Abstract
T-cell differentiation and induction of tolerance to self-antigens occurs mainly in the thymus. Thymic stromal cells, specifically medullary thymic epithelial cells, express a diverse set of genes encoding parenchymal organ-specific proteins. This phenomenon has been termed promiscuous gene expression (PGE) and has been implicated in preventing organ-specific autoimmunity by inducing T-cell tolerance to self antigens. Early thymopoiesis and the critical factors involved in T-cell differentiation can be reproduced in vitro by murine fetal thymus organ culture (FTOC), which mimics the natural thymic microenvironment. To evaluate the occurrence of PGE in FTOC, gene expression profiling during in vitro thymic development in BALB/c mice was performed using a set of nylon cDNA microarrays containing 9216 sequences. The statistical analysis of the microarray data (sam program) revealed the temporal repression and induction of 57 parenchymal and seven lymphoid organ-specific genes. Most of the genes analysed are repressed during early thymic development (15–17 days post-coitum). The expression of the autoimmune regulator (AIRE) gene at 16 days post-coitum marks the onset of PGE. This precedes the induction of parenchymal organ genes during the late developmental phase at 20 days post-coitum. The mechanism of T-cell tolerance induction begins during fetal development and continues into adulthood. Our findings are significant because they show a fine demarcation of PGE onset, which plays a central role in induction of T-cell tolerance.
Keywords: cDNA microarray, fetal thymus organ culture, promiscuous gene expression, thymus, T-cell tolerance
Introduction
The thymus is the key primary lymphoid organ that is mainly involved in the progressive differentiation of thymocytes to mature T cells in which all developmental stages are distinguishable by their expression of a combination of clusters of differentiation (CD) cell-surface markers. Studies with freshly obtained fetal thymus allowed for the demarcation of T-cell receptor β V(D)J recombination during in vivo thymus ontogeny among different inbred mouse strains; this contributed to the revealing of the effect of the genetic background on T-cell development.1–3 The control of gene expression during the development of this organ has gained priority among several research groups, including our own, leading to the identification of candidate genes involved in thymopoiesis. Using the cDNA microarray method, a dozen expressed sequence tags have been found that are modulated during the in vivo development of the thymus.4 Using the same kind of analysis, the in vivo modulation of several cell-signalling genes was identified, including those of the calcium cascade pathway, which is important for individual stages of T-cell maturation and the control of anergy during murine thymus ontogeny.5
Meanwhile, a better understanding of the central tolerance mechanism, that is, all the mechanisms by which T cells contribute to self-tolerance by intrathymic recognition of self-antigens, emerged from evidence that tissue-specific antigens are expressed in the thymus and contribute to the selection of the T-cell repertoire. These tissue-specific antigens are expressed as a normal physiological property of thymic epithelial cells (TECs).6 This phenomenon was termed promiscuous gene expression (PGE), which includes self antigens that represent most of the parenchymal organs.7–14 Evidence of PGE in the thymus is causing a reversal in our understanding of central tolerance, allowing for an unorthodox conception of the possible mechanism of self–non-self discrimination.15,16
The initial evidence for promiscuously expressed genes was biased towards antigens involved in autoimmune reactions, such as insulin, acetylcholine receptor or myelin basic protein. Today it is recognized that rather than being selective, the set of expressed genes is as broad as possible, estimated to include up to 5–10% of all currently known mouse genes.9 Thus, the main implication of this heterogeneous gene expression in the thymus is associated with maintenance of the immunological homeostasis in the body, controlling pathogenic autoimmune reactions.
Studies on thymus gene expression in the broadest aspect, including PGE, are normally designed to be performed immediately using samples collected by surgery, which must reflect the short-term in vivo situation.
