Abstract
Telomere length is maintained in species-specific equilibrium primarily through a competition between telomerase-mediated elongation and the loss of terminal DNA through the end-replication problem. Recombinational activities are also capable of both lengthening and shortening telomeres. Here we demonstrate that elongated telomeres in Arabidopsis Ku70 mutants reach a new length set point after three generations. Restoration of wild-type Ku70 in these mutants leads to discrete telomere-shortening events consistent with telomere rapid deletion (TRD). These findings imply that the longer telomere length set point is achieved through competition between overactive telomerase and TRD. Surprisingly, in the absence of telomerase, a subset of elongated telomeres was further lengthened, suggesting that in this background a mechanism of telomerase-independent lengthening of telomeres operates. Unexpectedly, we also found that plants possessing wild-type-length telomeres exhibit TRD when telomerase is inactivated. TRD is stochastic, and all chromosome ends appear to be equally susceptible. The frequency of TRD decreases as telomeres shorten; telomeres less than 2 kb in length are rarely subject to TRD. We conclude that TRD functions as a potent force to regulate telomere length in Arabidopsis.
Telomeres are dynamic nucleoprotein complexes at the end of eukaryotic chromosomes that consist of long stretches of a simple G-rich repeat and sequence-specific DNA binding proteins. The primary function of telomeres is to protect chromosome termini from being recognized as a double-strand break. The extreme 3′ terminus of the chromosome is single stranded and can undergo a protein-assisted conformational change, folding back upon and invading the duplex region to form a structure termed the t-loop (18). The t-loop is thought to physically sequester the chromosome end, masking the telomere from DNA repair machinery (13).
Telomeric DNA is maintained through a variety of mechanisms that compensate for loss of terminal DNA sequences that occurs as a consequence of nucleolytic processing or the end-replication problem (19, 38). Slow loss of telomeric sequences during DNA replication can be offset by the action of telomerase, a ribonucleoprotein reverse transcriptase that extends the 3′ overhang through reiterative copying of its internal RNA template (reviewed in reference 11). Telomerase is subjected to both positive and negative regulation in cis on the chromosome terminus, mitigating its ability to extend any given telomere (1, 31, 37, 42, 49, 50, 52).
The protein counting model posits that the primary means of telomere length regulation is through an ability to “count” the number of telomeric binding proteins (37). If too many proteins are bound, the telomere will be recalcitrant to extension by telomerase, while if too few proteins are bound, the telomere will be in a more open conformation and will be accessible to telomerase activity. Accordingly, telomerase extension results in an increase in the number of binding sites for telomere proteins and hence an increase in protein occupancy. On the other hand, telomere loss due to the end-replication problem or nuclease attack results in a decrease in the number of sites and fewer proteins bound. This model is strongly supported by studies in yeast (50), mammals (1, 21), and Arabidopsis (48), where telomerase has been shown to act preferentially on the shortest telomeres in the population. Competition between the end-replication problem and telomerase results in a range of telomere lengths that fluctuate between species-specific boundaries. For example, telomeres in Saccharomyces cerevisiae are approximately 300 bp (34), those in Arabidopsis are from 2 to 8 kb (48), and those in mice are from 10 to 60 kb (55).
Positive and negative regulators of telomere length include the double-strand telomere binding proteins TRF1 (49), Rap1 (36), and Taz1 (12) and the single-strand telomere binding protein Pot1 (31). Additionally, telomere length is influenced by KU, a heterodimer of 70- and 80-kDa subunits that is an integral component of the nonhomologous end-joining (NHEJ) DNA double-strand break repair pathway (44). KU is a strong negative regulator of telomerase in Arabidopsis (7, 16, 47); its deletion results in rapid telomerase-dependent extension of telomere tracts (16, 46). Interestingly, in S. cerevisiae and humans, deletion of KU leads to telomere shortening (5, 40), indicating that KU's influence on telomere length regulation is evolving.
In the absence of telomerase, telomeres progressively shorten until they reach a critical length that elicits a DNA damage checkpoint response (14). If cells are forced to continue dividing, telomeres will become uncapped and fuse together. The resulting dicentric chromosomes may then break during the next mitosis only to fuse in the next cell cycle. The resulting breakage-fusion-bridge cycle leads to genomic instability (39). Strong selective pressure against genome instability results in the formation of different types of survivors in yeast (33), whose chromosome ends are maintained through alternate means (28, 51). Several different types of survival have been identified, including recombinational elongation and rolling circle amplification (22, 32). In humans, this form of telomere maintenance is termed alternative lengthening of telomeres (ALT), and is characterized by extremely heterogenous telomeres and the presence of ALT-associated promyelocytic leukemia (PML) bodies.
Cells with elongated telomeres do not face the same selective pressure as cells with extremely short telomeres. Indeed, Arabidopsis ku70 mutants maintain telomeres much longer than wild type, with no apparent affect on growth, development, or genome stability (47). However, studies in yeast indicate that elongated telomeres are quickly returned to wild-type length in a single-step event termed telomere rapid deletion (TRD) (29). These deletion events are intrachromosomal and result in loss of the most terminal sequences (6). A similar phenomenon has been described in humans and Kluyveromyces lactis. Human cells expressing a mutant form of the telomere double-strand binding protein TRF2 undergo catastrophic telomere deletions, concomitant with the formation of extrachromosomal telomere circles (ECTCs) the size of t-loops (53). Similarly, in K. lactis mutants with elongated telomeres due to a mutation in Stn1p, reintroduction of Stn1p results in rapid loss of the elongated telomeres and a return to wild-type length (23).
It has been proposed that branch migration of the displacement loop formed by the invading G-overhang within the t-loop structure results in a Holliday junction (HJ). This structure is then resolved, leading to the formation of a shortened telomere and an extrachromosomal telomeric DNA fragment (35), which in mammals is a circle (ECTC). In S. cerevisiae, TRD and the two major types of survivors are dependent upon Rad52, indicating both processes are recombinational in nature (29, 33). Telomere lengthening in stn1 K. lactis mutants is similarly dependent upon Rad52 (23). Sequestration of the MRX complex in human ALT cells results in slow loss of telomeric DNA and repression of the ALT mechanism of elongation (24). Additionally, the Rad51 paralog Xrcc3, which may be a mammalian Holliday junction resolvase (30), is required for the TRD events observed in TRF2 mutants (53).
Arabidopsis is a genetically tractable model that has been exploited for studies of telomere dynamics (39). One important feature of this organism is that 8 of the 10 chromosome arms are abutted by unique subtelomeric sequences, making it possible to study the fate of individual telomeres in different genetic backgrounds. Here we examine the fate of ultralong telomeres in Arabidopsis ku70 mutants. We demonstrate that elongated telomeres in this background can be rapidly shortened by TRD, either upon reintroduction of KU70 or through loss of telomerase. In addition, we provide evidence for an ALT-like mechanism in plants with elongated telomeres, which we term telomerase-independent lengthening of telomeres (TILT). Finally, we show that wild-type-length telomeres are subject to both TRD and TILT, arguing that recombinational mechanisms play a role in regulating telomere length in wild-type plants.
