Abstract
ALR (MLL2) is a member of the human MLL family, which belongs to a larger SET1 family of histone methyltransferases. We found that ALR is present within a stable multiprotein complex containing a cohort of proteins shared with other SET1 family complexes and several unique components, such as PTIP and the jumonji family member UTX. Like other complexes formed by SET1 family members, the ALR complex exhibited strong H3K4 methyltransferase activity, conferred by the ALR SET domain. By generating ALR knockdown cell lines and comparing their expression profiles to that of control cells, we identified a set of genes whose expression is activated by ALR. Some of these genes were identified by chromatin immunoprecipitation as direct ALR targets. The ALR complex was found to associate in an ALR-dependent fashion with promoters and transcription initiation sites of target genes and to induce H3K4 trimethylation. The most characteristic features of the ALR knockdown cells were changes in the dynamics and mode of cell spreading/polarization, reduced migration capacity, impaired anchorage-dependent and -independent growth, and decreased tumorigenicity in mice. Taken together, our results suggest that ALR is a transcriptional activator that induces the transcription of target genes by covalent histone modification. ALR appears to be involved in the regulation of adhesion-related cytoskeletal events, which might affect cell growth and survival.
Epigenetic regulation of transcription is a major mechanism of gene regulation in eukaryotes. Methylation of histones on lysine residues is one such epigenetic mechanism (18, 42). At least five lysine residues of histone H3 (K4, K9, K27, K36, and K79) and single lysine residues of histones H4 and H1 (K20 and K26, respectively) have been shown to be sites for methylation (8, 24). The effects of methylation on gene expression are dependent on the particular lysine residue. For example, methylation of histone H3 on lysine K4 (H3K4 methylation) is linked to transcriptional activation, whereas H3K27 methylation is associated with transcriptional repression (24, 40).
The hallmark of proteins possessing lysine methyltransferase activity is the evolutionarily conserved catalytic SET domain. The SET domain methyltransferases can be divided into seven main families. Proteins within each family have a higher level of homology within the SET domain and in the SET domain-surrounding sequences and catalyze methylation of a specific lysine residue(s) (8). One such family is SET1, whose members methylate the K4 residue on histone H3. The family includes the Saccharomyces cerevisiae Set1 protein and its human homolog, hSet1; the Drosophila Trithorax (TRX) and TRR (Trithorax-related) proteins; and their closely related human homologs, designated MLL group proteins. Human SET1 family members are present in multiprotein complexes that appear to be structurally and functionally similar to the yeast Set1 complex (9, 13, 16, 25, 29, 48, 51).
The best-studied MLL group member is MLL (for mixed-lineage leukemia; also termed MLL1 and ALL-1). MLL is often involved in chromosome translocations in human acute leukemia and, like its Drosophila homolog TRX, is required for maintaining the expression of homeobox (Hox) genes during embryo development (2). Subsequent to the cloning of MLL, the human genome was found to contain at least four more genes encoding MLL-like proteins, each carrying some or most of the MLL motifs. These proteins include MLL4 (the former MLL2), which is highly similar to MLL (12, 17); the closely related ALR (MLL2) and HALR (MLL3) proteins (31, 33, 45); and MLL5 (11). Despite extensive research done on MLL, little is known about the functions of the other MLL-related proteins.
Recently, new insights were obtained regarding ALR (MLL2) and its Drosophila homolog, TRR. ALR is a gigantic 5,262-residue-long protein containing a SET domain, five PHD fingers, an HMG-I binding motif, a zinc finger, and FY-rich motifs (31). Analysis of its expression in the mouse embryo and in adult human tissues suggests that ALR might have multiple functions during development, as well as in the adult organism (31). Consistent with these data, some important aspects of TRR activity during Drosophila development were uncovered (36). TRR was shown to regulate ecdysone-dependent transcription of a principal developmental-factor gene, HEDGEHOG (HH). Upon ecdysone addition, TRR bound the ecdysone receptor and, together with the latter, was recruited to the promoter of the HH gene. In the promoter region, TRR methylated H3K4, thus activating HH transcription (36).
Like its Drosophila counterpart, ALR also appears to participate in nuclear receptor-dependent transcriptional activation. ALR was identified as a component of a stable protein complex formed by the nuclear receptor coactivator ASC-2. Besides ASC-2 and ALR, this complex, termed ASCOM, included the ALR homolog HALR (MLL3), ASH2, RBQ3, and α/β tubulins. ASCOM was shown to bind in a ligand-dependent fashion to the retinoic acid-responsive elements to induce gene expression (13). The subsequently purified estrogen receptor alpha (ERα) coactivator complex was reported to contain MLL2 (ALR), ASH2, RBQ3, and WDR5 proteins. Upon estrogen stimulation, the complex interacted with ERα and was recruited to promoters of ERα target genes (28).
While Drosophila TRR was clearly demonstrated to be an H3K4 methyltransferase (36), such enzymatic activity of the ALR protein remains questionable. Thus, although Goo and colleagues reported very weak H3K4 methylation by ASCOM, this activity was demonstrated only for the SET domain of the ASCOM component HALR, but not for the ALR SET domain (13). Further, Mo and colleagues were unable to show methyltransferase activity of the ALR SET domain or of the ERα coactivator complex, which contains ALR but not HALR (28). Additional unresolved issues include the identification of ALR target genes and elucidation of the biological function(s) of ALR.
Here, we report the purification of a stable multiprotein ALR complex whose composition is similar, but not identical, to those of the ERα-interacting MLL2 complex and ASCOM. The ALR complex exhibited strong H3K4 methyltransferase activity conferred by the ALR SET domain. By generating ALR knockdown cell lines and comparing their expression profiles to that of control cells, we identified a set of genes whose expression is activated by ALR. Some of these genes were identified by chromatin immunoprecipitation (ChIP) as direct ALR targets. We also show evidence that the ALR protein is involved in the regulation of adhesion-related cytoskeletal events and of cell growth.
MATERIALS AND METHODS
Some materials (antibodies [Abs]) and methods (immunoprecipitations, immunofluorescence, time-lapse video microscopy, terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling [TUNEL], cell growth, cell scratch, agar colony growth, and tumorigenicity assays) can be found in the supplemental material.
Biochemical fractionation of nuclear extracts.
All steps of the biochemical fractionation of nuclear extracts were carried out at 4°C. The presence of ALR during all fractionation steps was monitored by Western blotting. Nuclear extracts were obtained from 150 liters of K562 cells by a modified Dignam procedure as previously described (30) and dialyzed against CB buffer (25 mM Tris [pH 7.8], 0.2 mM EDTA, 10% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 2 μg/ml each of leupeptin, aprotinin, and pepstatin A) supplemented with 0.1 M KCl (CB100). An aliquot of 600 mg was loaded on a P11 phosphocellulose column of 100 ml and eluted sequentially with CB buffer supplemented with 0.1, 0.3, 0.5, and 1 M KCl (CB100, CB300, CB500, and CB1000, respectively). The majority of ALR was eluted at 0.3 M KCl, and after dialysis versus CB100, it was loaded onto a 10-ml Q Sepharose column and eluted with a gradient of 0.1 to 1 M KCl. The majority of ALR was eluted from Q Sepharose at 0.25 to 0.35 M KCl. The ALR peak fractions were pooled, concentrated with a BioMax centrifugal device (Millipore) to a volume of 3 ml, loaded onto a preparative Superose 6 column (270-ml bed volume), and eluted subsequent to the void volume in 4-ml fractions. The majority of ALR was eluted from the Superose 6 column at a volume of 100 to 124 ml (corresponding to fractions 25 to 30). The enriched ALR complex was further concentrated with a BioMax centrifugal device and subjected to immunoaffinity purification.
Immunoaffinity purification of the ALR complex and mass spectrometry.
The immunoaffinity purification procedure was performed as described previously (30). Five hundred micrograms of control rabbit anti-glutathione S-transferase (GST) or affinity-purified anti-ALR (αG or α492), anti-PTIP, or anti-UTX antibodies were cross-linked to 0.5 ml protein G-Sepharose 4 Fast Flow (Pharmacia Biotech) in the presence of 50 mM dimethyl pimelimidate (Pierce), blocked overnight in 0.2 M ethanolamine (pH 8.2), and pretreated with 0.1 M glycine (pH 2.8). An aliquot of enriched ALR preparation derived from 300 mg of K562 nuclear extract was used for each immunoaffinity column. Complex binding was done in CB150 containing 0.2% Tween 20 and was carried out at 4°C for 8 h on a rotation wheel. The beads were then extensively washed with CB500 containing 0.2% Tween 20 and transferred into a Poly-prep column (Bio-Rad) in CB100 containing 0.2% Tween 20. Bound proteins were eluted at room temperature with 0.1 M glycine (pH 2.8) and immediately neutralized with 1/10 volume of 1 M Tris (pH 9.0). The affinity-purified ALR complex was then adsorbed on Strataclean beads (Stratagene), eluted by the addition of 1× Laemmli sample buffer, boiled, and resolved on 4% to 15% linear-gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The protein bands were visualized by staining with Gelcode blue stain reagent (Bio-Rad), excised from the gel, and sent for mass spectrometry analysis, performed at the Smoler Protein Research Center, Technion, Israel.
