated no LOH at the p53 locus. Only one adenocarcinoma showed LOH (Fig. 6b). These data suggest that the loss of the wild-type p53 allele is not essential for tumor development and further supports the gain-of-function phenotype of mice inheriting the p53R172HΔg allele.
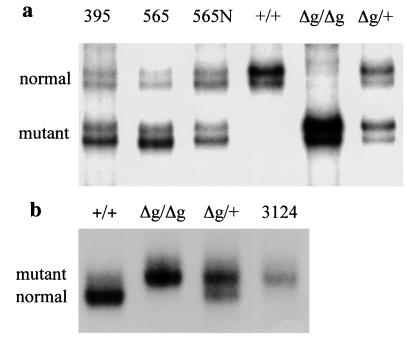
Figure 6.
A loss of heterozygosity study of tumors from p53R172HΔg/+ mice. SSCP analysis was performed on tumor samples obtained by using laser capture technology. (a) LOH analysis of DNA from osteosarcomas from mice numbered 395 and 565 using A3 and A4 primers (see Materials and Methods). 565N indicates DNA isolated from normal tissue adjacent to tumor of mouse 565. (b) LOH analysis of DNA from a carcinoma from mouse 3124 using primers A3 and Mie5R (see Materials and Methods). Tail DNA from wild type (+/+), homozygous (Δg/Δg), and heterozygous (Δg/+) mice were used as controls to distinguish the two alleles.
Table 2.
Summary of LOH results from 11 tumors that developed from p53R172HΔg/+ mice by SSCP analysis
| Mouse no. | Age, weeks | Tumor | LOH |
|---|---|---|---|
| 565 | 81 | Osteosarcoma | − |
| 565 | 81 | Adjacent normal tissue | − |
| 395 | 79 | Osteosarcoma | − |
| 528 | 79 | Fibrosarcoma | − |
| 3100 | 73 | Fibrosarcoma | − |
| 3124 | 79 | Adenocarcinoma | + |
| 320 | 88 | Adenocarcinoma | − |
| 606 | 87 | Hepatocellular carcinoma | − |
| 606 | 87 | Adjacent normal tissue | − |
| 3135 | 71 | Squamous cell carcinoma | − |
| 5220 | 73 | Squamous cell carcinoma | − |
| 5238 | 66 | Lymphoma | − |
| 328 | 80 | Lymphoma | − |
+, LOH; −, no LOH.
Discussion
p53R172HΔg mice have a targeted p53 missense mutation, an arg-to-his substitution at amino acid 172, and an additional splicing mutation in intron 2. This allele expressed a small amount of p53 mutant protein. The tumor spectrum of mice heterozygous for this mutation differed from that of p53+/− mice, as did the metastatic potential. Thus, we have established mice with a p53 mutant allele by homologous recombination. The differences in tumor spectrum and the increased frequency of metastasis in the p53R172HΔg/+ mice suggest that this allele represents a gain-of-function. Unlike transgenic models used to study the functional significance of overexpression of p53 missense mutations in vivo (20–22), this mouse generated by homologous recombination expressed only a small amount of protein. Therefore, the observed phenotypes most likely represent the intrinsic properties of the mutant protein.
The increased incidence of metastasis in p53R172HΔg/+ mice does not appear to be caused by differences in tumor spectrum. Increased metastasis is observed in osteosarcomas of p53R172HΔg/+ mice as compared with p53+/− mice, yet the numbers of sarcomas arising in both genotypes are identical (10, 17). Previous studies using cell lines lacking p53 or expressing a mutant p53 showed that those cells expressing mutant p53 readily metastasized to the lung of syngeneic mice (23), further supporting the gain-of-function phenotype of p53 missense mutations.
Another important observation is that the fibrosarcomas as opposed to osteosarcomas and carcinomas that develop in p53R172HΔg/+ mice did not metastasize. Of 10 fibrosarcomas that were identified, none metastasized, whereas of 16 osteosarcomas, 11 did. In contrast, Nf-2 heterozygous mice showed a high frequency of metastasis from both osteosarcomas and fibrosarcomas (24). This finding suggests that the determinants of metastasis are different for different tissue types.
LOH studies were also performed on 11 different tumors from p53R172HΔg/+ mice to determine whether the wild-type p53 allele had been lost. Only 1 of 11 tumors showed loss of heterozygosity, suggesting that loss of wild-type p53 was not important to tumor development. This is in contrast to LOH studies in p53+/− mice, in which 50% of the mice under 18 months of age were shown to have LOH at the wild-type p53 allele whereas 85% of mice over 18 months of age showed no LOH (25). The age of the mice when tumors were diagnosed varied in our experiments and did not seem to have an impact on loss of the wild-type allele. These data indicate a gain-of-function phenotype for the p53R172HΔg allele. The mutant protein may disrupt other cellular processes in the cell that cannot be compensated for by the presence of wild-type p53. Alternatively, the data may also suggest that this allele functions as a dominant negative. The mutant protein has the ability to form tetramers with the wild-type p53, effectively removing it from the cell.