MATERIALS AND METHODS
Plant growth conditions and mutants.
Plants were grown in EGC growth chambers (Chagrin Falls, OH) with a 16-h photoperiod at 22°C. Generation of ku70 and tert mutants was previously described (45, 47). The ku70/rad51 paralog double mutants were generous gifts from Karel Riha. Characterization of the rad51 mutants was performed as described before (3). The primers and genotyping conditions used for tert, ku70, mre11, and rad51 paralogs were described previously (3, 20, 45, 47).
TRF and FIGE analysis.
For terminal restriction fragment (TRF) analysis, genomic DNA was extracted using a cetyltrimethylammonium bromide (CTAB)-based method (4). For bulk telomere analysis, approximately 1 μg of genomic DNA was digested with 20 U of Tru1I overnight in 200 μl at 65°C. For subtelomere analysis, approximately 1.5 μg of genomic DNA was digested overnight with 10 units of SpeI and PvuII in 200 μl at 37°C. Digested DNA was precipitated and subjected to electrophoresis through 0.7% agarose. Field inversion gel electrophoresis (FIGE) was performed with a contour-clamped homogeneous electric field (CHEF) Mapper XA (Bio-Rad Hercules, CA). DNA was separated through 1% agarose in 0.5× Tris-borate-EDTA (TBE) at 14°C. Conditions were determined using the auto-algorithm function to separate 4- to 50-kb molecules. Conditions were 9 V/cm forward and 6 V/cm reverse, with a linear ramp from 0.08 s to 0.92 s with a total run time of 19 h 2 min. Transfer to nylon membranes and hybridization were performed as previously described (48). Average telomere length was measured using Telometric (17).
Constructs and transformation.
Transfer DNA (T-DNA) constructs were previously described (47). Briefly, the overexpression construct pCBK21 consists of the CaMV35S promoter driving the cDNA of KU70. The genomic construct, pCBK22, consists of 6.7 kb of the KU70 gene along with 1.6 kb of putative promoter sequence. Primary transformants were designated T1, with successive generations being numbered sequentially.
PETRA analysis.
Primer-extension telomere repeat amplification (PETRA) analysis was performed as discussed in reference 20, with slight modifications. CTAB-extracted DNA (4) from a single flower or leaf was resuspended in 30 μl of water. Primer extension was carried out in a 20-μl reaction mixture containing 8 μl DNA, 1× Ex-Taq buffer (TAKARA), 125 μM deoxynucleoside triphosphates (dNTPs), 1 μM PETRA-T, 2 U Ex-Taq polymerase. This reaction was incubated at 65°C for 5 min, 55°C for 1 min, and 72°C for 10 min. One microliter of the reaction mixture was used in a 20-μl reaction mixture containing 1× Ex-Taq buffer, 200 μM dNTPs, 0.25 μM PETRA-A, 0.25 μM telomere-specific primer, and 0.5 U Ex-Taq. These samples were incubated at 96°C for 2 min followed by 16 to 18 cycles of 96°C for 30 s, 60°C for 30 s, and 72°C for 2 min 30 s, with a final incubation at 72°C for 5 min. PCR products were subjected to Southern blotting and hybridization with a 32P 5′-end-labeled (T3AG3)4 probe. Signals were visualized using a STORM phosphorimager (GE Healthcare) and were quantified using Imagequant (Molecular Dynamics). To measure telomere length, a 6th order polynomial equation was fit using Excel to the distance migrated of a 1-kb+ DNA ladder (Invitrogen) and the length of a given PETRA signal was then converted to DNA size using this equation. Finally, the distance of the PETRA primer to the telomere was subtracted from the total length measured by PETRA to give the actual length of the telomere tract.
RESULTS
Telomeres establish a new set point length in the absence of KU70.
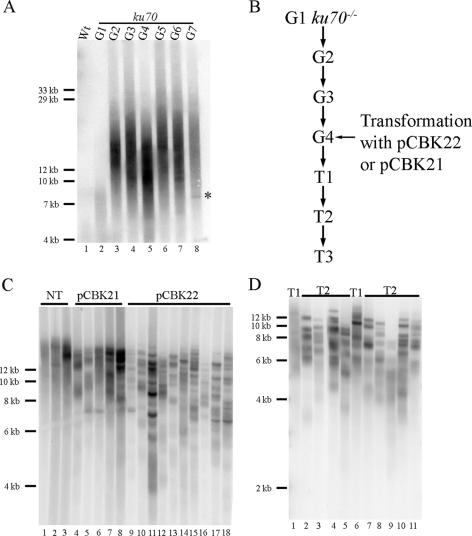
We previously showed that telomeres in ku70 mutants become rapidly elongated and within two generations run near the limit of mobility in regular agarose gels (47). To more accurately determine the size of telomeres in these mutants, we performed TRF analysis on successive generations of ku70 plants. Genomic DNA was digested with Tru1I, which cuts DNA in the subtelomere region and releases the terminal telomere tract. The products were separated using FIGE and hybridized with a telomere-specific probe. As expected, telomeric DNA from wild-type plants migrated near the bottom of the gel consistent with its known size of 2 to 5 kb (Fig. 1A, lane 1). In contrast, first generation (G1) ku70 mutants displayed elongated telomeres reaching 9 kb (Fig. 1A, lane 2), while in G2 bulk telomeres ranged from approximately 10 to 25 kb (Fig. 1A, lane 3). No further dramatic lengthening was observed in subsequent generations, and the average length of telomeres in G2 to G7 mutants was 15.9 ± 2.1 kb (Fig. 1A, lanes 4 to 8). While bulk telomeres in ku70 plants ran as a heterogeneous smear, a smaller discrete telomeric fragment was visible at approximately 7 kb in the G7 ku70 DNA sample (Fig. 1A, lane 8). Of 48 plants analyzed individually by TRF, 21 had similar hybridizing products (data not shown). We were unable to determine the exact nature of these products, although they appeared to be inherited in a Mendelian fashion (data not shown). One possibility is that they reflect point mutations in the elongated telomere tracts that produce novel Tru1I restriction sites. A mutation of the wild-type repeat, TTTAGGG, to either TTTAAGG or TTAAGGG would allow cleavage by Tru1I.
FIG. 1.
Telomere length homeostasis and TRD in ku70 mutants and rescued plants. (A) FIGE of the wild type (Wt) and successive generations of ku70 mutants. TRF analysis was carried out on DNA extracted from ∼50 seedlings. An asterisk denotes a specific hybridizing signal in the G7 line. (B) Scheme for creating KU70 rescued plants. G4 ku70 mutant plants were transformed with either pCBK21 or pCBK22. Plants selected in the next generation correspond to T1. (C) TRF analysis of nontransformed (NT) and selected plants. (D) Parent-progeny TRF analysis of two independent T1 transformants.
Restoration of KU70 leads to TRD.