Histone methylation assays.
ALR complex and ALR SET polypeptides were immunoprecipitated as described in the supplemental material. The immunoprecipitates were then washed four times with CB150 buffer supplemented with 0.5% NP-40 and one time with histone methyltransferase (HMTase) buffer (25 mM Tris [pH 8.0] 75 mM NaCl, 0.1 mM EDTA, 1 mM dithiothreitol, and 10% glycerol). The methyltransferase assay was performed essentially as described previously (30). The reactions were carried out in HMTase buffer at a final volume of 20 μl. Each reaction mixture contained 2 μl of S-adenosyl-l-[methyl-3H]methionine (Amersham Biosciences; approximately 1 μM final concentration) and 2 μg of recombinant histone H3 (Upstate Biotechnology) or 5 μg of synthetic peptides corresponding to the first 21 N-terminal histone H3 residues with or without dimethylation on K4 and/or K9 (Upstate Biotechnology). The qualities and methylation statuses of the peptides were ascertained by mass spectrometry. Reaction mixtures were incubated for 2 h at room temperature. To stop the reaction, Laemmli sample buffer was added to the samples to 1× concentration. Following 5 min of boiling, the samples were fractionated on 15% SDS-PAGE (when the recombinant H3 was used as a substrate) or on 20% SDS-PAGE (when the H3 peptides were used) and stained with Gelcode blue stain reagent. The gel was then amplified for 30 min (Amplify; Amersham Biosciences), dried, and exposed to film.
Retroviral shRNA expression constructs and retroviral infection.
For the expression of short hairpin RNAs (shRNAs) targeting ALR, we used the pLPCXL-GFP retroviral siRNA expression vector kindly provided by E. Feinstein (QBI, Kiryat Weizmann, Rehovot, Israel). This vector is a relative of the pLPCX retroviral vector, which contains within the U3 region of the right long terminal repeat an H1 RNA promoter to drive small interfering RNA expression, as well as a cytomegalovirus promoter-driven green fluorescent protein marker for tracking transfection efficiency and viral infection. Five 19-bp ALR sequences were used for generation of the vector-based shRNA constructs. Two ALR sequences, 5′-CCTAGCAGAAACCCAGAAG and 5′-GCAGTTTGTGCACTCCAAG, were used for the generation of constructs termed shALR D (the control construct) and shALR A10, respectively. Three additional ALR sequences used for the generation of constructs termed shALR SP1, shALR SP3, and shALR SP4 were obtained from Dharmacon. For retroviral infection, amphotropic Phoenix packaging cells were transfected with 20 μg DNA of the appropriate retroviral constructs by a standard calcium phosphate procedure. Transfection efficiency was measured by green fluorescent protein fluorescence and was estimated at ≥90%. Culture supernatants were collected 24 h after transfection and filtered. HeLa and A549 cells were infected with the filtered viral supernatants in the presence of 4 μg/ml Polybrene (Sigma). Fresh viral suspensions were added to the cells every 8 h during the subsequent 48 h. Infected cells were selected in a growth medium containing 1 μg/ml puromycin for 7 days.
DNA microarray analysis.
Affymetrix microarray analysis of the ALR knockdown and of the control HeLa cells was performed at the Microarray Unit of the Weizmann Institute of Science, Rehovot, Israel. Total RNAs were extracted from subconfluent proliferating cells by using Tri reagent (Sigma). Ten-microgram aliquots of total RNA were used to prepare biotin-labeled cRNA according to the Affymetrix (Santa Clara, CA) protocol. The labeled cRNA was fragmented and subsequently hybridized to human HG-U133A oligonucleotide arrays corresponding to 22,000 probe sets for known human genes and expressed sequence tags (ESTs) (Affymetrix). The arrays were scanned, and the expression value for each gene was calculated by using Affymetrix software. These raw expression data were rescaled to compensate for variations in hybridization intensity between arrays. The filtering criteria are described in Results below.
Semiquantitative reverse transcription (RT)-PCR and ChIP.
First-strand cDNA template was generated from 2 μg of total RNA in a final volume of 20 μl using oligo(dT)15 primer (Promega) and Superscript II reverse transcriptase (Invitrogen). PCRs were performed in a total volume of 50 μl containing 1 μl of cDNA template, 2 μM (each) specific primers, and 25 μl of Ready Mix PCR master mix (BioLab). PCR cycle numbers were individually optimized for each gene so that each reaction fell into the linear range of product amplification. PCR conditions and primer sequences are available upon request.
ChIP assays were performed as previously described (30). An equivalent of 1 × 107 to 1.5 × 107 cells were used for each ChIP. Antibodies against ALR (αG and α492), PTIP, and UTX, as well as normal rabbit immunoglobulin G (IgG), were used at 5 μg per reaction. Anti-trimethylated H3K4 Ab was used at 2 μg per reaction. For PCR, 1 to 3 μl out of 40 μl DNA extracts was used. To allow comparison among primer sets, unprecipitated input samples were serially diluted and used as a standard for all PCRs. Each PCR mixture contained 1 to 3 μl DNA template, 2 μM of each primer (the primer sequences are available upon request), and 25 μl of Ready Mix PCR master mix (BioLab) in a total volume of 50 μl. After 36 cycles of amplification, the PCR products were separated on a 1.5% agarose gel and identified by ethidium bromide staining.
RESULTS
Purification and characterization of the ALR complex.
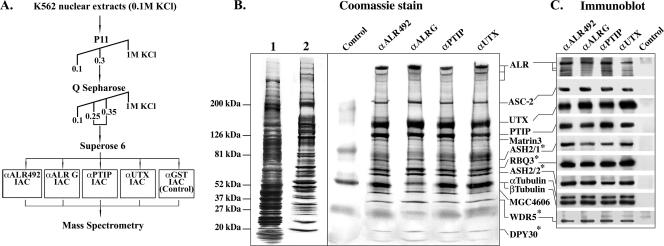
For purification of the ALR complex, a combination of conventional column chromatography and immunoaffinity chromatography was used (Fig. 1A). Nuclear extracts prepared from the K562 cells were subjected to two purification steps on Phosphocellulose P11 and Q Sepharose columns, followed by size fractionation on a preparative Superose 6 column. This enriched preparation was divided and applied to two immunoaffinity columns composed of rabbit polyclonal antibodies directed against ALR residues 3020 to 3192 and 4121 to 4284 (anti-ALR G and anti-ALR 492, respectively). An anti-GST immunoaffinity column was used as a control. The eluted proteins were electrophoretically separated, visualized by Coomassie blue staining, and subsequently subjected to mass spectrometry analysis. As seen in Fig. 1B, the ALR complex eluted from the immunoaffinity columns was about 6,000-fold purified over nuclear extract. Twelve proteins were unambiguously identified by mass spectrometry analysis (listed alongside the Coomassie blue-stained polyacrylamide gel in Fig. 1B). Six of them (ALR, ASC-2, ASH2, RBQ3 [RbBP5], and tubulins α and β) were previously reported to constitute the activation signal cointegrator (ASC-2) complex, ASCOM (13). The only ASCOM protein that was not detected in the ALR complex was HALR. However, this is consistent with the results of a very recent study demonstrating that ASCOM represents a pool of similar complexes, each containing only a single MLL group protein (either ALR or HALR) (22). This revised version of ASCOM was now shown to contain the WDR5 protein, also present in our complex.
FIG. 1.
Purification of the ALR complex and identification of ALR-associated proteins. (A) Scheme for four-step purification of the ALR complex. IAC, immunoaffinity column. (B) Coomassie staining of proteins eluted from anti-ALR, anti-PTIP, anti-UTX, and control immunoaffinity columns. About 50 μg of affinity-purified complex was eluted from each IAC (except for the control column); 98% of this amount was used for Coomassie staining and subsequent mass spectrometry analysis. For comparison of protein pattern complexities before, during, and after complex purification, Coomassie staining patterns of 50 μg of crude K562 nuclear extract (lane 1) and 50 μg of the ALR-enriched Superose 6 pooled fractions (lane 2) are shown, corresponding to 1/6,000 and 1/60, respectively, of the total material used for complex purification and subjected to IAC. The positions of molecular mass markers are indicated on the left. *, components of SET1-like histone methyltransferase complexes. (C) The identities of ALR complex components were verified by immunoblotting.
Additional proteins that were recognized in our complex included UTX (GenBank accession no. 108639430), PTIP (accession no. 27481075), matrin 3 (accession no. 21626466), hypothetical protein MGC 4606 (accession no. 13375654), and hDPY30 (the human homolog of the Caenorhabditis elegans protein DPY-30, accession no. 14211889).
To ascertain the authenticity of the identified proteins as genuine constituents of the ALR complex, we generated rabbit polyclonal antibodies directed against the UTX and PTIP proteins and used them for preparation of immunoaffinity columns. The enriched ALR complex was subjected to purification on these two columns, and the eluted proteins were identified by mass spectrometry (Fig. 1B) and immunoblot analysis, using the available antibodies (Fig. 1C). The pattern of proteins eluted from the anti-PTIP and anti-UTX immunoaffinity columns was nearly identical to that obtained from the anti-ALR columns; moreover, all columns bound the same proteins, supporting our contention that these proteins are integral components of the ALR complex.