Nevertheless, fetal thymus organ culture (FTOC), introduced by DeLuca’s group,17 which is based on the use of early fetal thymus tissue, preserves the original architecture of the thymus, with its cellularity formed of stromal TECs and homogeneous thymocyte population double-negative cells. This is a singular, powerful culture model system reproducing intrathymic T-cell development in vivo, making it easier to study the candidate genes implicated in normal thymus development. Using the cDNA microarray method, our group recently showed the modulation of gene expression in murine FTOCs at the transcriptome level and revealed an overlap between genes associated with T-cell receptor V(D)J recombination and DNA repair. We showed that the association of FTOC and cDNA microarray technology allows sufficient accuracy to uncover the participation of essential genes implicated in thymus development.18
In the present study, cDNA microarrays were employed to observe the onset of PGE during the in vitro development of murine thymus in FTOCs. Comparing the time of thymus gestation as a result of fetal age and culture duration, it was possible to discover the onset of gene induction, representing 57 different parenchymal and seven lymphoid organs. Some coded proteins are considered to be tissue-specific antigens, thus indicating that FTOCs reproduce promiscuous gene expression.
The FTOC transcriptome profiling was assessed using a set of six nylon cDNA microarrays containing a total of 9216 image thymus sequences. PGE was identified on the basis of gene expression data for different parenchymal organs. To our knowledge, these findings represent the first demonstration of the temporal beginning of PGE in murine FTOC. Moreover, the importance of this event is associated with the fact that this model system could be a useful tool for testing cytokines, RNA interference or other compounds that directly control gene expression in the thymus, as possible modulators of PGE.
Material and methods
FTOCs and total RNA preparation
BALB/c mice were bred in an isolator with 0·45-μm pore-size air filtered in our own breeding facility. To obtain timed pregnancies, the mice were mated and the day when the vaginal plug was observed at 7:00 hr was considered to be day zero of gestation post-coitum (p.c.). The pregnant mice were killed by ether inhalation and the fetuses were collected via surgery of the uterus. The p.c. age of the fetuses was confirmed by the morphological characteristics of each developmental phase.18,19 The thymi were removed from the fetuses under a stereomicroscope and cultured according to the organ culture technique previously described.17
Briefly, the organs were dissected and cleaned from the adjacent tissue and placed on the surface of Millipore filters (0·45 μm pore size) pre-embedded with culture medium. These filters were supported on plastic grids in 2 ml Dulbecco’s modified Eagle’s medium/HAM F-10 culture medium (Gibco, Gaithersburg, MD, USA) supplemented with 20% heat inactivated fetal bovine serum (Biobrás, Montes Claros, MG, Brazil) in 24-well tissue culture plates. The medium was also supplemented with 100 μg/ml streptomycin, 250 U/ml penicillin, 10 μg/ml gentamicin, 1 mm sodium pyruvate, 20 μm 2-mercaptoethanol and 3·4 g/l sodium bicarbonate. The incubation time of the organ culture varied from 2 to 5 days at 37° in a 5% CO2 incubator.
Thymi were dissected from fetuses obtained at 13, 14 and 15 days of gestation and cultured for 2 days, so mimicking 15, 16 and 17 days p.c. of in vivo development, respectively. Thymi from 15-day p.c. fetuses were also cultured for 5 days, mimicking the 20th day of in vivo development, and in this case, the culture medium was changed on the third day of incubation.
To evaluate the viability of FTOCs, three randomly selected thymi from each group were used for the single cell suspension preparations, which were separately stained with acridine orange (100 μg/ml) or with ethidium bromide (100 μg/ml) and analysed on an Axiophot II fluorescence microscope (Carl Zeiss, Oberkochen, Germany) to evaluate the frequency of apoptosis and necrosis, respectively. Total RNA samples were prepared from FTOCs using Trizol® reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA).
cDNA microarray method
A set of six microarrays were used containing a total of 9216 cDNA sequences (1536 sequences each) in the form of polymerase chain reaction (PCR) products, spotted in duplicate on 2·5 × 7·5 cm Hybond N + nylon membranes (GE Healthcare, Amersham, UK). The arrays were prepared in our laboratory using a Generation III Array Spotter (Amersham Biosciences-Molecular Dynamics, Sunnyvale, CA, USA).
Mouse thymus expressed sequence tag cDNA clones were obtained from the Soares thymus 2NbMT normalized library, prepared from the thymus of a C57BL/6J 4-week-old male mouse, and available from the IMAGE Consortium (http://image.llnl.gov/image/html/iresources.shtml) and from the RZPD Deutsches Resourcenzentrum für Genomforschung GmbH (http://www.rzpd.de).