We have previously shown that telomere elongation in the absence of KU70 is telomerase dependent (46). Thus, our data indicate that telomerase is capable of extending telomeres by as much as 15 kb within a single generation (Fig. 1A, compare lanes 2 and 3). Telomeres reach a maximum size of approximately 25 kb and do not continue to elongate. This new set point could be established through telomerase inhibition at the elongated telomeres. Alternatively, ultralong telomeres may reach homeostasis through competition between the action of telomerase and TRD. We reasoned that reintroduction of KU70 would ultimately restore telomeres to their wild-type length, allowing us to examine the dynamics of reestablishing the wild-type telomere-length set point. If length equilibrium is achieved through telomerase inhibition, telomeres should slowly drift back down to the wild-type length, losing 200 to 500 bp per generation (15) as a consequence of the end-replication problem. Alternatively, if TRD was operational, telomeres should shorten much more rapidly. As tert mutants lose 200 to 500 bp/generation, we define TRD as any telomere-shortening event that leads to a loss of more than 500 bp in a single plant generation.
To examine the fate of elongated telomeres, we transformed G4 ku70 plants with either a construct overexpressing KU70 cDNA (35S::KU70 pCBK21) or a genomic copy of the KU70 gene (pCBK22) and then selected for T1 transformants (Fig. 1B). As expected, telomeres from nonselected siblings migrated near the limit of mobility as a heterogeneous smear (Fig. 1C, lanes 1 to 3). In contrast, plants transformed with pCBK21 (Fig. 1C, lanes 4 to 8) produced a TRF pattern with shortened telomeres. Bulk telomeres in these plants shortened by an average of 0.5 ± 0.3 kb. Strikingly, all of the T1 plants transformed with pCBK22 displayed shortened telomeres. In contrast to the pCBK21 transformants, telomeres in these plants showed a discrete banding profile reminiscent of telomerase mutants (15). The shortest telomeres in the pCBK22 transformants approached the wild-type length of 4 kb in a single generation, with an average loss of 2.3 ± 0.8 kb of telomeric DNA (Fig. 1C, lanes 9 to 18). The appearance of discrete hybridizing bands in the pCBK22 transformants implies that they were resistant to telomerase-mediated elongation. Furthermore, this sharp banding pattern is not consistent with the action of exonucleases, which would likely produce a much more heterogeneous profile. Notably, several telomeric fragments were not shortened in the pCBK22 transformants and instead migrated near the length of the telomeres in their mutant siblings. This observation indicates that individual telomeres are differentially processed.
We conclude that a subpopulation of elongated telomeres shorten much more rapidly than can be accounted for by the end-replication problem, implying that they have been subjected to TRD. Since most of the pCBK22 transformants displayed evidence for TRD, all subsequent work was carried out on these lines.
To further examine the dynamics of telomere shortening, TRF analysis was carried out on T1 plants and their T2 progeny. From T1 to T2, the longest telomeres continued to shorten in a stochastic manner (Fig. 1D). Some T2 plants exhibited dramatic telomere shortening relative to their parent (Fig. 1D, compare lane 5 with lane 1 and lane 9 with lane 6), while other telomeres remained relatively unchanged (Fig. 1D, compare lane 2 with lane 1). On average, telomeres in T2 shortened by 1.9 ± 1.2 kb. This stochastic shortening continued for the two subsequent generations that were analyzed. Strikingly, the frequency of obvious TRD events decreased as telomeres returned to the wild-type length. The average rate of shortening also declined, with a loss of 0.45 ± 0.36 kb from the T2 to T3 generation (data not shown). Telomeres in T3 generation plants averaged 7 ± 0.6 kb (data not shown), within the wild-type range of this ecotype of Arabidopsis. Thus, over three generations, telomeres in plants where KU70 had been restored lost almost 9 kb of telomeric DNA (15.9 kb in G2 to G7 ku70 to 7 kb in T3 transformants).
TRD is not dependent upon KU.
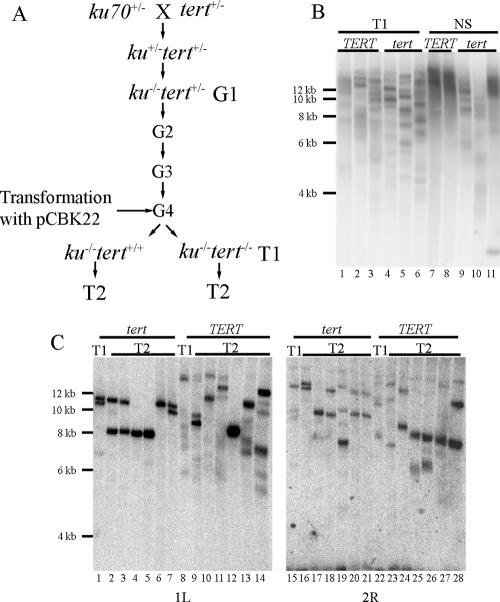
We asked whether TRD was dependent upon reintroduction of KU70. If TRD functions to limit telomere size in ku70 mutants, any telomere shortened by TRD would likely be reextended by telomerase, thus masking TRD. We therefore examined the fate of elongated telomeres in the absence of both TERT and KU70. To accomplish this, plants heterozygous for TERT, the gene encoding the catalytic subunit of telomerase, and homozygous for ku70 were propagated for three generations to elongate telomeres. Plants were transformed with pCBK22 and then segregated for tert in G4, generating a population of TERT+/+ ku70−/− and tert−/− ku70−/− progeny, with or without the KU70 transgene (Fig. 2A).
FIG. 2.
KU70 is not required for TRD. (A) Schematic diagram for generating ku70 tert double mutants. A plant heterozygous for ku70 was crossed to a plant heterozygous for tert. Double heterozygotes for both genes were genotyped in F1. Self-fertilized progeny of the F1 plant were genotyped to identify ku70−/− tert+/− (designated G1). These plants were self-fertilized, and progeny were maintained as ku70−/− tert+/− until G4. G4 plants were transformed with pCBK22 prior to segregation for tert. (B) TRF analysis of T1 and nonselected (NS) progeny of a G4 plant transformed with pCBK22. (C) Subtelomere analysis of T1 parents and their T2 progeny. The subtelomere probe used for the experiments is indicated below each blot. The panels represent sequential hybridization of a single membrane.
Telomeres in both telomerase-positive and telomerase-negative T1 plants shortened to similar lengths, though telomeres in tert mutants appeared as somewhat sharper bands than in telomerase-positive plants (Fig. 2B, lanes 5 and 6). The average telomere length in pCBK22-transformed TERT+/+ plants was 12.3 ± 0.2 kb, and in tert−/− plants it was 12 ± 0.3 kb. Telomerase-positive nonselected plants (genotype ku70−/− tert+/+) displayed the elongated telomeres of ku70 mutants, and telomeres averaged 13.4 ± 0.3 kb, several kilobases less than the average measured by FIGE. This discrepancy is likely due to the poor resolution at high molecular weight under these gel conditions (Fig. 2B, lanes 7 and 8). Nonselected plants that were homozygous null for tert also showed TRD (Fig. 2B, lanes 9 and 10), although one plant of this genotype retained most of its telomeres at an elongated length (Fig. 2B, lane 11). The average size of telomeres in these plants was 11.9 ± 1.4 kb. Taken together, the data indicate that TRD does not require Ku70 and can occur in the absence of telomerase.