Further evidence in support of the observed composition of the ALR complex was obtained by size-dependent gel filtration chromatography. The distribution of the ALR-associated proteins (subsequent to enrichment of the ALR complex on P11 and Q Sepharose columns) across a Superose 6 elution profile was assayed by immunoblotting. The ALR protein was distributed from the void to fraction 31, peaking in fractions 25 to 29. All the ALR complex components tested were identified in these fractions as well (Fig. 2A). This finding reinforces the results of immunoaffinity analysis. Further, while ASH2, RBQ3, WDR5, and α/β tubulins were also detected in other (ALR-negative) Superose 6 fractions, implying their association with additional protein complexes, three proteins—ASC-2, PTIP, and UTX—comigrated exclusively with the ALR peak, suggesting that, at least in the ALR-enriched preparation, these proteins are present mainly within the ALR complex. Similar stoichiometries of the ALR complex components precipitated by anti-ALR, -PTIP, and -UTX immunoaffinity columns (Fig. 1B and C) further support this suggestion.
FIG. 2.
Confirmation of ALR complex composition. (A) Size fractionation of the enriched ALR complex. The ALR-enriched K562 nuclear extracts, obtained from subsequent purification steps on P11 and Q-Sepharose columns, were applied to a preparative Superose 6 column. One percent of each fraction collected from Superose 6 was analyzed by immunoblotting using Abs directed to various ALR complex components. fr#, fraction number. The positions of molecular mass markers determined in a parallel run are indicated at the top. (B) Coprecipitation of ALR with other ALR complex components from A549 cells. Crude nuclear extracts obtained from A549 cells were utilized in reciprocal coimmunoprecipitations. Proteins coprecipitated (IP) with anti-ALR, anti-PTIP, anti-UTX, and anti-RBQ3 Abs, as well as with control IgG, were separated on SDS-PAGE and immunoblotted (IB) with the indicated Abs. Since different amounts of Abs were used in some IPs, the analysis is qualitative rather than quantitative.
To confirm that components of the ALR complex purified from the K562 cells are also associated with ALR in other cell types, reciprocal coimmunoprecipitation experiments using A549 nuclear extracts were performed. As seen in Fig. 2B, anti-ALR, anti-PTIP, anti-UTX, and anti-RBQ3 antibodies, but not the control rabbit IgG, specifically coprecipitated all of the complex components tested. Similar results were obtained with nuclear extracts prepared from 293T, ML2, U2OS, and HeLa cells (it should be noted that the amount of the UTX protein present in HeLa cells was lower than in other cell types tested).
Taken together, these results demonstrate that ALR-interacting proteins identified initially by mass spectrometry are genuine constituents of the ALR complex. From its composition, the ALR complex appears to be a classical SET1-like H3K4 methyltransferase complex. Such complexes contain a subset of proteins (they include ASH2, RBQ3, WDR5, and sometimes hDPY30) homologous to the components of the S. cerevisiae Set1 complex but with a different Set1 homolog. It should be emphasized that the composition of the ALR complex resembles, but is not identical to, that of ASCOM. Thus, it is possible that ASCOM is a variant of the ALR complex.
H3K4 methyltransferase activity of the ALR complex.
We further found that, similar to other SET1-like complexes, the ALR complex possesses HMTase activity. As is apparent from Fig. 3A, the ALR complex, precipitated from K562 nuclear extracts with anti-ALR, anti-PTIP, or anti-UTX antibodies, specifically methylated histone H3 within core histones. To confirm that this enzymatic activity was conferred by the SET domain of ALR, recombinant ALR SET domain polypeptides were used. Since the polypeptides expressed in bacterial cells were insoluble, the ALR SET domain was expressed in mammalian cells. Two GST-fused ALR SET domain polypeptides of different lengths overexpressed in 293T cells were immunoprecipitated from the cell lysates with anti-GST Ab and incubated with synthetic H3 peptides in the presence of the methyl donor H3-SAM. The H3 peptides used in the reaction were 21-residue-long H3 N-terminal tails containing unmodified K4 and K9, dimethylated K4, dimethylated K9, or trimethylated K9. Both the longer and the shorter versions of the ALR SET-GST fusions (designated SET a and SET b, respectively), but not the control GST protein, exhibited HMTase activity (Fig. 3B). Since the ALR SET polypeptides methylated only H3 substrates in which K4 was unmodified, our conclusion was that ALR, like other members of the SET1 family, methylates the K4 residue on histone H3. (It should be noted that the absence of signal when the H3K4-dimethylated peptide is used as a substrate [Fig. 3B] excludes unmodified K9 as a target for ALR SET. We have not ruled out the possibility that mono- and dimethyl H3K9 may serve as substrates for the SET domain of ALR. However, the latter possibility is contradicted by the similar signal intensities obtained with the H3 peptides containing di- or trimethylated K9 [Fig. 3B]).
FIG. 3.
Histone methyltransferase activities of the ALR complex and of the ALR SET domain. (A) The ALR complex methylates histone H3. Anti-ALR, anti-PTIP, anti-UTX, or control immunoprecipitate (IP) was incubated with core histones and the methyl donor H3-SAM. Samples were resolved on 15% SDS-PAGE, stained with Coomassie blue (right), amplified, dried, and fluorographed (left). Histone subunits are indicated on the right. (B) ALR SET domain methylates H3K4. Two ALR SET domain-containing fragments, encompassing the ALR amino acids 5009 to 5261 and 5089 to 5261 (designated SET a and SET b, respectively), were fused to GST and overexpressed in 293T cells. Cells overexpressing GST only were used as a control. The overexpressed polypeptides were immunoprecipitated from cell lysates using anti-GST Ab and incubated with N-terminal H3 peptides containing unmodified K4 and K9, dimethylated K4, dimethylated K9, or trimethylated K9. Samples were resolved on 20% SDS-PAGE, stained with Coomassie blue (bottom), and fluorographed (top). (C) The recombinant ALR SET domain does not interact with the ASH2, RBQ3, and WDR5 proteins. GST-SET fusions or control GST was precipitated from 293T cell lysates with anti-GST Ab and analyzed by immunoblotting using anti-GST, -ASH2, -RBQ3, or -WDR5 Abs. Five percent of each GST-SET- or control GST-overexpressing lysate was loaded as an input control. (D) The ALR complex methylates H3 peptides containing either unmodified or dimethylated K4. Anti-ALR, anti-PTIP, anti-UTX, or control immunoprecipitate was incubated with unmodified or appropriately methylated H3 peptides. The reaction was analyzed as described above.
Although lysine residues may be mono-, di-, or trimethylated, in vivo trimethyl H3K4 is preferentially associated with promoter and transcribed regions of active genes (34, 35). Recent studies have demonstrated that efficient dimethylation, and especially trimethylation, of H3K4 by SET1-related complexes requires the presence of the complex-associated conserved core components ASH2, RBQ3, and WDR5 (10, 41, 49). This explains previous observations demonstrating that dimethylated H3K4 peptide can be further methylated by endogenous SET1-like complexes, such as hSet1 and MLL complexes (9, 48), but not by recombinant SET domains (26, 30). Our finding that the recombinant ALR SET domain failed to methylate the dimethylated H3K4 peptide is fully consistent with these data. As expected, the GST-ALR SET domain also did not interact with endogenous ASH2, RBQ3, and WDR5 (Fig. 3C).
We next examined whether the endogenous ALR complex, which does contain the components mentioned above, could further methylate the H3 peptide dimethylated on K4. For this, the ALR complex, precipitated from the K562 nuclear extracts with anti-ALR, anti-PTIP, or anti-UTX antibodies, was incubated with nonmethylated or appropriately modified synthetic H3 peptides. In contrast to the ALR SET polypeptides, the endogenous ALR complex was indeed capable of methylating the H3 peptide containing dimethylated K4 (as well as H3 peptides with unmodified K4); the only H3 peptide that failed to be methylated by the ALR complex was trimethylated H3K4 (Fig. 3D). These results further demonstrate the K4 specificity of the ALR complex and imply that H3K4 modification carried out by the complex is trimethylation, similar to the modification carried out by other SET1-related complexes.
Generation and gene expression analysis of cell lines with ALR knockdown.
As previously shown, ALR is involved in nuclear hormone receptor-dependent transcriptional activation (13, 28). Therefore, many ALR target genes are supposedly expressed in a hormone-inducible manner. In this work, we wished to determine whether ALR regulates the transcription of constitutively expressed hormone-independent genes. The expression of ALR in HeLa cells was knocked down by small interfering RNA methodology using an efficient retrovirus-derived gene delivery system. A retroviral vector for short hairpin RNA expression was utilized to make several constructs encoding ALR-targeting shRNAs. Four constructs termed shALR SP1, shALR SP3, shALR SP4, and shALR A10 were used to generate virus stocks for infection of HeLa cells and subsequent generation of four stable ALR shRNA-expressing cell lines (termed SP1, SP3, SP4, and A10). In addition, two control cell lines were generated: one infected with an empty vector and the second infected with an ALR shRNA construct, termed shALR D, impaired in elimination of the ALR protein (these control cell lines were designated Vector and D, respectively).