The cDNA inserts were homogeneous in size (near 1 kb), cloned in three vectors (pT7T3D, pBluescript and Lafmid) and were amplified in 384- or 96-well plates using vector-PCR amplification with the following primers, which recognize the three vectors: LBP 1S GTGGAATTGTGAGCGGATACC forward and LBP 1AS GCAAGGCGATTAAGTTGG reverse. This set of cDNAs included sequences of some known autoantigens, such as small nuclear ribonucleoprotein SMD1, lupus Ku protein p70, p80 and p86 and PM-SCL; the full list of cDNA sequences used to prepare the microarrays can be retrieved online at http://rge.fmrp.usp.br/passos/mtb_library.
The membranes were first hybridized with LBP 1AS [γ-33P]dCTP-labelled oligonucleotide (vector hybridization). Quantification of the signals obtained allowed for the estimation of the amount of cDNA insert fixed in each spot. After stripping, the membranes were used for hybridization with cDNA complex probes (sample hybridization). The characterization of each cDNA sequence was updated using the Eloge® (Ipsogen, Marseille, France, http://www.ipsogen.fr) software, which runs on our local server and links each clone ID with the genome data banks (http://genome-www5.stanford.edu/cgi-bin/source/sourceSearch), providing information such as DNA and protein sequences, biological and molecular functions and chromosomal location.
Complex cDNA probe preparation and hybridization
In this study, we refer to the radioactive cDNA originated from the thymus RNA samples (FTOCs) as complex probe and to the PCR product originated from the clones and deposited on nylon microarrays as target.
The 33P-labelled cDNA complex probes were prepared by reverse transcription of 10 μg thymus total RNA using oligo-dT(12-18) as a primer; 100 μl 33P–cDNA complex probe containing 30–50 million counts per minute was hybridized with nylon microarrays as previously described.20–22
cDNA microarray image acquisition
Imaging plates and a phosphor imager apparatus (Packard Phosphor Imager, model Cyclone, Packard Instruments, Downers Grove, IL, USA) were used to capture the hybridization signals and the BZScan software23 was used to quantify the signals with local background subtraction, whose spots were matched with a template grid. The ratio between vector probe hybridization values and complex probe hybridization values for each spot was used as a reference normalization value. Total intensity normalization was also employed, using the median expression value.24
Significance analysis of microarray data
The gene expression data analysed here were obtained from six independent determinations for each day of fetal development in FTOC. The sam method25 (Significance Analysis of Microarrays, available at http://www.stat.stanford.edu/~tibs/SAM/index.html) was used to analyse significant variations in gene expression. This method is based on t-test statistics, specially modified for high throughput analysis. The significant variations in the gene expression of developing thymus in FTOCs were compared using the 15-day p.c. as reference (15 versus 16, 15 versus 17 and 15 versus 20 days of development). In this study, only those genes presenting a two-fold change of expression (either repressed or induced) and a gene error chance (q-value) equal to 0·01 (see Supplementary Table S1) were considered.
Determination of PGE in the thymus
PGE was identified on the basis of data from microarray analysis of the different mouse organs using combined information from the public database GNF Gene Expression Atlas26 (http://symatlas.gnf.org/SymAtlas). This data bank shows gene expression in more than 60 mouse tissues/organs, as assessed by gene array analysis using Affymetrics microarrays. Data information includes GenBank accession, chromosomal location and the molecular/biological function of each gene analysed (Table S1).
In this study, only the promiscuous genes of which the expression was detected in different organs or tissues, besides the thymus, and of which the expression levels were greater than median in relation to all other organs which appear in the GNF Atlas were considered. The modulation of transcription levels (repression or induction) of these genes was evaluated during FTOC development.