TRD is a stochastic process.
To more accurately gauge the rate of telomere shortening, we monitored the fate of individual telomeres in the T2 progeny of these transformants (Fig. 2A [both tert−/− and tert+/+ plants monitored]). Genomic DNA was digested with restriction enzymes that cut several kilobases internal to the telomere and then hybridized with probes directed to specific chromosome arms. As seen in Fig. 2C, dramatic, stochastic changes in telomere length were detected between the parent plants and their progeny in both genetic backgrounds. Again, telomerase-positive plants showed more heterogeneous signals at shorter lengths, consistent with telomerase acting on these shorter telomeres.
The clearest example of telomere shortening was seen with the 1L telomere in tert mutant plants. The parental plant had two prominent hybridizing signals at approximately 10.5 and 11 kb (Fig. 2C, lane 1). Additionally, a much less intense signal of approximately 7 kb was observed. Four of the six progeny of this plant displayed a telomere of approximately 8 kb (Fig. 2C, lanes 2 through 5), and two plants had completely lost the 10.5- and 11-kb signals (Fig. 2C, lanes 4 and 5). We can envision two ways in which the 1L telomere shown in lanes 4 and 5 arose. First, TRD could shorten one of the prominent hybridizing signals in the parent, leading to a decrease of 2.5 kb. Alternatively, telomeres in the progeny could arise through a telomerase-independent lengthening of the weakly hybridizing 7-kb signal in the parent. We consider the latter possibility less likely; the very weak hybridization of the 7-kb telomere in the parent is more consistent with a somatic TRD event that occurred during plant development. If this is true, the 7-kb fragment arose from a TRD event that shortened the 10.5-kb telomere by 3.5 kb, implying that TRD is capable of shortening an individual telomere by several kilobases in a single generation.
The subtelomeric analysis also revealed a surprisingly complex array of products. A plant can inherit a maximum of two telomeres of different lengths (on the homologous chromosomes) from its parents. Thus, the presence of more than two hybridizing bands for an individual subtelomere arm argues that shortened telomeres in the progeny are not simply due to the inheritance of an undetectable subset of shorter telomeres from the parent. Rather, these telomeres must be derived from discrete telomere-processing events in the progeny.
Plants displaying multiple signals for one chromosome arm do not necessarily have multiple signals at other chromosome ends. The tert mutant plant analyzed in Fig. 2C, lanes 5 and 19, has a single hybridizing signal for the 1L telomere and four hybridizing signals for the 2R telomeres. Similarly, the telomerase-positive plant analyzed in Fig. 2C, lanes 14 and 28, gives rise to six hybridizing signals for chromosome 1L and only three signals for chromosome 2R. Thus, the number of TRD events that occur upon restoration of Ku70 is relatively small. The presence of six hybridizing signals indicates that only four or five TRD events occurred at that telomere throughout the life span of this plant. We conclude that TRD functions stochastically on different telomeres and can shorten telomeres by at least 2.5 kb in a single generation. In budding yeast, the size of the deleted products is largely governed by the size of the majority of the telomeres in the cell. This does not appear to be the case in this background, as all telomeres are grossly elongated. However, a direct test of this aspect of TRD would require the generation of a plant with only a subset of elongated telomeres.
TRD proceeds in Arabidopsis in the absence of genes required for TRD in other organisms.
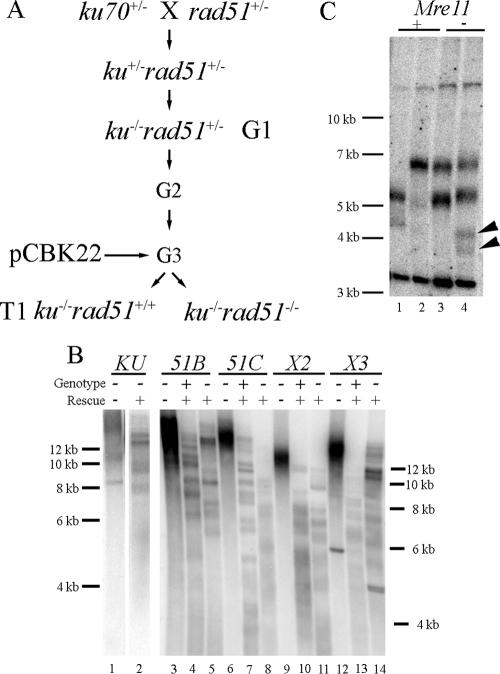
We tested whether MRE11 and the available RAD51 paralogs are required for TRD in Arabidopsis. Plants homozygous null for ku70 and heterozygous for an additional mutation in MRE11, RAD51B, RAD51C, XRCC2, or XRCC3 were propagated for several generations before transformation with pCBK22. The selected T1 plants were then genotyped for the presence of the additional mutant allele. Figure 3B shows the TRF profile of T1 plants mutant for XRCC2, XRCC3, RAD51B, and RAD51C. Deletion of any of these four genes did not inhibit TRD upon reintroduction of KU70. This finding was verified through subtelomere analysis, with all four mutants showing multiple signals for at least one of two tested subtelomeric probes (data not shown).
FIG. 3.
TRD in Arabidopsis is not dependent upon known recombinases. (A) Genetic scheme for obtaining rad51 mutants with elongated telomeres. Plants null for ku70 and heterozygous for the indicated genotypes were transformed with pCBK22, and the transformed progeny were genotyped to identify transformants homozygous null for the indicated genotype. (B) TRF analysis of T1 progeny of the indicated genotypes. Transformants (+) and nontransformants (−) are indicated. (C) Parent progeny subtelomere analysis of a single T1 mre11+/− plant. Self-fertilized progeny of this plant were genotyped for MRE11 and for the presence of the pCBK22-derived T-DNA. Arrowheads denote additional products in one of the mre11 mutants. The probe is 2R.
In three independent experiments, and in contrast to the other mutants tested, we were unable to select plants that were null for MRE11 in the T1 population (we recovered a total of 14 mre11+/+ and 22 mre11+/− plants in the three separate transformations). This could indicate that MRE11 is required for T-DNA integration in this background. Therefore, we genotyped T2 progeny of a T1 plant heterozygous for MRE11 and isolated individuals mutant for MRE11 (Fig. 3C). Of the two plants mutant for MRE11, one showed four hybridizing signals for chromosome 2R (Fig. 3C, lane 5), demonstrating that MRE11 is also not required for TRD. From these data, we conclude that genes previously shown to be required for TRD in other organisms are not necessary for TRD in Arabidopsis.