Immunoblotting (Fig. 4A) and immunostaining (see Fig. S1 in the supplemental material) analyses indicated that the abundance of the ALR protein was significantly reduced in all cell lines expressing the ALR shRNA constructs SP1, -3, and -4 and A10. The abundances of other representative components of the ALR complex were not affected (Fig. 4A).
FIG. 4.

(A) Effects of ALR shRNAs on the level of ALR. Four stable cell lines generated by infection of HeLa cells with retroviruses carrying four different ALR shRNA constructs (lanes shALR SP1 to shALR A10) and two control HeLa cell lines, infected with the retroviral vector or with noncompetent ALR shRNA D, were examined by immunoblotting for expression of the ALR protein and some ALR complex components. (B) Confirmation of the microarray results by RT-PCR analysis. RNAs isolated from the above-described cell lines were used for semiquantitative RT-PCR analysis to monitor the expression of selected genes. PCR cycle numbers were individually optimized for each gene so that each reaction fell into the linear range of product amplification. The genes analyzed included 11 downregulated genes (CSPG4, CRIP1, CRIP2, DKK1, FABP3, ENO3, LOXL1, NT5E, GPR56, LAMB3, and MEOX1), 1 upregulated gene (DPT), and 2 control genes (β-actin and HoxA9).
To identify genes regulated by ALR directly or indirectly, microarray analysis using RNAs isolated from the ALR-underexpressing cell line SP3 and from the control vector-infected cells was carried out. Three independent experiments (i.e., separate RNA isolations and microarray assays) were performed. Genes were considered as up- or downregulated if they met the following criteria: (i) differential expression was at least twofold in all three experiments and (ii) the corresponding P values were significant, as judged by application of an Affymetrix algorithm.
Overall, 74 genes (including ESTs) were downregulated, whereas only 2 were upregulated, in the ALR-deprived cells. These results suggest that generally ALR acts to activate transcription. The 20 genes most strongly downregulated by ALR knockdown are shown in Table 1. The downregulated genes comprised various functional categories (see Table S1 in the supplemental material). Notable among them were groups of genes involved in cell adhesion, in cytoskeleton organization, or in transcriptional regulation. Many of these genes have a role in development (see Table S1 in the supplemental material), suggesting that ALR might be important during embryogenesis.
TABLE 1.
The 20 genes most strongly downregulated in SP3 cellsa
| GenBank accession no. | Gene product symbol and name | Change (fold)b | Function, biological process |
|---|---|---|---|
| U36190 | CRIP2, cysteine-rich protein 2 | −25.9 ± 4.8 | Unknown |
| NM_005576 | LOXL1, lysyl oxidase-like 1 | −22.6 ± 2.4 | Formation and repair of extracellular matrix, aging |
| X96753 | CSPG4/MCSP, chondroitin-sulfate proteoglycan 4 (melanoma associated) | −21.3 ± 2.3 | Cell polarization, spreading, motility and proliferation, tumor invasion |
| AL554008 | GPR56, G protein-coupled receptor 56 | −19.1 ± 2.7 | Cell adhesion, cell-cell signaling, brain development |
| NM_002526 | NT5E, 5′-nucleotidase, ecto (CD73) | −17.6 ± 3.5 | Nucleotide metabolism |
| L25541 | LAMB3, laminin beta-3 chain | −17.6 ± 5.1 | Extracellular matrix component; cell adhesion, epidermis development |
| NM_001976 | ENO3, enolase 3 (beta) | −17.2 ± 0.6 | Glycolisis, muscle development and regeneration |
| NM_012242 | DKK1, Dickkopf (Xenopus) homolog 1 | −16.9 ± 3.1 | Signal transducer; development (extracellular Wnt receptor signaling antagonist) |
| X56549 | FABP3, fatty acid binding protein 3 | −15.0 ± 2.3 | Lipid transporter; regulation of cell proliferation |
| NM_004527 | MEOX 1, mesenchyme homeobox 1 | −12.7 ± 2.5 | Transcription factor; development (mesoderm induction and regional specification) |
| NM_001311 | Cystine-rich protein 1 (intestinal) | −11.6 ± 2.9 | Cell proliferation, antimicrobial humoral response |
| NM_005756 | GPR64, G protein-coupled receptor 64 | −11.3 ± 3.0 | Neuropeptide signaling pathway, spermatogenesis |
| AF020769 | TNNC1, troponin C type 1 (cardiac) | −9.4 ± 1.8 | Regulation of muscle contraction, muscle development |
| X79857 | TNNT2, troponin T type 2 | −9.1 ± 2.5 | Regulation of muscle contraction, muscle development |
| NM_000638 | VTN, vitronectin (serum spreading factor, complement S protein) | −9.1 ± 2.0 | Cell adhesion, spreading, immune response |
| NM_002589 | PCDH7, BH-protocadherin (brain, heart) | −7.7 ± 1.3 | Homophilic cell adhesion, regulation of neuronal connections |
| NM_000908 | NPR3, natriuretic peptide receptor C, guanylate cyclase C | −6.9 ± 1.7 | Skeletal development |
| M64930 | PPP2R2B, protein phosphatase 2, regulatory subunit B (PR 52), beta isoform | −5.9 ± 1.7 | Signal transduction |
| NM_005585 | MADH6, MAD (mother against decapentapledgic) Drosophila homolog 6, SMAD6 | −5.5 ± 2.2 | Intracellular antagonist of transforming growth factor β/BMP receptor signaling, regulation of transcription |
| NM_001449 | FHL1, four and a half LIM domains 1 protein | −5.0 ± 1.9 | Muscle development and differentiation, cell adhesion (effector of integrin signaling), cell growth |
Not including ESTs. Two genes overexpressed in SP3 cells are those encoding DPT, dermatopontin (accession number NM_001937), and ABCA5, ATP-binding cassette, subfamily A (ABC1), member 5 (accession number AY028897), giving 8.00- ± 3.57- and 3.20- ± 1.12-fold change, respectively.
Mean ± SE of three independent experiments.
A semiquantitative RT-PCR analysis of the top 11 downregulated genes and of the upregulated DPT gene further confirmed the changes in expression level of these genes in SP3 cells, as well as in other ALR-deficient cell lines (SP1, SP4, and A10), in comparison to the control cells (Fig. 4B). The abundances of control transcripts, such as β-actin or HoxA9, were not affected by ALR knockdown.
Chromatin immunoprecipitation analysis reveals genes directly regulated by ALR.
Genes whose expression is downregulated by ALR knockdown represent both direct and indirect ALR targets. To identify genes directly regulated by ALR, we focused on the 10 most strongly downregulated genes and examined the association of the ALR complex with these genes by ChIP analysis. Chromatin from SP3 and control cells was cross-linked, sheared to DNA fragments of sizes averaging several hundred nucleotides, and immunoprecipitated with two anti-ALR antibodies (αG and α492), as well as with antibodies directed against two additional complex components: UTX and PTIP. Protein G-purified normal rabbit IgG was used as a negative control. Also included in the analysis were antibodies against trimethylated H3K4. Following immunoprecipitation, sets of primers bordering DNA segments of 200 to 300 nucleotides, positioned upstream from or spanning the transcription start sites of the genes of interest, were amplified in PCR.
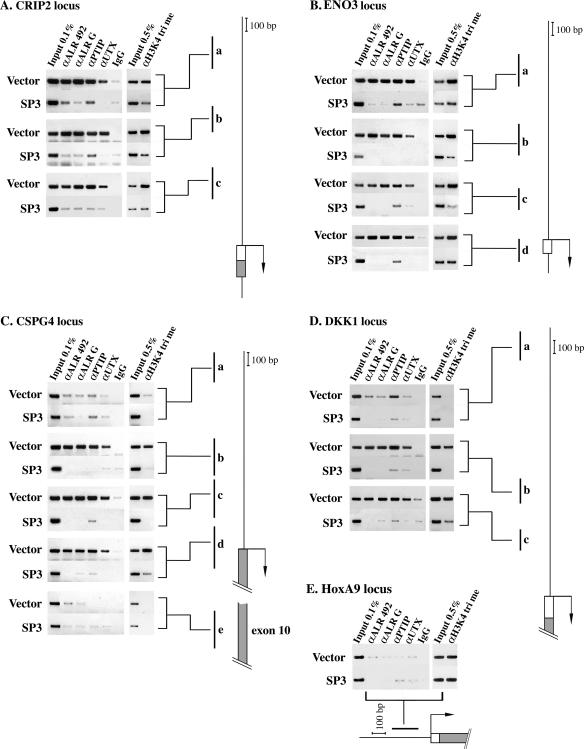
The most clear-cut and reproducible results were obtained in analysis of the cysteine-rich protein 2 (CRIP2), melanoma-associated chondroitin sulfate proteoglycan 4 (CSPG4), enolase 3 (ENO3), and Drosophila Dickkopf homolog 1 (DKK1) genes. As seen in Fig. 5A to D, in the vector-infected ALR-positive cell line, the ALR, PTIP, and UTX proteins were consistently bound to DNA regions adjacent to the transcription initiation sites (regions a and c on the CRIP2 and ENO3 loci [Fig. 5A and B, respectively] and regions b and c on the CSPG4 and DKK1 loci [Fig. 5C and D, respectively]). Examination of regions encompassing the beginnings of the open reading frames of the ENO3 and CSPG4 genes (regions d in Fig. 5B and C, respectively) indicated that ALR, PTIP, and UTX occupied these domains, as well. Taken together, our results demonstrated that the ALR complex is bound to the promoter and to the first exon of the CRIP2, CSPG4, ENO3, and DKK1 genes. H3K4 trimethylation, generally associated with transcriptional activation, was detected in these regions, as well.