Semi-quantitative reverse transcription-PCR (SQ RT-PCR)
The zinc finger protein 369 gene (ZNF 369; acc NM_178364, clone ID 573600) that was found to be induced in the microarray experiments and the AIRE gene (acc NM_009646) were tested by SQ RT-PCR; 1 μg total RNA from each developmental phase in FTOC was copied to cDNA by reverse transcription using oligo-dT(12-18) as a primer. One microlitre of successive dilutions of the cDNA (1:2 to 1:128) were used as a PCR template. PCRs were performed in a final volume of 25 μl containing 100 μm each dNTP, 1 μm each primer and 1 U Taq DNA polymerase (Amersham Biosciences).
The amplification protocol for the ZNF 369 and AIRE genes assayed was as follows; one cycle at 94° for 5 min, 30 cycles of 94° for 1 min, 55° for 1 min, then 72° for 1 min, and then one cycle at 72° for 10 min then 4° for 10 min. The amplification was terminated with a final incubation step at 72° for 10 min.
Aliquots of the PCR products were mixed with 1 μl of a diluted solution of ethidium bromide, resolved on 2% agarose gel electrophoresis using 1 × TAE buffer. The gels were visualized on a UV transilluminator and the images were captured using a digital camera.
The PCR primers were identified using the Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) on the cDNA sequences retrieved from GenBank (http://www.ncbi.nlm.nih.gov) for each gene. The forward and reverse sequences are given in the 5′ to 3′ orientation; ZNF 369 (clone ID 573600, GenBank acc NM_178364) (GGGAGGACCTTATGTGTGT and GGGGCTCTCCTTATCCAAAAG) and the autoimmune regulator gene, AIRE (GenBank acc NM_009646), although not included in the microarrays, was also assayed with the following primers (CATCCTGGATTTCTGGAGGA and GCTCTTTGAGGCCAGAGTTG).
Results
Genes differentially expressed during FTOC development
To identify significant changes in gene expression during fetal development in FTOCs, the early thymus at 15 days was compared with those observed on the subsequent days (15 versus 16, 15 versus 17 and 15 versus 20 days p.c. FTOC). A scatter plot of the observed relative difference d(i) against the expected relative difference dE(i) was used, as shown by the sam program.
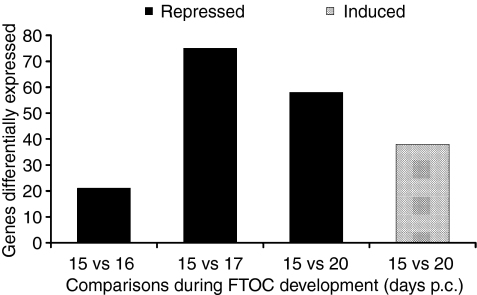
Most of the 9216 sequences tested presented d(i) ≌ dE(i), indicating that their expression pattern remained unchanged; however, some genes were significantly repressed (154 genes) or induced (38 genes) during FTOC development (Fig. 1).
Figure 1.
Differential gene expression during the development of FTOC showing that induction of tissue-specific genes emerges at 20 days. Staging corresponds to development of fetal thymus in culture; 15 days FTOC = 13 days gestation thymus plus 2 days in FTOC.
Interestingly the induction of the 38 genes that include representation of parenchymal organs in the thymus, thus characterizing PGE, was observed at late FTOC (20 days p.c.) (Table S1).
Gene expression assessed by SQ RT-PCR
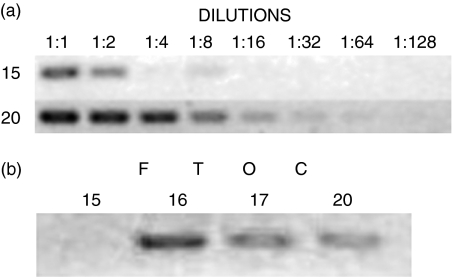
The ZNF 369 gene (acc NM_178364) was selected to confirm the respective result obtained with the cDNA microarray. SQ RT-PCR evaluation confirmed that this gene was induced at 20 days p.c. FTOC (Fig. 2a). Moreover, the AIRE gene (acc NM_009646) began its expression at 16 days p.c. FTOC (Fig. 2b).
Figure 2.