Telomerase-independent lengthening occurs at elongated Arabidopsis telomeres.
To date, no direct evidence for telomere elongation in the absence of telomerase has been observed in Arabidopsis (54). However, recent data from K. lactis indicate that telomere lengthening can be driven by ECTCs generated as a byproduct of TRD, and circular molecules are present in human cells undergoing TRD (8, 23, 53). Although we found no evidence for ECTCs by two-dimensional gel analysis of pCBK22 transformants (data not shown), this could simply reflect the low frequency of TRD events. Therefore, we looked for telomere elongation in plants lacking telomerase and undergoing TRD using the genetic approach described in Fig. 2A. Specifically, we performed parent-progeny subtelomere analysis on T1 and T2 pCBK22 transformants that were mutant for ku70 and tert.
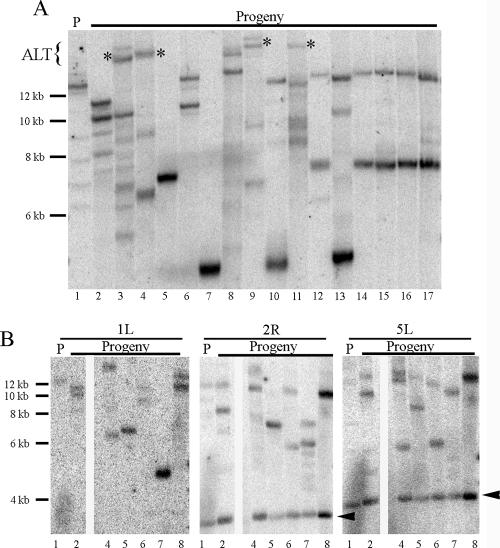
In one of three lines examined, the 1L telomere of several progeny plants was 5 to 10 kb longer than the longest telomere in its parent (Fig. 4A, lanes 3, 4, 9, and 11). A trivial explanation for this finding is that subtelomeric DNA was rearranged, changing the restriction profile of this telomere to make it appear elongated. Several observations are inconsistent with this conclusion. First, the elongated products hybridized with a telomeric probe (data not shown), suggesting they are in fact terminal. Second, digestion of the DNA with other restriction enzymes that cleave in the subtelomeric region generated products of expected sizes (data not shown). Third, other subtelomere arms were elongated (see below). Taken together, these data argue that the subtelomeric sequence of 1L has not been grossly rearranged.
FIG. 4.
TILT in tert mutants with elongated telomeres. (A) Parent progeny subtelomere analysis of a T1 pCBK22 transformant homozygous for tert. Mutants were generated as described in the legend to Fig. 2A. Asterisks denote telomeres that were elongated relative to the parent (P). The hybridizing probe is 1L. (B) Sequential hybridization of three probes to a blot containing a subset of samples from panel A. Lane numbers correspond to numbers from the gel in panel A. Arrowheads denote interstitial hybridizing signals.
Further analysis of individual telomeres in these plants conducted with three different subtelomere-specific probes showed that telomeres behaved independently (Fig. 4B). For example, telomeres from the plants analyzed in Fig. 4B, lanes 5 and 6, showed no lengthening relative to their parent for any of the arms examined. In contrast, telomeres from the plant analyzed in lane 3 were all extended relative to their parent, while some telomeres from the plants in lanes 2, 4, and 7 were elongated, and others were shortened. Rare elongation events in other lines were also observed (Fig. 2C, lane 16). These data indicate that Arabidopsis is capable of elongating telomeres in the absence of telomerase. We term this process TILT, since we currently have no evidence of cytological markers consistent with ALT, nor have we been able to directly demonstrate that TILT occurs through a recombinational mechanism as described for recombinational telomere elongation in K. lactis (41).
TRD functions at telomeres with lengths in the wild-type range.
We next asked whether telomeres in the wild-type range are subjected to TRD. For this analysis, we employed PETRA, a sensitive method for accurately measuring telomere length at individual chromosome arms (20). Although the ultralong telomeres in ku70 mutants are not good substrates for PETRA, this is the preferred method for individual-telomere analysis in plants with wild-type-length telomeres as minimal quantities of DNA are required (a single Arabidopsis leaf is sufficient), and seven chromosome arms can be measured at the same time. If TRD occurs in telomerase-positive tissues, the newly shortened telomere is likely to be efficiently elongated by telomerase. To avoid this confounding factor, we examined the rate of telomere shortening in G1, G2, and G6 tert mutants.
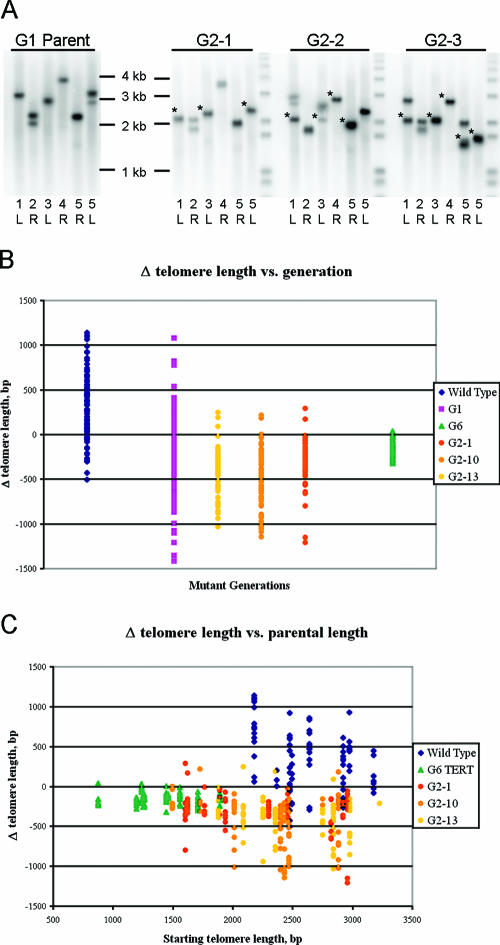
A representative gel with PETRA products is shown in Fig. 5A. The parent is a G1 tert mutant (Fig. 5A, left panel). Notably, in the heterozygous parent of this G1 progeny, only the 2R telomere had undergone TRD (data not shown). However, in the three G2 progeny shown (Fig. 5A, right panel), several examples of TRD were detected, as noted by asterisks.
FIG. 5.
TRD occurs in telomeres within the wild-type range. (A) Representative PETRA data. Changes defined as TRD are indicated by asterisks. The telomere that was monitored is indicated below each lane. (B) Graph depicting the change in telomere length versus generation for different genotypes and their progeny. (B) Graph depicting the change in telomere length from parent to progeny relative to the length of the telomere in the parent.