FIG. 5.
ChIP analysis of CRIP2 (A), ENO3 (B), CSPG4 (C), DKK1 (D), and HoxA9 (E) loci in vector-infected and SP3 cells. The presence of the ALR, PTIP, and UTX proteins, as well as of histone H3K4 trimethylation, on several regions of CRIP2, ENO3, CSPG4, and DKK1 loci and on the 5′ end of the HoxA9 locus was examined. Chromatin was prepared from the control vector-infected cells and from the ALR-deprived SP3 cell line. The analyzed portions of the genes are depicted to the right of panels A to D and at the bottom of panel E. The boxes shown on the schemes indicate the first exons, and the dark areas correspond to the coding regions. The arrows point to the transcription initiation sites; a to e correspond to the sequences amplified by PCR following immunoprecipitation with each of the six Abs (two anti-ALR Abs, anti-PTIP, anti-UTX, control IgG, and anti-trimethylated H3K4). For input, 0.1% or 0.5% of the amount of sonicated chromatin to be subsequently processed for ChIP was removed and PCR amplified.
Both H3K4 trimethylation and ALR binding were strongly reduced in the ALR-deficient SP3 cells (Fig. 5A to D, lanes SP3). In parallel, the PTIP and UTX proteins were largely eliminated from the four genes examined in SP3 cells. This further supported our contention that ALR binds to the target genes as a complex.
To demonstrate that the binding of the ALR complex is gene/promoter specific, ChIP analysis was also performed on the promoter of the HoxA9 gene, which, as suggested from the microarray and RT-PCR data, is not regulated by ALR. The association of the ALR complex with the HoxA9 promoter was hardly detected in both vector-infected and SP3 cells. In addition, the level of promoter H3K4 methylation of this actively transcribed gene was not affected by ALR knockdown (Fig. 5E).
In summary, we found the following. (i) The CRIP2, CSPG4, ENO3, and DKK1 genes are direct targets of the ALR complex. (ii) The complex is associated with upstream regions adjacent to the transcription start sites (probably promoter regions) of these genes and with the first exons of two of the genes examined. We note that the ChIP analysis was not extended into the downstream transcribed regions, which might also be occupied by the ALR complex. (iii) The recruitment of the complex to target genes is ALR-dependent. (iv) H3K4 trimethylation parallels ALR occupancy.
ALR-deficient cells exhibit different modes and dynamics of spreading.
As noted above, a large group of genes, whose transcription was downregulated in ALR knockdown cells (according to the DNA microarray data), comprised genes involved in adhesion-related processes, especially in the regulation and organization of the actin cytoskeleton (see Table S1 in the supplemental material).
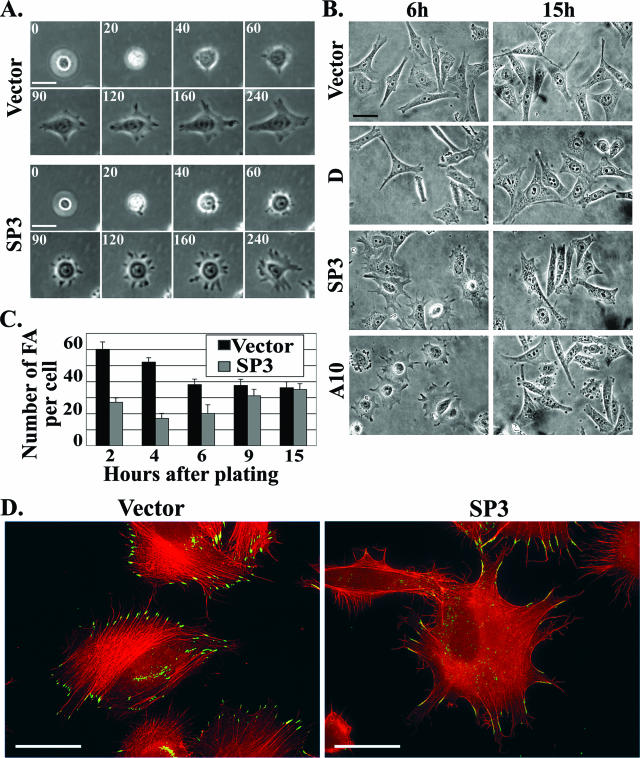
Examination of cell shape and immunofluorescence analysis of focal adhesions and actin cytoskeleton in fully spread control and ALR-depleted cells did not reveal prominent morphological differences (Fig. 6B, right, and data not shown). Therefore, we decided to examine whether ALR deficiency could affect cell morphology during spreading. To this end, we used time-lapse photography of spreading ALR-deprived and control cells during the 4 h subsequent to replating (Fig. 6A). Alternatively, the cells were seeded on fibronectin-coated coverslips and fixed at different time points during the subsequent 15 h. Representative photographs of the control and ALR-deficient cell lines at the 6-h and 15-h time points are shown in Fig. 6B. The video recording of cells revealed that the initial phases of adhesion were not notably affected by the ALR knockdown: after being plated, about 20 min was required for most of the cells to adhere, and about 40 to 50 min was required for the cells to spread out and become nonrefractile (Fig. 6A). However, at later stages, significant morphological differences were observed. At 1.5 to 2 h after being plated, the control cells became polarized and started to assume the rhomboidal shape characteristic of the fully spread HeLa cells. At the same time, the ALR-deficient cells were still radial, with multiple large protrusions, which are seldom seen in the control cells. Time-lapse video recording revealed that these protrusions were highly dynamic, continuously disappearing and reforming. Furthermore, whereas most of the control cells acquired the characteristic rhomboidal shape at 3 to 4 h after being plated (Fig. 6A), the ALR-deficient cells were multipolar even after 6 h (Fig. 6B, left). At later stages, these morphological differences gradually disappeared, and at 13 to 15 h after being plated, both the control and ALR-depleted cells had the typical bi- or tripolar shape (Fig. 6B, right).
FIG. 6.
Changes in the morphology of ALR knockdown cells during spreading. (A and B) Differences in dynamics and modes of cell spreading. (A) Control (Vector) and ALR-deficient (SP3) cells were plated on fibronectin-coated dishes and analyzed by time-lapse photography during the subsequent 4 h. The numbers represent the time after plating in min. (B) Two control (Vector and D) and two ALR-underexpressing (SP3 and A10) cell lines were seeded on fibronectin-coated coverslips and fixed with 3.7% paraformaldehyde at different time points during the subsequent 15 h. Typical photographs of the cells at 6 and 15 h after being plated are shown. (C and D) Decreased numbers of FA and alterations in actin cytoskeleton organization in spreading ALR-deficient cells. (C) Control (Vector) and ALR knockdown (SP3) cells were processed as described for panel B, and FA were double stained with anti-vinculin and anti-paxillin Abs. The numbers of FA formed by each cell type at the indicated time points were calculated. The data represent the average FA number of 30 cells plus standard deviation. (D) The cells described above, fixed at 3 h after being plated, were stained for filamentous actin with tetramethyl rhodamine isothiocyanate-phalloidin (red) and for FA with anti-paxillin Ab (green). Bars (A, B, and D), 50 μm.
Next, we examined the properties of focal adhesion contacts (FA) in the cells with ALR knockdown. Double staining of FA with anti-paxillin and anti-vinculin antibodies revealed that in the spreading ALR-deficient cells, the number of FA per cell was significantly reduced in comparison to the control cells. This difference was particularly significant at 2 and 4 h after plating (correlating with the difference in cell shape and the percentage of cells containing long protrusions) and disappeared as the cells assumed the typical polarized shape (Fig. 6C). Staining with anti-phosphotyrosine antibodies revealed that the levels of tyrosine phosphorylation at FA sites were similar in both cell types (data not shown), suggesting that signaling events leading to FA tyrosine phosphorylation were not affected by ALR knockdown. We also found that ALR knockdown dramatically altered the organization of the actin cytoskeleton during cell spreading. Whereas in control cells the actin filaments formed well-organized FA-originated stress fibers, ALR-deficient cells had fewer stress fibers (consistent with reduced FA numbers). Instead, these cells formed an intense meshwork of cortical actin, reflective of the more dynamic cell anchoring and delayed polarity acquisition (Fig. 6D). Taken together, our results suggest that ALR deficiency affects cytoskeleton organization during cell spreading and polarization.
ALR knockdown impairs cell growth in vitro and decreases cell tumorigenicity in vivo.