Semi-quantitative reverse transcription-PCR. Confirmation of ZNF gene as induced at 20 days FTOC (a) and emergence of AIRE gene expression at 16 days FTOC (b).
Parenchymal organ representation in the thymus
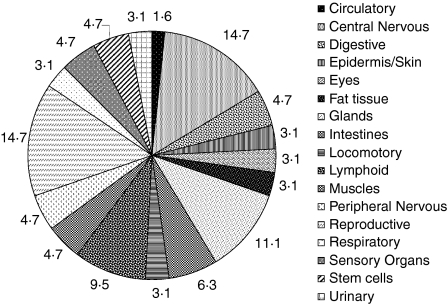
The 38 genes identified as significantly induced at 20 days p.c. in FTOC were assigned to 57 parenchymal and seven lymphoid organs according to their predominant expression, which were subgrouped in 17 systems. Interestingly, central nervous and reproductive systems and glands are the predominant parenchymal organs represented in the thymus at this phase of development, followed by the lymphoid system (Fig. 3). Four genes; small nuclear ribonucleoprotein D1, acc NM_009226; CD27 antigen, acc L24495; Riken cDNA 1110065L07, acc AA119642; and d-amino-acid oxidase, acc AI447515; code for tissue-specific antigen proteins, because they are expressed in only five or six tissues (Table S1).
Figure 3.
Representation of tissue/organ system-specific gene expression in 20 days FTOC.
Chromosomal location of differentially expressed genes
The genomic distribution of the 192 genes modulated (repressed or induced) during the 5 days of FTOC development allowed for the co-ordinated expression of organizing chromosomal clusters.
Figure 4 shows the frequency distribution of the repressed and induced genes among chromosomes. Taking all the developmental phases together, the number of repressed genes (154) was greater than that of induced genes (38). All chromosomes, except Y, harboured differentially expressed genes, with a slightly biased distribution on chromosomes 2, 5, 11 and 19 for repressed genes and on chromosomes 3, 4, 11, 15 and 19 for induced genes.
Figure 4.
Chromosomal distribution of the repressed and induced genes considering all FTOC development (15–20 days). NF, not found.
Discussion
PGE in the thymus is a complex phenomenon observed in humans and mice, which is exhibited by TECs (mTECs) and involves 5–10% of all known genes of these species. This process is a guarantee of self-representation of most parenchymal tissues and organs during the central tolerance of T cells.
Using PCR, Derbinski’s group previously demonstrated that the expression of five tissue-specific genes, such as thyroglobulin, CRP, GAD67, insulin and albumin, was detectable only in mTECs purified from ex vivo thymic tissue at embryonic day 15 (E15).7
Accordingly, the complexity of PGE increases in, from cTECs to immature mTECs to mature CD80hi mTECs. These different gene pools are not complementary, but additive, that is, there is no apparent association between the respective molecular/biological functions of the genes in parenchymal organs. The significance of PGE in the thymus is associated with the central tolerance of T cells.9–14
While PGE by mTECs was well documented by the authors cited above, many aspects of this phenomenon remain unexplored, for example, its evaluation in the thymus of autoimmune strains and/or knockout mice, its modulation by means of cytokines or other molecules interfering in gene expression, such as RNA interference, and its time–course during the ontogenetic development of the thymus.
Molecular characterization of PGE in the FTOC model system is a relevant approach in immunobiology, because the functional analysis of T-cell development has become possible after the introduction of techniques in which the thymic microenvironment is mimicked, such as FTOC. This method is based on the use of early fetal thymus tissue, which is composed of a homogeneous thymocyte population (double-negative cells).
In this study, the occurrence of PGE in vitro in FTOC is described for the first time. To evaluate whether PGE in this model system is a development-dependent phenomenon, we regarded the differential gene expression during ontogeny by comparing days of gestation (p.c.), as the most informative in the delineation of the gene pool. Use of the FTOC model system was chosen, rather than compare the changes in expression profiles in thymus tissue obtained at different gestational days, to approach three important aspects simultaneously. Since FTOC reproduces the in vivo thymus development, this model system represented a useful tool first, for the fine demarcation of PGE onset and, second, to demonstrate that PGE is a phenomenon that can be reproduced in vitro. The third aspect was a beneficial possibility of the FTOC model system, comparing the same pool of thymus tissue at day 15 with later time-points.