Table 1 shows the mean rate of telomere length change for all generations analyzed. Individual data points are displayed graphically in Fig. 5B. In wild-type plants, there is a broad distribution of the change in telomere length, with most telomeres showing a net increase relative to their parent. Strikingly, of 88 telomere changes measured in telomerase-positive plants, only a single telomere was shortened by more than 500 bp. Similar degrees of stochastic telomere length changes are observed in G1 and G2 tert mutants. However, as expected for telomerase mutants, the net change in telomere length is negative. In contrast, there was very little change in telomere length for G6 tert mutants; the maximal shortening observed in the G6 population was 254 bp. The 500-bp cutoff for TRD is twice the maximal shortening observed in the G6 population. Telomeres in both G1 and G2 tert mutants shortened by well over 1 kb in a single generation. In a total of 355 telomere length changes measured in G1 and G2 tert mutants, 19 telomeres shortened by over 1 kb and 66 shortened by more than 500 bp, twice the maximal shortening (254 bp) observed in the G6 tert mutants. These data imply that telomeres in the G6 mutants shorten only via the end-replication problem, while telomeres in G1 and G2 tert mutants are also substrates for TRD. We note that many of the telomeres in the G6 tert mutants are capable of losing more than 500 bp and would still remain above the absolute minimal telomere length of ∼350 bp (20). This observation suggests that our inability to detect TRD in G6 tert mutants is not due to attrition of these plants because of critically shortened telomeres. The dramatic increase in TRD frequency in G1 and G2 tert mutants relative to the wild type could indicate that TERT protects against TRD. Alternatively TRD products may simply be rapidly reextended by telomerase. Taken together, these data support the conclusion that telomeres in early generation tert mutants, despite being within the wild-type-length range overall, are subjected to TRD.
TABLE 1.
Mean changes in telomere length from parents to their progeny
| Genotype (n) | Mean ± SD change in length (bp) |
|---|---|
| Wild type (88) | 301 ± 382 |
| G1 tert (88) | −246 ± 476 |
| G2-1 tert (85) | −285 ± 212 |
| G2-10 tert (96) | −450 ± 322 |
| G2-13 tert (86) | −391 ± 222 |
| G6 tert (82) | −154 ± 77 |
Interestingly, there are several examples of TILT events in tert mutants (Fig. 5B). In G6 tert mutants, only a very small degree of elongation was observed (<73 bp), which may be due to errors in the accuracy of measurement. Telomere elongation in G1 mutants is difficult to assess as telomeres could have been extended by telomerase in the previous generation. In contrast, 5 of 265 telomeres in G2 tert mutants were lengthened from 90 to 288 bp; the greatest differential likely represents true elongation events. Notably, the G2 tert mutants had a much higher frequency of these elongation events relative to the G6 tert mutant, supporting the notion of a mechanistic link between TRD and TILT.
Finally, to specifically address whether TRD is length dependent, we plotted the change in telomere length versus the parental telomere length (Fig. 5C). In telomerase-positive plants, and consistent with previous reports (48), the shorter telomeres were more likely to be elongated than the longer telomeres (compare the shortest wild-type telomere to the longest). Moreover, the shortest telomeres were elongated to a greater extent than the longest telomeres. In G6 tert mutants, all telomeres shortened by the same amount, regardless of the initial telomere length. Strikingly, telomeres in G2 tert mutants shortened by a much larger amount if the parental telomere was longer than approximately 2 kb. Telomeres below 2 kb displayed a rate of shortening similar to that of G6 tert mutants. The frequency of TRD by initial telomere length is shown in Table 2. When queried by chromosome arm, all telomeres underwent TRD with approximately equal frequencies (between 16% and 34%; data not shown). While there is no clear relationship between telomere lengths above 2 kb and the incidence of TRD, the frequency of TRD drops dramatically for telomeres that are less than 2 kb, which is the minimal size of telomeres in wild-type plants.
TABLE 2.
Frequency of telomere length changes in G1 and G2 tert progeny
| Initial length (kb) (no. with length) | Frequency of telomeres with:
|
Frequency of >500-bp lossb | |||
|---|---|---|---|---|---|
| Gaina | Loss (bp)
|
||||
| 0-500 | 500-1,000 | >1,000 | |||
| >3 (2) | 0 | 1 | 1 | 0 | 0.5 |
| 2.5-3 (116) | 14 | 67 | 27 | 8 | 0.30 |
| 2-2.5 (167) | 13 | 107 | 36 | 11 | 0.27 |
| <2 (70) | 3 | 65 | 2 | 0 | 0.03 |
| Total (355) | 30 | 240 | 66 | 19 | |
Represents any telomere that was elongated relative to the parent.
The sum of all length changes losing >500 bp divided by the total number examined.
DISCUSSION
Telomere dynamics in ku70 mutants.
Cells must maintain a minimal telomere length to provide full protection for chromosome ends and to distinguish them from double-strand breaks. Regulation of a maximal telomere length is also likely to be important to inhibit recombination and to reduce the total amount of DNA synthesis. In the absence of KU70, telomeres in Arabidopsis are dramatically elongated (7, 16, 47). This feature, along with the unique subtelomeric sequences on most Arabidopsis chromosome arms, allowed us to examine the dynamics of ultralong telomeres. We find that a new, longer telomere length set point is established, and this set point is likely maintained by a competition between a highly active telomerase and a TRD-like mechanism that shortens grossly elongated telomeres. While the elongated G-overhang in ku70 mutants (46) may lead to an increase in the loss of telomeric DNA due to the end-replication problem, reintroduction of KU70 would be expected to restore the appropriate overhang length, effectively negating this increase in telomere loss. Reintroduction of KU70 results in dramatic telomere shortening, at a rate much greater than can be accounted for by the end-replication problem. A loss of at least 2.5 kb is readily apparent in Fig. 4C, and several telomeres in the T2 generation appear to have shortened by up to 6 kb over two generations. This rate of telomere attrition is not sufficient to offset 15 kb of telomerase-dependent elongation observed in G1 to G2 ku mutants, suggesting either that telomerase is inhibited at extremely long telomeres, or that TRD can shorten telomeres by even larger amounts than those we observed. We further demonstrated that TRD is not dependent upon reintroduction of KU70 itself, as segregation of tert from ku70 mutants with long telomeres is sufficient to dramatically shorten telomeres.
Our data are consistent with the dynamics of elongated telomeres in other organisms (11, 26, 27, 29) and argue that TRD is a highly conserved mechanism for telomere size control. The role of the KU heterodimer in Arabidopsis TRD is unknown. In yeast (43), and perhaps humans (35), KU inhibits TRD. In contrast, TRD events in ku70 mutants rescued with pCBK22 were indistinguishable from plants doubly deficient for ku70 and tert. Current models of the mechanism of TRD posit that the 3′ G-overhang invades the duplex telomeric DNA, forming a displaced loop at the site of invasion (8, 35, 53). Branch migration would then convert this structure into a recombination intermediate resembling an HJ, which could be resolved into a shortened telomere and an extrachromosomal telomeric circle. The genetic requirements for TRD in Arabidopsis are not clear. Surprisingly, deletion of either MRE11 or XRCC3, which are shown to be required for TRD in mammals and yeast, shows that they are not required for TRD in Arabidopsis. Several explanations for these findings can be considered. First, there is extensive redundancy in the Arabidopsis genome (as much as 60% of the genome is present in duplications), and thus there may be another enzyme capable of resolving HJs in Arabidopsis (2). Alternatively, the terminal structure formed at Arabidopsis telomeres may be slightly different from that formed in yeast or mammals. Interestingly, MUS81, an enzyme that resolves HJ-like structures, has two homologues in Arabidopsis. A third consideration is that our assays can not fully distinguish between TRD events that occur in meiosis or mitosis. In budding yeast, TRD occurs with a much higher frequency in meiosis than in mitosis (25), making the distinction between meiotic and mitotic requirements important. It may be necessary to disrupt both processes to observe inhibition of TRD.