To further assess the consequences of ALR deficiency, we examined whether ALR knockdown affects cell growth kinetics in vitro. The growth rate was measured in six-well plates for up to 4 days. As seen in Fig. 7A, depletion of ALR resulted in a substantial decrease in cell growth compared to the control HeLa cells. Examination of the cell cycle profile did not reveal prominent differences between the control and ALR-deficient cells, except for some increase in the sub-G1 population in the ALR knockdown cells, indicative of higher cell death (see Fig. S2 in the supplemental material); this cell death appeared to be apoptotic, as assessed by nuclear condensation (not shown) and TUNEL assay (Fig. 7B). Increased intrinsic apoptosis might contribute to the slower growth of the ALR-deprived cells.
FIG. 7.
ALR knockdown affects cell growth, apoptosis, and migration. (A) Cell growth assay. The growth rates of three ALR-deprived HeLa cell lines (SP3, SP4, and A10) and of the control cells (Vector and D) were determined by plating 105 cells in triplicate on six-well plates and counting the cells every 24 h during the subsequent 4 days. The error bars represent the standard deviations of three independent experiments. (B) Increased apoptosis in the ALR-deprived cells. Control (Vector and D) and ALR-deprived (SP1, SP3, SP4, and A10) HeLa cells were stained with the TUNEL assay kit. Percentages of TUNEL-positive (apoptotic) populations are shown. The error bars represent the standard deviations of three independent experiments. (C) ALR knockdown inhibits soft-agar growth of HeLa cells. Five hundred and 1,000 cells/3-cm dish were seeded on 0.3% agar in triplicate. Three weeks later, the dishes were photographed and the colonies were counted. The experiment was repeated twice. Typical photographs of plates seeded with control (Vector) and ALR-deficient (SP3) cells are shown on the left. The histogram shows the numbers of colonies formed in each cell line at both plating concentrations as a percentage of the control vector-expressing cells. Absolute colony numbers are shown above the bars. (D) Effect of ALR knockdown on cell migration. A scratch injury was introduced with a micropipette tip into confluent cultures of the control (Vector) and ALR-depleted (SP3) A549 cells. Phase-contrast images of cultures were taken either immediately after scratching (0 h) or 16 h later.
We next evaluated the effect of ALR underexpression on the ability of cells to form colonies on plastic supports. The cells were seeded on 10-cm plates. Two weeks later, the formed colonies were stained and counted. Cells with ALR knockdown produced 35 to 40% fewer colonies than the controls. The ALR deficiency also resulted in considerable reduction of colony size: colonies formed by the ALR-underexpressing cells were 2.5- to 3-fold smaller then those formed by the control cells (see Fig. S3 in the supplemental material). Further, we examined the effect of ALR depletion on anchorage-independent colony formation in soft agar. ALR knockdown dramatically abrogated anchorage-independent HeLa cell growth, causing reduction in both colony number and size (Fig. 7C).
Next, we tested the effect of ALR deficiency on cell motility/migration. For this, a monolayer scratch assay was performed. Since HeLa cells do not migrate efficiently into the scratched area (our observations) and therefore are not suitable for such an assay, ALR shRNA constructs were transduced by retroviruses into A549 lung carcinoma cells to generate stable ALR-underexpressing cell lines. The growth properties of A549 and HeLa cells with ALR knockdown were quite similar (data not shown). A549 cells were seeded on six-well plates, and a scratch was introduced into a confluent monolayer by the use of a pipette tip. To ensure that differences in cell migration were not due to differences in cell growth, the cells were incubated overnight in serum-free medium prior to scratch injury, and the assay was performed in the presence of mitomycin C to limit cell proliferation. As seen in Fig. 7D, the ALR-deprived cells migrated to the scratched area much less efficiently than the control cells.
Taken together, these results indicate that ALR knockdown affects cell growth and motility in vitro. To determine whether reduced ALR expression also influences the growth characteristics of cells in vivo, a tumorigenicity assay was performed. The ALR-deficient and control HeLa cells were injected subcutaneously into athymic nude mice, and tumor development was monitored. As seen in Fig. 8A, animals injected with ALR-deficient cells showed delayed kinetics of tumor development compared to the control groups. The tumors originating from the cells with ALR knockdown were also much smaller and lighter in weight than the tumors formed in the control groups (Fig. 8A to C). Similar results were obtained in two independent experiments. These data provide strong evidence that ALR knockdown reduces cell growth in vivo.
FIG. 8.
Decreased tumorigenicity of ALR knockdown cells in nude mice. (A) Ten animals in each group were injected subcutaneously with 106 cells of either control (Vector and D) or ALR-deficient (SP3 and A10) HeLa cell lines. Tumor volumes were determined at regular time intervals. The experiment was stopped after 4 weeks, when the tumors in the control groups reached 1 to 1.5 cm in diameter. The results represent the means ± standard errors (SE). (B) Photographs and (C) mean weights of tumors formed in each group at 4 weeks after injection. Error bars, SE.
DISCUSSION
ALR is a H3K4 methyltransferase that forms a conserved SET1-like complex.
Using a biochemical approach, we have established that ALR is present within a multiprotein complex that shares several characteristics with the S. cerevisiae Set1 and human SET1-like HMTase complexes. (i) All these complexes contain enzymes with carboxy-terminal SET domains that are highly conserved and define a specific branch of the larger SET domain protein superfamily of HMTases (8). (ii) All human SET1-like complexes share three core proteins: ASH2 (also termed ASH2L), RBQ3 (RbBP5), and WDR5, related to the components of the yeast Set1 complex (summarized in reference 10). These core proteins appear to constitute a structural platform that facilitates enzyme-substrate interactions and regulates the enzymatic processivity of the SET domain (10, 41, 49). (iii) Some human SET1-like complexes (e.g., the MLL4 complex [16] and the ALR complex [our study]) contain homologs of the C. elegans DPY-30 protein, required for X chromosome dosage compensation (15). The yeast Set1 complex also contains a DPY-30-related protein, Sdc1/Cps25, which plays a role in determining the efficiency of H3K4 trimethylation carried out by Set1 (20, 27).
We further demonstrated that the similarity between the ALR complex and other SET1-related complexes is not only compositional but also functional: the ALR complex shows a robust H3K4 methyltransferase activity on H3 peptides and on recombinant histone H3. Although lysine residues may be mono-, di-, and trimethylated in vivo, usually H3K4 trimethylation (sometimes also dimethylation) is correlated with transcriptional competence in both yeast and mammals (27, 34, 35). In S. cerevisiae, Set1 is the only H3K4 HMTase and can methylate H3K4 to all three levels. In contrast, higher eukaryotes contain several H3K4 HMTases with various specificities, suggesting a more complex picture for the regulation and function of H3K4 methylation (40). Recent studies have demonstrated that at least three proteins, ASH2, RBQ3, and WDR5, which, along with the SET domain-containing catalytic subunit, constitute a functional core of all SET1-related complexes, are essential for efficient dimethylation and especially trimethylation of H3K4 by MLL (10, 41, 49). Our findings that only the entire ALR complex, and not the recombinant ALR SET domain, is capable of methylating the H3 substrates with dimetylated K4 are consistent with these data and provide further experimental support for the importance of the complex context in regulating the enzymatic activities of SET1-related HMTases.
UTX and PTIP, the unique components of the ALR complex.
While sharing some core subunits described above, human SET1-like complexes nevertheless contain unique or partially shared sets of proteins (9, 30, 48, 51). The unique proteins suggest nonoverlapping functions of these complexes. The ALR complex also contains several unique components. Some of these proteins (e.g., α/β tubulins and ASC-2) are associated with the previously purified ASCOM complex (13), whereas others are found only in the ALR complex. Among these unique components, the major proteins are UTX and PTIP. UTX is encoded by a gene present on the X chromosome that escapes X inactivation and has an expressed homolog on the Y chromosome (14). UTX belongs to the jumonji family of transcription and chromatin regulators containing the highly conserved JmjC domain (44). Very recently, members of the jumonji family were found to comprise a novel class of histone demethylases that demethylate mono-, di-, and trimethylated K9 and/or K36 residues on histone H3 in an enzymatic reaction utilizing the JmjC domain-bound catalytic cofactors: 2-oxoglutarate (2OG) and Fe(II) (6, 46, 47, 50). Although it remains to be elucidated whether UTX also possesses such enzymatic activity, it is tempting to speculate that in the ALR complex, UTX and ALR could induce transcriptionally active chromatin conformation by opposite mechanisms: whereas ALR places an activating methylation mark on H3K4, UTX might remove the repressive H3K9 methylation.