We demonstrated that PGE in FTOC, which is characterized by the overexpression of parenchymal organ and tissue-specific antigen genes, emerges at 20 days p.c. (Fig. 1). Since this in vitro organ culture mimicked the thymus gestation in vivo, including the maturation of T cells,18 these results strongly suggest that PGE is dependent on thymus maturation. The significance of these findings is associated with the timing of T-cell tolerance induction during ontogeny. T-cell receptor β rearrangements emerge at 16 days p.c. in vivo and in FTOC in BALB/c mice.1–3,18
The early fetal thymus, by 13–15 days p.c., is composed of homogeneous double-negative CD4− CD8− T-cell precursors; however, by day 18 p.c. this population gradually acquires the CD4 marker resembling the adult CD4low precursor.26,27 These features allow for T-cell receptor–major histocompatibilty complex peptide recognition and enable the positive/negative selection of T cells in fetal thymus. Our evidence for the occurrence of PGE in late fetal thymus is associated with the timing of the molecular events of T-cell tolerance induction during ontogeny.
The data collected here were obtained using the cDNA microarray method, with the expression of the 9216 sequences analysed by the sam algorithm.25 Considering 15 days p.c. FTOC as a reference, it was possible to make comparisons with the subsequent days of development. A statistically significant set of 154 repressed genes were found between 15 and 17 days p.c. and 38 genes were considered as overexpressed at 20 days p.c. FTOC, indicating the emergence of PGE (Table S1).
Moreover, these findings are important to the PGE differentiation model in the thymus and the concomitant increased AIRE gene expression in the most mature mTECs. In the FTOC model system studied, AIRE gene expression begins at 16 days p.c., before a significant induction of parenchymal and tissue-specific antigen genes (Fig. 2b), suggesting that this gene could be associated with the control of the parenchymal organ gene expression observed.28
Although the experiments were not conducted with purified mTECs, this caused no problems regarding PGE detection in the thymus. To bypass this potential difficulty, a cDNA microarray method was used, including a dedicated statistical algorithm for data analysis (sam algorithm), which presented sufficient accuracy to distinguish and quantify parenchymal organ gene expression originating from thymic epithelial cells, especially mTECs.8,9,13
The cDNA microarray data mining has permitted the virtual dissection of the mouse thymus into its principal cellular components by means of the identification of the specific cellular transcripts (mRNAs).29 These observations demonstrate the feasibility for the use of whole thymus as starting material in PGE studies.
Moreover, we selected a gene found in the microarray experiments (ZNF 369) that is representative of parenchymal organs, whose modulation in the expression was confirmed by SQ-RT-PCR (Fig. 2a).
In agreement with previous observations,13 the molecular/biological function of promiscuously expressed genes found in the present model system showed no interrelationship (Table S1).
Regarding the chromosomal localization of the repressed and induced genes, no important preferential distribution was identified. All chromosomes harbour promiscuously expressed genes with a slightly biased distribution on chromosomes 2, 5, 11 and 19 among the repressed genes and on chromosomes 3, 4, 11, 15 and 19 among the induced genes. The exceptions were chromosome Y, on which no repressed or induced genes were identified, and chromosome 2, on which no induced genes were positioned (Fig. 4).
This feature of random PGE distribution in the genome strongly suggests an uncommon model of gene regulation found in the thymus, which requires further investigation, including the elucidation of the molecular mechanism of the AIRE gene action.
The model system presented here was important to demonstrate that PGE is a differentially controlled phenomenon,9 beginning in BALB/c mice at 20 days p.c. (FTOC), and because this phenomenon can be reproduced in vitro, these findings raise the possibility of testing the induction of gene expression alteration caused by FTOC cytokine treatment or selected gene transfections that could modulate PGE. Recent evidence for a second thymus in mice30 increase the possibility of further research into their contribution to self-tolerance mechanisms, including determination of occurrence of the PGE in this organ.