Surprisingly, even though differences in tumor spectrum and metastasis are observed, there was no significant difference in the survival of these mice. It is possible that other alterations were rate limiting in mice or that, for osteosarcomas and carcinomas, metastasis is not a contributing factor to survival in mice. The mating of p53R172HΔg mice to other tumor models will help address some of these issues. Our study clearly establishes that the p53R172H mutant represents a gain-of-function allele. These data support clinical studies showing a high correlation of this specific p53 mutation, an arg-to-his substitution at amino acid 175, and extremely poor prognosis in humans (26).
Acknowledgments
We thank Larry Donehower for the p53 data set, Howard Thames for help with statistical analysis, and Lisa Amelse for help with the mice. This work was supported by grants from the National Institutes of Health.
Abbreviations
- ES cell
embryonic stem cell
- SSCP
single strand conformation polymorphism
- LOH
loss of heterozygosity
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Greenblatt M S, Bennett W P, Hollstein M, Harris C C. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 2.Wolf D, Harris N, Rotter V. Cell. 1984;38:119–126. doi: 10.1016/0092-8674(84)90532-4. [DOI] [PubMed] [Google Scholar]
- 3.Eliyahu D, Michalovitz D, Oren M. Nature (London) 1985;316:158–160. doi: 10.1038/316158a0. [DOI] [PubMed] [Google Scholar]
- 4.Finlay C A, Hinds P W, Levine A J. Cell. 1989;57:1083–1093. doi: 10.1016/0092-8674(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 5.Hinds P W, Finlay C A, Quartin R S, Baker S J, Fearon E R, Vogelstein B, Levine A J. Cell Growth Differ. 1990;1:571–580. [PubMed] [Google Scholar]
- 6.Dittmer D, Pati S, Zambetti G, Chu S, Teresky A K, Moore M, Finlay C, Levine A J. Nat Genet. 1993;4:42–46. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- 7.Slingerland J M, Jenkins J R, Benchimol S. EMBO J. 1993;12:1029–1037. doi: 10.1002/j.1460-2075.1993.tb05744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gualberto A, Aldape K, Kozakiewicz K, Tlsty T D. Proc Natl Acad Sci USA. 1998;95:5166–5171. doi: 10.1073/pnas.95.9.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A. Nature (London) 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 10.Jacks T, Remington L, Williams B O, Schmitt E M, Halachmi S, Bronson R T, Weinberg R A. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 11.Lowe S W, Schmitt E M, Smith S W, Osborne B A, Jacks T. Nature (London) 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 12.Wu H, Liu X, Jaenisch R. Proc Natl Acad Sci USA. 1994;91:2819–2823. doi: 10.1073/pnas.91.7.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMahon A P, Bradley A. Cell. 1990;62:1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- 14.Haiguchi R. Recombinant PCR Protocols: 1990. San Diego: Academic; 1990. [Google Scholar]
- 15.Evans S C, Mims B, McMasters K M, Foster C J, deAndrade M, Amos C I, Strong L C, Lozano G. Hum Genet. 1998;102:681–686. doi: 10.1007/s004390050761. [DOI] [PubMed] [Google Scholar]
- 16.Kapoor M, Lozano G. Proc Natl Acad Sci USA. 1998;95:2834–2837. doi: 10.1073/pnas.95.6.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harvey M, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A, Donehower L A. Nat Genet. 1993;5:225–229. doi: 10.1038/ng1193-225. [DOI] [PubMed] [Google Scholar]
- 18.Taverna D, Ullman-Cullere M, Rayburn H, Bronson R T, Hynes R O. Cancer Res. 1998;58:848–853. [PubMed] [Google Scholar]
- 19.McDonnell T, Montes De Oca Luna R, Cho S, Amelse L L, Chavez-Reyes A, Lozano G. J Pathol. 1999;188:322–328. doi: 10.1002/(SICI)1096-9896(199907)188:3<322::AID-PATH372>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 20.Lavigueur A, Maltby V, Mock D, Rossant J, Pawson T, Bernstein A. Mol Cell Biol. 1989;9:3982–3991. doi: 10.1128/mcb.9.9.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X J, Greenhalgh D A, Jiang A, He D, Zhong L, Medina D, Brinkley B R, Roop D R. Oncogene. 1998;17:35–45. doi: 10.1038/sj.onc.1201890. [DOI] [PubMed] [Google Scholar]
- 22.Li B, Murphy K L, Laucirica R, Kittrell F, Medina D, Rosen J M. Oncogene. 1998;16:997–1007. doi: 10.1038/sj.onc.1201621. [DOI] [PubMed] [Google Scholar]
- 23.Pohl J, Goldfinger N, Radler-Pohl A, Rotter V, Schirrmacher V. Mol Cell Biol. 1988;8:2078–2081. doi: 10.1128/mcb.8.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McClatchey A I, Saotome I, Mercer K, Crowley D, Gusella J F, Bronson R T, Jacks T. Genes Dev. 1998;12:1121–1133. doi: 10.1101/gad.12.8.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venkatachalam S, Shi Y P, Jones S N, Vogel H, Bradley A, Pinkel D, Donehower L A. EMBO J. 1998;17:4657–4667. doi: 10.1093/emboj/17.16.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goh H S, Yao J, Smith D R. Cancer Res. 1995;55:5217–5221. [PubMed] [Google Scholar]