PTIP is another component present in the ALR complex, but not in ASCOM. This BRCT domain-containing protein was originally identified on the basis of its interaction with the transcription factor PAX2 in mice (21). Recently, human PTIP was isolated in a screen for proteins capable of interacting with peptides phosphorylated by ATM/ATR protein kinases in response to ionizing radiation (23). PTIP was shown to be recruited to the sites of double-stranded DNA breaks and to be a key component of the DNA damage response (19, 23). Analysis of mouse embryos deficient for PTIP showed extensive unrepaired DNA ends, implicating the protein in DNA repair during replication (4). The presence of PTIP in the ALR complex raised the possibility of the involvement of the latter in DNA damage response and in replication. However, while upon treatment of cells with ionizing radiation, PTIP was recruited to the sites of DNA damage, the ALR protein could not be detected at these sites (see the supplemental material). Similarly, during S phase transition, PTIP, but not ALR, was found to localize to discrete nuclear foci (see the supplemental material), which probably correspond to the sites of DNA breaks arising during DNA replication. These data, in conjunction with the results of biochemical fractionation demonstrating that the elution profile of PTIP on P11 phosphocellulose is broader than that of ALR (see the supplemental data), lead us to suggest that (i) ALR is not involved in the DNA damage response and (ii) PTIP is present in the cell in at least two different cellular pools with nonoverlapping activities: the PTIP pool present in the ALR complex is involved in transcriptional regulation, as was previously shown for mouse PTIP and its Xenopus homolog, Swift (21, 38), whereas another pool of PTIP, not bound to ALR, has a role in the DNA damage response and/or DNA repair.
ALR target genes.
Like its Drosophila homolog, TRR, the ALR protein was reported to participate in nuclear hormone receptor-dependent transcription activation (13, 28, 36). In the present study, we wanted to establish whether ALR is also involved in transcriptional regulation of constitutively expressed genes. By using DNA microarray methodology, we identified a set of genes whose expression was downregulated due to knockdown of endogenous ALR in HeLa cells. These genes include both direct and indirect targets of ALR. In an attempt to identify primary ALR targets, we focused on a small number of genes that were most strongly downregulated by ALR knockdown (genes showing a higher differential in expression are more likely to correspond to direct targets). By applying ChIP methodology, we found that four of these genes are direct targets of ALR. They are CRIP2, ENO3, CSPG4/MSCP, and DKK1. The conclusions we drew from the ChIP experiments were the following. (i) ALR, together with all complex components tested (and presumably with the entire complex), is recruited to the target genes. (ii) The presence of ALR on target genes correlates with H3K4 trimethylation. In conjunction with the demonstration that the ALR complex and the ALR SET domain methylate H3K4 in vitro, the most likely interpretation of these data is that H3K4 trimethylation on target gene loci is executed by ALR. Since none of the ALR complex components appears to contain known sequence-specific DNA binding sites, the mechanism by which the complex is recruited to the regulatory elements of the target genes identified here is unresolved. However, our observation that, following knockdown of ALR, other complex components are not associated with the target genes implies that ALR is instrumental for the binding.
Further studies, such as ChIP-on-chip analysis, are required to identify additional ALR target genes and to characterize more precisely the ALR binding regions in the current subset of targets. Identification of a larger pool of primary ALR targets may also provide a statistically reliable data set to determine whether such genes contain common regulatory elements and to gain insight into the mechanism(s) of the recruitment of the ALR complex to target DNA sequences.
ALR knockdown affects adhesion-related processes and suppresses cell growth.
We demonstrated that reduction in the level of the ALR protein resulted in prolonged dynamics and different modes of cell spreading/polarization, accompanied by decreased FA formation and aberrant actin cytoskeleton organization. The migration capacity of ALR knockdown cells was reduced as well. It is likely that these changes were due to decreased expression of ALR-dependent genes involved in cell adhesion and motility, adhesion-dependent signal transduction, and actin-filament rearrangements (according to the microarray expression data). Some of the genes we identified are associated with malignant transformation and are often overexpressed in highly aggressive metastatic cancers. Examples include the extracellulr matrix (ECM)-degrading enzyme heparanase (39), the urokinase-type plasminogen activator uPA/PLAU (7), adhesion molecules (BH-protocadherin [3], L1CAM, and ALCAM [43]), the cytoskeletal protein keratin 17 (3, 5), the F-actin binding protein transgelin (32, 37), and the melanoma-associated integrin coreceptor CSPG4/MSCP (1). Consistent with decreased expression profiles of these and several other cancer-related genes in the ALR-underexpressing cells, we found that the growth characteristics of these cells were significantly altered in comparison to their wild-type counterparts. Thus, ALR knockdown cells demonstrated reduced growth kinetics, higher levels of intrinsic apoptosis, and, most importantly, impaired anchorage-independent growth in vitro. Moreover, the in vivo growth of these cells was compromised, as well. These differences might be due to alterations in cell adhesion and motility caused by ALR knockdown.
Supplementary Material
Acknowledgments
We are very grateful to A. Bershadsky for thoughtful and enlightening suggestions and to B. Geiger for valuable advice. We thank W. Herr, J. W. Lee, and G. R. Dressler for generous gifts of anti-WDR5, anti-ASC-2, and chicken anti-PTIP Abs, respectively. We also thank A. Greenfield and R. Romero for their kind help in providing us with anti-UTX antisera for the initial experiments.
This work was supported by grants from the U.S.-Israel Binational Science Foundation, the Israel Cancer Research Fund, the Israel Science Foundation, and the National Institutes of Health.
Footnotes
Published ahead of print on 18 December 2006.
Supplemental material for this article may be found at http://mcb.asm.org.
REFERENCES
- 1.Campoli, M. R., C. C. Chang, T. Kageshita, X. Wang, J. B. McCarthy, and S. Ferrone. 2004. Human high molecular weight-melanoma-associated antigen (HMW-MAA): a melanoma cell surface chondroitin sulfate proteoglycan (MSCP) with biological and clinical significance. Crit. Rev. Immunol. 24:267-296. [DOI] [PubMed] [Google Scholar]
- 2.Canaani, E., T. Nakamura, T. Rozovskaia, S. T. Smith, T. Mori, C. M. Croce, and A. Mazo. 2004. ALL-1/MLL1, a homologue of Drosophila TRITHORAX, modifies chromatin and is directly involved in infant acute leukemia. Br. J. Cancer 90:756-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, J. J., K. Peck, T. M. Hong, S. C. Yang, Y. P. Sher, J. Y. Shih, R. Wu, J. L. Cheng, S. R. Roffler, C. W. Wu, and P. C. Yang. 2001. Global analysis of gene expression in invasion by a lung cancer model. Cancer Res. 61:5223-5230. [PubMed] [Google Scholar]
- 4.Cho, E. A., M. J. Prindle, and G. R. Dressler. 2003. BRCT domain-containing protein PTIP is essential for progression through mitosis. Mol. Cell. Biol. 23:1666-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu, Y. W., P. C. Yang, S. C. Yang, Y. C. Shyu, M. Hendrix, R. Wu, and C. W. Wu. 1997. Selection of invasive and metastatic subpopulations from a human lung adenocarcinoma cell line. Am. J. Respir. Cell Mol. Biol. 17:353-360. [DOI] [PubMed] [Google Scholar]
- 6.Cloos, P. A., J. Christensen, K. Agger, A. Maiolica, J. Rappsilber, T. Antal, K. H. Hansen, and K. Helin. 2006. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 442:307-311. [DOI] [PubMed] [Google Scholar]
- 7.Dano, K., N. Behrendt, G. Hoyer-Hansen, M. Johnsen, L. R. Lund, M. Ploug, and J. Romer. 2005. Plasminogen activation and cancer. Thromb. Haemost. 93:676-681. [DOI] [PubMed] [Google Scholar]
- 8.Dillon, S. C., X. Zhang, R. C. Trievel, and X. Cheng. 2005. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 6:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dou, Y., T. A. Milne, A. J. Tackett, E. R. Smith, A. Fukuda, J. Wysocka, C. D. Allis, B. T. Chait, J. L. Hess, and R. G. Roeder. 2005. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121:873-885. [DOI] [PubMed] [Google Scholar]
- 10.Dou, Y., T. A. Milne, A. J. Ruthenburg, S. Lee, J. W. Lee, G. L. Verdine, C. D. Allis, and R. G. Roeder. 2006. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 13:713-719. [DOI] [PubMed] [Google Scholar]