Acknowledgments
This research was funded by the Brazilian agencies FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) through thematic project (99/12135–9) and doctorate fellowships to R.S.C., D.A.R.M., C.M.J., and M.M.C.M. and the CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) to C.M. The IMAGE cDNA library used was kindly ceded by Dr Catherine Nguyen of the Unité INSERM ERM 206, Marseille, France.
Supplementary material
The following supplementary material is avaible online for this article:
Promiscuously induced genes in 20 days p.c. Balb-c thymus (FTOC).
This material is available as part of the online article from http://www.blackwell-synergy.com
References
- 1.Junta CM, Passos GAS. Emergence of TCRαβ V (D) J recombination and transcription during thymus ontogeny of inbred mouse strains. Mol Cell Biochem. 1998;187:67–72. doi: 10.1023/a:1006807021251. [DOI] [PubMed] [Google Scholar]
- 2.Macedo C, Junta CM, Passos GAS. Onset of T-cell receptor Vβ8.1 and Dβ2.1, V(D)J recombination during thymus development of inbred mouse strains. Immunol Lett. 1999;69:371–3. doi: 10.1016/s0165-2478(99)00110-8. [DOI] [PubMed] [Google Scholar]
- 3.Espanhol AR, Macedo C, Junta CM, et al. Gene expression profiling during thymus ontogeny and its association with TRVB8-DB2.1 rearrangements of inbred mouse strains. Mol Cell Biochem. 2003;252:223–8. doi: 10.1023/a:1025556510001. [DOI] [PubMed] [Google Scholar]
- 4.Espanhol AR, Cardoso RS, Junta CM, Victorero G, Loriod B, Nguyen C, Passos GAS. Large scale gene expression analysis of CBA/J mouse strain fetal thymus using cDNA-array hybridizations. Mol Cell Biochem. 2004;206:65–8. doi: 10.1023/b:mcbi.0000026055.24084.98. [DOI] [PubMed] [Google Scholar]
- 5.Magalhães DA, Macedo C, Junta CM, et al. Hybridization signatures during thymus ontogeny reveals modulation of genes coding for T-cell signaling proteins. Mol Immunol. 2005;42:1043–8. doi: 10.1016/j.molimm.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 6.Jolicoeur C, Hanahan D, Smith KM. T-cell tolerance toward a transgenic β-cell antigen and transcription of endogenous pancreatic genes in thymus. Proc Natl Acad Sci USA. 1994;91:6707–11. doi: 10.1073/pnas.91.14.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol. 2001;2:1032–9. doi: 10.1038/ni723. [DOI] [PubMed] [Google Scholar]
- 8.Gotter J, Brors B, Hergenhahn M, Kyewski B. Medullary epithelial cells of the human thymus express a highly diverse selection of tissue-specific genes colocalized in chromosomal clusters. J Exp Med. 2004;199:155–66. doi: 10.1084/jem.20031677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kyewski B, Derbinski J. Self-representation in the thymus: an extended view. Nat Rev Immunol. 2004;4:688–98. doi: 10.1038/nri1436. [DOI] [PubMed] [Google Scholar]
- 10.Sospedra M, Ferrer-Francesch X, Dominguez O, Juan M, Foz-Sala M, Pujol-Borrel R. Transcription of a broad range of self-antigens in human thymus suggests a role for central mechanisms in tolerance toward peripheral antigens. J Immunol. 1998;161:5918–29. [PubMed] [Google Scholar]
- 11.Bruno R, Sabater L, Sospedra M, Ferrer-Francesch X, Escudero D, Martínez-Cáceres E, Pujol-Borrel R. Multiple sclerosis candidate autoantigens except myelin oligodendrocyte glycoprotein are transcribed in the human thymus. Eur J Immunol. 2002;32:2737–47. doi: 10.1002/1521-4141(2002010)32:10<2737::AID-IMMU2737>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 12.Bruno R, Sabater L, Tolosa E, et al. Different patterns of nicotinic acetylcholine receptor subunit transcription in human thymus. J Neuroimmunol. 2004;149:147–59. doi: 10.1016/j.jneuroim.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 13.Derbinski J, Gäbler J, Brors B, Tierling S, Jonnakuty S, Hergenhahn M, Peltonenh L, Walter J, Kyewski B. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med. 2005;202:33–45. doi: 10.1084/jem.20050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallegos A, Bevan MJ. Central tolerance: good but imperfect. Immunol Rev. 2006;209:290–6. doi: 10.1111/j.0105-2896.2006.00348.x. [DOI] [PubMed] [Google Scholar]