ECTCs and telomerase-independent telomere elongation.
ECTCs have been shown to drive recombinational telomere elongation in K. lactis (41) and have been associated with ALT in mammals (8, 53). In previous experiments designed to select for telomerase-negative Arabidopsis cells that can maintain telomeres, no telomere lengthening was observed (54). However, the cells used in these experiments were derived from plants that had extremely short telomeres and thus would be unlikely substrates for TRD. As a consequence, we speculate there would not be an accumulation of ECTCs to serve as substrates for telomere elongation and TILT through a roll-and-spread mechanism. Although we failed to detect ECTCs in plants with elongated telomeres where we had restored KU70, we did find evidence for TILT events in telomerase-deficient plants with elongated telomeres. TRD was also acting on telomeres in these plants, suggesting a model in which TRD and TILT are mechanistically linked through the formation of ECTCs by TRD and their use as substrates for telomere elongation. Further work will be required to demonstrate that ECTCs can drive telomere lengthening in Arabidopsis.
TRD as a means to regulate telomere length.
One important observation from our study is that the frequency of TRD is proportional to the length of telomeres, arguing that TRD can function as a form of length regulation. Although a role for TRD has been established in budding yeast (29), here we show that TRD not only shortens grossly elongated Arabidopsis telomeres, but also acts on telomeres within the wild-type size range. Notably, the extent of telomere shortening in G1 and G2 tert mutants is much greater than that in G6 tert mutants. Furthermore, the amount of DNA lost varies dramatically between different telomeres within the same cellular population. These two findings indicate that TRD can function stochastically at wild-type-length telomeres in early generation tert mutants. We found that the frequency of TRD decreases as telomere length declines, with a very sharp decrease when telomeres drop below 2 kb in length. Intriguingly, the lower range of telomeres in wild-type Arabidopsis is 2 kb. Thus, TRD might play a role in determining the minimal telomere length.
Work in budding yeast has demonstrated that TRD can occur at telomeres within the wild-type range of some strains (6). In Candida albicans, loss of RAD52, which is required for TRD in other organisms (29), resulted in telomere lengthening (10). This observation implies that TRD plays a role in regulating telomere length within wild-type Candida. Our findings argue that TRD can act in concert with telomerase and the end-replication problem as a potent force for controlling telomere length in Arabdidopsis. How could TRD regulate telomere length? A protein-counting model similar to one that regulates telomerase activity is attractive, but what protein is counted? KU is an interesting possibility. KU is associated with telomeres in all organisms studied, where it contributes to telomere length regulation as well as chromosome end protection. It is possible that KU serves as a roadblock to branch migration and that as more KU binds to the telomere tract, roadblocks are more frequent. One prediction of this model is that an increase in the amount of KU can prevent TRD. While this model will require more extensive study, we note that overexpression of KU70 in ku70 mutant plants results in a telomere profile distinct from restoration of the wild-type construct (Fig. 1C).
TRD events at telomeres in wild-type plants place tremendous pressure on telomerase to extend the truncated telomeres. An unlucky TRD event could result in a telomere that falls below the critical length threshold, leading to telomere dysfunction. It is possible that telomerase actively inhibits TRD. Such a model is supported by work in Caenorhabditis elegans, where loss of mrt-2 results in an ever-shorter-telomere phenotype, while loss of telomerase results in sudden telomere-shortening events (9). Thus, telomerase appears to be critical either for extending telomeres subjected to TRD or for protecting them from TRD in the first place.
Acknowledgments
We thank Barbara Zellinger and Karel Riha for their invaluable help in providing unpublished materials and sharing unpublished data. We are also grateful to Elizabeth Summer for the use of her CHEF Mapper and Laurent Vespa, Michelle Heacock, and Yulia Surovtseva for critically reading the manuscript.
This work was supported by NIH GM65383 to D.E.S.
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Ancelin, K., M. Brunori, S. Bauwens, C.-E. Koering, C. Brun, M. Ricoul, J.-P. Pommier, L. Sabatier, and E. Gilson. 2002. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol. Cell. Biol. 22:3474-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanc, G., A. Barakat, R. Guyot, R. Cooke, and M. Delseny. 2000. Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12:1093-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bleuyard, J. Y., M. E. Gallego, F. Savigny, and C. I. White. 2005. Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J. 41:533-545. [DOI] [PubMed] [Google Scholar]
- 4.Borevitz, J. O., D. Liang, D. Plouffe, H. S. Chang, T. Zhu, D. Weigel, C. C. Berry, E. Winzeler, and J. Chory. 2003. Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res. 13:513-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulton, S. J., and S. P. Jackson. 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bucholc, M., Y. Park, and A. J. Lustig. 2001. Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:6559-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bundock, P., H. van Attikum, and P. Hooykaas. 2002. Increased telomere length and hypersensitivity to DNA damaging agents in an Arabidopsis KU70 mutant. Nucleic Acids Res. 30:3395-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cesare, A. J., and J. D. Griffith. 2004. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol. Cell. Biol. 24:9948-9957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung, I., M. Schertzer, A. Rose, and P. M. Lansdorp. 2006. High incidence of rapid telomere loss in telomerase-deficient Caenorhabditis elegans. Nucleic Acids Res. 34:96-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciudad, T., E. Andaluz, O. Steinberg-Neifach, N. F. Lue, N. A. Gow, R. A. Calderone, and G. Larriba. 2004. Homologous recombination in Candida albicans: role of CaRad52p in DNA repair, integration of linear DNA fragments and telomere length. Mol. Microbiol. 53:1177-1194. [DOI] [PubMed] [Google Scholar]
- 11.Cohen, S., and M. Mechali. 2002. Formation of extrachromosomal circles from telomeric DNA in Xenopus laevis. EMBO Rep. 3:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper, J. P., E. R. Nimmo, R. C. Allshire, and T. R. Cech. 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385:744-747. [DOI] [PubMed] [Google Scholar]