- 11.Emerling, B. M., J. Bonifas, C. P. Kratz, S. Donovan, B. R. Taylor, E. D. Green, M. M. Le Beau, and K. M. Shannon. 2002. MLL5, a homologue of Drosophila trithorax located within a segment of chromosome band 7q22 implicated in myeloid leukemia. Oncogene 21:4849-4854. [DOI] [PubMed] [Google Scholar]
- 12.Fitzgerald, K. T., and M. O. Diaz. 1999. MLL2: a new mammalian member of the trx/MLL family of genes. Genomics 59:187-192. [DOI] [PubMed] [Google Scholar]
- 13.Goo, Y.-H., Y. C. Sohn, D.-H. Kim, S.-W. Kim, M.-J. Kang, D.-J. Jung, E. Kwak, N. A. Barlev, S. L. Berger, W. T. Chow, R. G. Roeder, D. O. Azorsa, P. S. Meltzer, P.-G. Suh, E. J. Song, K.-J. Lee, Y. C. Lee, and J. W. Lee. 2003. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol. Cell. Biol. 23:140-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenfield, A., L. Carrel, D. Pennis, C. Phillippe, N. Quaderi, P. Siggers, K. Steiner, P. P. L. Tam, T. P. Monaco, H. F. Willard, and P. Koopman. 1998. The UTX gene escapes X inactivation in mice and humans. Hum. Mol. Genet. 7:737-742. [DOI] [PubMed] [Google Scholar]
- 15.Hsu, D. R., P. T. Chuang, and B. J. Meyer. 1995. DPY-30, a nuclear protein essential early in embryogenesis for Caenorhabditis elegans dosage compensation. Development 121:3323-3334. [DOI] [PubMed] [Google Scholar]
- 16.Hughes, C. M., O. Rozenblatt-Rosen, T. A. Milne, T. D. Copeland, S. S. Levine, J. C. Lee, D. N. Hayes, K. S. Shanmugam, A. Bhattacharjee, C. A. Biondi, G. F. Kay, N. K. Hayward, L. J. Hess, and M. Meyerson. 2004. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol. Cell 13:587-597. [DOI] [PubMed] [Google Scholar]
- 17.Huntsman, D. G., S.-F. Chin, M. Muleris, S. J. Batley, V. P. Collins, L. M. Wiedemann, S. Aparicio, and C. Caldas. 1999. MLL2, the second human homologue of the Drosophila trithorax gene, maps to 19q13.1 and is amplified in solid tumor cell lines. Oncogene 18:7975-7984. [DOI] [PubMed] [Google Scholar]
- 18.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 19.Jowsey, P. A., A. J. Doherty, and J. Rouse. 2004. Human PTIP facilitates ATM-mediated activation of p53 and promotes cellular resistance to ionizing radiation. J. Biol. Chem. 279:55562-55569. [DOI] [PubMed] [Google Scholar]
- 20.Krogan, N. J., J. Dover, S. Khorrami, J. F. Greenblatt, J. Schneider, M. Johnston, and A. Shilatifard. 2002. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 277:10753-10755. [DOI] [PubMed] [Google Scholar]
- 21.Lechner, M. S., I. Levitan, and G. R. Dressler. 2000. PTIP, a novel BRCT domain-containing protein interacts with Pax2 and is associated with active chromatin. Nucleic Acids Res. 28:2741-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, S., D. K. Lee, Y. Dou, J. Lee, B. Lee, E. Kwak, Y. Y. Kong, S. K. Lee, R. G. Roeder, and J. W. Lee. 2006. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc. Natl. Acad. Sci. USA 103:15392-15397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manke, I. A., D. M. Lowery, A. Nguyen, and M. B. Yaffe. 2003. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302:636-639. [DOI] [PubMed] [Google Scholar]
- 24.Martin, C., and Y. Zhang. 2005. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6:838-849. [DOI] [PubMed] [Google Scholar]
- 25.Miller, T., N. J. Krogan, J. Dover, H. Erdjument-Bromage, P. Tempst, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2001. Compass: a complex of proteins associated with a trithorax-related set domain protein. Proc. Natl. Acad. Sci. USA 98:12902-12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milne, T. A., S. D. Briggs, H. W. Brock, M. E. Martin, D. Gibbs, C. D. Allis, and J. L. Hess. 2002. MLL targets SET domain methyltransferase activity to HOX gene promoters. Mol. Cell 10:1107-1116. [DOI] [PubMed] [Google Scholar]
- 27.Morillon, A., N. Karabetsou, A. Nair, and J. Mellor. 2005. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol. Cell 18:723-734. [DOI] [PubMed] [Google Scholar]
- 28.Mo, R., S. M. Rao, and Y. J. Zhu. 2006. Identification of the MLL2 as a coactivator for estrogen receptor alpha. J. Biol. Chem. 281:15714-15720. [DOI] [PubMed] [Google Scholar]
- 29.Nagy, P. L., J. Griesenbeck, R. D. Kornberg, and M. L. Cleary. 2002. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl. Acad. Sci. USA 99:90-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura, T., T. Mori, S. Tada, W. Krajewski, T. Rozovskaia, R. Wassell, G. Dubois, A. Mazo, C. M. Croce, and E. Canaani. 2002. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell 10:1119-1128. [DOI] [PubMed] [Google Scholar]
- 31.Prasad, R., A. B. Zhadanov, Y. Sedkov, F. Bullrich, T. Druck, R. Rallapalli, T. Yano, H. Alder, C. M. Croce, K. Huebner, A. Mazo, and E. Canaani. 1997. Structure and expression pattern of human ALR, a novel gene with strong homology of ALL-1 involved in acute leukemia and to Drosophila trithorax. Oncogene 15:549-560. [DOI] [PubMed] [Google Scholar]
- 32.Qi, Y., J. F. Chiu, L. Wang, D. L. Kwong, and Q. Y. He. 2005. Comparative proteomic analysis of esophageal squamous cell carcinoma. Proteomics 5:2960-2971. [DOI] [PubMed] [Google Scholar]
- 33.Ruault, M., M. E. Brun, M. Ventura, G. Roizes, and A. De Sario. 2003. MLL3, a new human member of the TRX/MLL gene family, maps to 7q36, a chromosome region frequently deleted in myeloid leukemia. Gene 284:73-81. [DOI] [PubMed] [Google Scholar]
- 34.Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. Emre, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407-411. [DOI] [PubMed] [Google Scholar]
- 35.Schneider, R., A. J. Bannister, F. A. Myers, A. W. Thorne, C. Crane-Robinson, and T. Kouzarides. 2004. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 6:73-77. [DOI] [PubMed] [Google Scholar]
- 36.Sedkov, Y., E. Cho, S. Petruk, L. Cherbas, S. T. Smith, R. S. Jones, P. Cherbas, E. Canaani, J. B. Jaynes, and A. Mazo. 2003. Methylation at lysine 4 of histone H3 in ecdysone-dependnet development of Drosophila. Nature 426:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi, Y. Y., H. C. Wang, Y. H. Yin, W. S. Sun, Y. Li, C. Q. Zhang, Y. Wang, S. Wang, and W. F. Chen. 2005. Identification and analysis of tumour-associated antigens in hepatocellular carcinoma. Br. J. Cancer 92:929-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimizu, K., P. Y. Bourillot, S. J. Nielsen, A. M. Zorn, and J. B. Gurdon. 2001. Swift is a novel BRCT domain coactivator of Smad2 in transforming growth factor beta signaling. Mol. Cell. Biol. 21:3901-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simizu, S., K. Ishida, and H. Osada. 2004. Heparanase as a molecular target of cancer chemotherapy. Cancer Sci. 95:553-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sims, R. J., III, K. Nishioka, and D. Reinberg. 2003. Histone lysine methylation: a signature for chromatin function. Trends Genet. 19:629-639. [DOI] [PubMed] [Google Scholar]
- 41.Steward, M. M., J. S. Lee, A. O'Donovan, M. Wyatt, B. E. Bernstein, and A. Shilatifard. 2006. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat. Struct. Mol. Biol. 13:852-854. [DOI] [PubMed] [Google Scholar]
- 42.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 43.Swart, G. W., P. C. Lunter, J. W. Kilsdonk, and L. C. Kempen. 2005. Activated leukocyte cell adhesion molecule (ALCAM/CD166): signaling at the divide of melanoma cell clustering and cell migration? Cancer Metastasis Rev. 24:223-236. [DOI] [PubMed] [Google Scholar]
- 44.Takeuchi, T., Y. Watanabe, T. Takano-Shimizu, and S. Kondo. 2006. Roles of jumonji and jumonji family genes in chromatin regulation and development. Dev. Dyn. 235:2449-2459. [DOI] [PubMed] [Google Scholar]
- 45.Tan, Y. C., and V. T. K. Chow. 2001. The novel human HALR gene encodes a protein homologues to ALR and to ALL-1 involved in leukemia, and maps to chromosome 7q36 association with leukemia and developmental defects. Cancer Detect. Prev. 25:454-469. [PubMed] [Google Scholar]
- 46.Tsukada, Y., J. Fang, H. Erdjument-Bromage, M. E. Warren, C. H. Borchers, P. Tempst, and Y. Zhang. 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439:811-816. [DOI] [PubMed] [Google Scholar]
- 47.Whetstine, J. R., A. Nottke, F. Lan, M. Huarte, S. Smolikov, Z. Chen, E. Spooner, E. Li, G. Zhang, M. Colaiacovo, and Y. Shi. 2006. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125:467-481. [DOI] [PubMed] [Google Scholar]
- 48.Wysocka, J., M. P. Myers, C. D. Laherty, R. N. Eisenman, and W. Herr. 2003. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17:896-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wysocka, J., T. Swigut, T. A. Milne, Y. Dou, X. Zhang, A. L. Burlingame, R. G. Roeder, A. H. Brivanlou, and C. D. Allis. 2005. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121:859-872. [DOI] [PubMed] [Google Scholar]
- 50.Yamane, K., C. Toumazou, Y. Tsukada, H. Erdjument-Bromage, P. Tempst, J. Wong, and Y. Zhang. 2006. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125:483-495. [DOI] [PubMed] [Google Scholar]
- 51.Yokoyama, A., Z. Wang, J. Wysocka, M. Sanyal, D. J. Aufiero, I. Kitabayashi, W. Herr, and M. L. Cleary. 2004. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell. Biol. 24:5639-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.