- 15.Kyewski B, Derbinski J, Gotter J, Klein L. Promiscuous gene expression and central T-cell tolerance: more than meets the eye. Trends Immunol. 2002;23:364–71. doi: 10.1016/s1471-4906(02)02248-2. [DOI] [PubMed] [Google Scholar]
- 16.Mathis D, Benoist C. Back to central tolerance. Immunity. 2004;20:509–16. doi: 10.1016/s1074-7613(04)00111-6. [DOI] [PubMed] [Google Scholar]
- 17.DeLuca D, Bluestone JA, Schultz LD, Sharoow SO, Tatsumi Y. Programmed differentiation of murine thymocytes during fetal thymus organ culture. J Immunol Meth. 1995;178:13–29. doi: 10.1016/0022-1759(94)00236-p. [DOI] [PubMed] [Google Scholar]
- 18.Cardoso RS, Junta CM, Macedo C, et al. Hybridization signatures of gamma-irradiated murine fetal thymus organ culture (FTOC) reveal modulation of genes associated with T-cell receptor V(D)J recombination and DNA repair. Mol Immunol. 2006;43:464–72. doi: 10.1016/j.molimm.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 19.Rugh R. The MouseIts Reproduction and Development. 1. Edina, MN: Burgess Publishing Co.; 1968. [Google Scholar]
- 20.Nguyen C, Rocha D, Granjeaud S, Baldit M, Bernard K, Naquet P, Jordan BR. Differential gene expression in the murine thymus assayed by quantitative hybridization of arrayed cDNA clones. Genomics. 1995;29:207–15. doi: 10.1006/geno.1995.1233. [DOI] [PubMed] [Google Scholar]
- 21.Bertucci F, Houlgatte R, Granjeaud S, et al. Prognosis of breast cancer and gene expression profiling using DNA arrays. Ann N Y Acad Sci. 2002;975:217–31. doi: 10.1111/j.1749-6632.2002.tb05954.x. [DOI] [PubMed] [Google Scholar]
- 22.Verdeil G, Puthier D, Nguyen C. Gene profiling approach to establish the molecular basis for partial versus full activation of naïve CD8 T lymphocytes. Ann N Y Acad Sci. 2002;975:68–76. doi: 10.1111/j.1749-6632.2002.tb05942.x. [DOI] [PubMed] [Google Scholar]
- 23.Lopez F, Rougemont J, Loriod B, et al. Feature extraction and signal processing for nylon DNA microarrays. BMC Genomics. 2004;29:38. doi: 10.1186/1471-2164-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quackenbush J. Microarray data normalization and transformation. Nat Genet Suppl. 2002;32:496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- 25.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su AI, Wiltshire T, Batalov S, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci. 2004;101:6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shortman K, Wu L. Early T lymphocyte progenitors. Annu Rev Immunol. 1996;14:29–47. doi: 10.1146/annurev.immunol.14.1.29. [DOI] [PubMed] [Google Scholar]
- 28.Anderson MS, Venanzi ES, Klein L, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 29.Puthier D, Joly F, Irla M, Saade M, Victorero G, Loriod B, Nguyen C. A general survey of thymocyte differentiation by transcriptional analysis of knockout mouse models. J Immunol. 2004;173:6109–18. doi: 10.4049/jimmunol.173.10.6109. [DOI] [PubMed] [Google Scholar]
- 30.Terszowski G, Müller S, Bleul CC, et al. Evidence for a functional second thymus in mice. Science. 2006;312:284–7. doi: 10.1126/science.1123497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Promiscuously induced genes in 20 days p.c. Balb-c thymus (FTOC).