- 13.de Lange, T. 2002. Protection of mammalian telomeres. Oncogene 21:532-540. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira, M. G., K. M. Miller, and J. P. Cooper. 2004. Indecent exposure: when telomeres become uncapped. Mol. Cell 13:7-18. [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald, M. S., K. Riha, F. Gao, S. Ren, T. D. McKnight, and D. E. Shippen. 1999. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl. Acad. Sci. USA 96:14813-14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallego, M. E., N. Jalut, and C. I. White. 2003. Telomerase dependence of telomere lengthening in Ku80 mutant Arabidopsis. Plant Cell. 15:782-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant, J. D., D. Broccoli, M. Muquit, F. J. Manion, J. Tisdall, and M. F. Ochs. 2001. Telometric: a tool providing simplified, reproducible measurements of telomeric DNA from constant field agarose gels. BioTechniques 31:1314-1316, 1318. [DOI] [PubMed] [Google Scholar]
- 18.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. de Lange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97:503-514. [DOI] [PubMed] [Google Scholar]
- 19.Harley, C. B., A. B. Futcher, and C. W. Greider. 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345:458-460. [DOI] [PubMed] [Google Scholar]
- 20.Heacock, M., E. Spangler, K. Riha, J. Puizina, and D. E. Shippen. 2004. Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J. 23:2304-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hemann, M. T., M. A. Strong, L. Y. Hao, and C. W. Greider. 2001. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107:67-77. [DOI] [PubMed] [Google Scholar]
- 22.Henson, J. D., A. A. Neumann, T. R. Yeager, and R. R. Reddel. 2002. Alternative lengthening of telomeres in mammalian cells. Oncogene 21:598-610. [DOI] [PubMed] [Google Scholar]
- 23.Iyer, S., A. D. Chadha, and M. J. McEachern. 2005. A mutation in the STN1 gene triggers an alternative lengthening of telomere-like runaway recombinational telomere elongation and rapid deletion in yeast. Mol. Cell. Biol. 25:8064-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang, W.-Q., Z.-H. Zhong, J. D. Henson, A. A. Neumann, A. C.-M. Chang, and R. R. Reddel. 2005. Suppression of alternative lengthening of telomeres by Sp100-mediated sequestration of the MRE11/RAD50/NBS1 complex. Mol. Cell. Biol. 25:2708-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joseph, I., D. Jia, and A. J. Lustig. 2005. Ndj1p-dependent epigenetic resetting of telomere size in yeast meiosis. Curr. Biol. 15:231-237. [DOI] [PubMed] [Google Scholar]
- 26.Kilian, A., C. Stiff, and A. Kleinhofs. 1995. Barley telomeres shorten during differentiation but grow in callus culture. Proc. Natl. Acad. Sci. USA 92:9555-9559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larson, D. D., E. A. Spangler, and E. H. Blackburn. 1987. Dynamics of telomere length variation in Tetrahymena thermophila. Cell 50:477-483. [DOI] [PubMed] [Google Scholar]
- 28.Le, S., J. K. Moore, J. E. Haber, and C. W. Greider. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, B., and A. J. Lustig. 1996. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 10:1310-1326. [DOI] [PubMed] [Google Scholar]
- 30.Liu, Y., J. Y. Masson, R. Shah, P. O'Regan, and S. C. West. 2004. RAD51C is required for Holliday junction processing in mammalian cells. Science 303:243-246. [DOI] [PubMed] [Google Scholar]
- 31.Loayza, D., and T. de Lange. 2003. POT1 as a terminal transducer of TRF1 telomere length control. Nature 423:1013-1018. [DOI] [PubMed] [Google Scholar]
- 32.Lundblad, V. 2002. Telomere maintenance without telomerase. Oncogene 21:522-531. [DOI] [PubMed] [Google Scholar]
- 33.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell 73:347-360. [DOI] [PubMed] [Google Scholar]
- 34.Lundblad, V., and J. W. Szostak. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57:633-643. [DOI] [PubMed] [Google Scholar]
- 35.Lustig, A. J. 2003. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat. Rev. Genet. 4:916-923. [DOI] [PubMed] [Google Scholar]
- 36.Lustig, A. J., S. Kurtz, and D. Shore. 1990. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science 250:549-553. [DOI] [PubMed] [Google Scholar]
- 37.Marcand, S., E. Gilson, and D. Shore. 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275:986-990. [DOI] [PubMed] [Google Scholar]
- 38.Maringele, L., and D. Lydall. 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 16:1919-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKnight, T. D., and D. E. Shippen. 2004. Plant telomere biology. Plant Cell 16:794-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myung, K., G. Ghosh, F. J. Fattah, G. Li, H. Kim, A. Dutia, E. Pak, S. Smith, and E. A. Hendrickson. 2004. Regulation of telomere length and suppression of genomic instability in human somatic cells by Ku86. Mol. Cell. Biol. 24:5050-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Natarajan, S., and M. J. McEachern. 2002. Recombinational telomere elongation promoted by DNA circles. Mol. Cell. Biol. 22:4512-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pennock, E., K. Buckley, and V. Lundblad. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387-396. [DOI] [PubMed] [Google Scholar]
- 43.Polotnianka, R. M., J. Li, and A. J. Lustig. 1998. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol. 8:831-834. [DOI] [PubMed] [Google Scholar]
- 44.Riha, K., M. L. Heacock, and D. E. Shippen. 2006. The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu. Rev. Genet. 40:237-277. [DOI] [PubMed] [Google Scholar]
- 45.Riha, K., T. D. McKnight, L. R. Griffing, and D. E. Shippen. 2001. Living with genome instability: plant responses to telomere dysfunction. Science 291:1797-1800. [DOI] [PubMed] [Google Scholar]
- 46.Riha, K., and D. E. Shippen. 2003. Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proc. Natl. Acad. Sci. USA 100:611-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riha, K., J. M. Watson, J. Parkey, and D. E. Shippen. 2002. Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 21:2819-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shakirov, E. V., and D. E. Shippen. 2004. Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell 16:1959-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smogorzewska, A., B. van Steensel, A. Bianchi, S. Oelmann, M. R. Schaefer, G. Schnapp, and T. de Lange. 2000. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 20:1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teixeira, M. T., M. Arneric, P. Sperisen, and J. Lingner. 2004. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117:323-335. [DOI] [PubMed] [Google Scholar]
- 51.Teng, S. C., J. Chang, B. McCowan, and V. A. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 6:947-952. [DOI] [PubMed] [Google Scholar]
- 52.van Steensel, B., and T. de Lange. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385:740-743. [DOI] [PubMed] [Google Scholar]
- 53.Wang, R. C., A. Smogorzewska, and T. de Lange. 2004. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119:355-368. [DOI] [PubMed] [Google Scholar]
- 54.Watson, J. M., P. Bulankova, K. Riha, D. E. Shippen, and B. Vyskot. 2005. Telomerase-independent cell survival in Arabidopsis thaliana. Plant J. 43:662-674. [DOI] [PubMed] [Google Scholar]
- 55.Zijlmans, J. M., U. M. Martens, S. S. Poon, A. K. Raap, H. J. Tanke, R. K. Ward, and P. M. Lansdorp. 1997. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl. Acad. Sci. USA 94:7423-7428. [DOI] [PMC free article] [PubMed] [Google Scholar]