Abstract
Erythroid Krüppel-like factor (EKLF) is an essential transcription factor for mammalian β-like globin gene switching, and it specifically activates transcription of the adult β globin gene through binding of its zinc fingers to the promoter. It has been a puzzle that in the mouse, despite its expression throughout the erythroid development, EKLF activates the adult βmaj globin promoter only in erythroid cells beyond the stage of embryonic day 10.5 (E10.5) but not before. We show here that expression of the mouse βmaj globin gene in the aorta-gonad-mesonephros region of E10.5 embryos and in the E14.5 fetal liver is accompanied by predominantly nuclear localization of EKLF. In contrast, EKLF is mainly cytoplasmic in the erythroid cells of E9.5 blood islands in which βmaj is silenced. Remarkably, in a cultured mouse adult erythroleukemic (MEL) cell line, the activation of the βmaj globin gene by dimethyl sulfoxide (DMSO) or hexamethylene-bis-acetamide (HMBA) induction is also paralleled by a shift of the subcellular location of EKLF from the cytoplasm to the nucleus. Blockage of the nuclear import of EKLF in DMSO-induced MEL cells with a nuclear export inhibitor repressed the transcription of the βmaj globin gene. Transient transfection experiments further indicated that the full-sequence context of EKLF was required for the regulation of its subcellular locations in MEL cells during DMSO induction. Finally, in both the E14.5 fetal liver cells and induced MEL cells, the β-like globin locus is colocalized the PML oncogene domain nuclear body, and concentrated with EKLF, RNA polymerase II, and the splicing factor SC35. These data together provide the first evidence that developmental stage- and differentiation state-specific regulation of the nuclear transport of EKLF might be one of the steps necessary for the switch-on of the mammalian adult β globin gene transcription.
In mammals, the β-like and α-like families are individually clustered on different chromosomes. Functional members of both gene clusters are arranged in the order of their expression during erythroid development (globin switch). In the mouse, the β-like globin locus is arranged as follows: 5′-ɛy (embryonic)-βh1 (embryonic)-βmaj (fetal and adult)-βmin (fetal and adult, minor species)-3′ (21, 2a) (top diagram in Fig. 1B). The hematopoietic cells, including the erythroid ones in the embryo, initially arise in the blood island that forms at embryonic day 7.5 (E7.5) (17). Within the primitive erythroblasts derived from the yolk sac are the embryonic genes, including ɛy and βh1, as transcribed previously (26). The adult βmaj globin gene, on the other hand, is silenced in E9.5 yolk sac, but it starts to be expressed on E10.5 in the erythroblasts in the aorta-gonad-mesonephros (AGM) region and the fetal liver of the embryo (26, 38, 54, 56). During development, the major site of the erythropoiesis shifts to the fetal liver and then again to the bone marrow at the time of birth. The erythroid cells generated in the latter-named two tissues represent a definitive lineage (2).
FIG. 1.
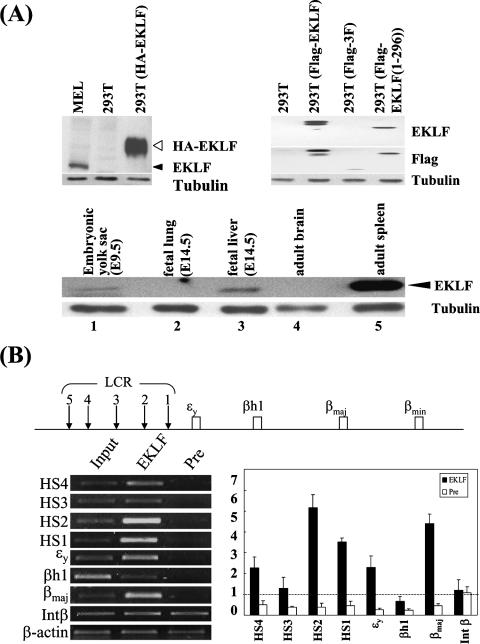
(A) Upper panels, specificity test of the AEK-1 antibody. Left panel, AEK-1 was used to probe Western blots containing whole-cell extracts prepared from MEL cells, 293T cells, and 293T cells transfected with pHA-EKLF. As a control, the blot was also probed with antitubulin. The positions of HA-EKLF and EKLF are indicated with open and black arrowheads, respectively. Right panel, Western blot analysis of whole-cell extracts prepared from 293T cells with or without transfection with pFlag-EKLF, pFlag-EKLF (1-296), and pFlag-3F (282-376), with anti-AEK-1, anti-Flag, and antitubulin used as the probes. Note the lack of AEK-1 signals in the lanes of 293T and 293T (Flag-3F). Shown in the lower panel are the expression patterns of EKLF in the mouse tissues. Whole-cell extracts were prepared from different mouse tissues and analyzed by Western blotting using AEK-1 as the probe. Note the presence of EKLF in the extracts of E9.5 yolk sac, E14.5 fetal liver, and adult spleen (lanes 1, 3, and 5, respectively). See reference 48 for more Western blotting data. (B) ChIP assay of β-like globin locus in murine E14.5 fetal liver. The physical map of the mouse β-like globin locus is shown on top. Shown in the lower left panel is the agarose gel pattern of PCR products from the immunoprecipitation reactions using the antibody AEK-1 (lane “EKLF”) or preimmune serum (lane “Pre”). The lane “Input” consists of PCR bands using the input DNAs as the standard. The relative intensities of the positive signals are given as the severalfold increases over those from the preimmune samples, and the severalfold differences are given as means ± standard errors of the means.
Like other gene systems, both cis-acting DNA elements and trans-acting factors are involved in the control of globin switch. The cis-acting sequences include the locus control regions (LCRs) and the upstream promoters of the individual genes (30). The mouse β locus control region (β-LCR) spans 5 to 25 kb upstream of ɛy (Fig. 1B), and it consists of five erythroid-specific DNase I-hypersensitive sites (HS), HS1 through HS5. The structure and function of the human β-LCR is similar to that of the mouse β-LCR (5, 15). Through binding to the cis-acting DNA elements in the LCRs and the globin promoters, directly or indirectly, the trans-acting factors are also required for regulation of the mammalian globin switch (references 1, 9, 24, 44, 48, 55, and 59 and references therein). In particular, the functions of two erythroid-enriched factors, NF-E2 and GATA-1, in the modulation of the modification patterns of histones across the globin gene clusters have been addressed by the chromatin immunoprecipitation (ChIP) assay (1, 24, 25, 44, 48). Overall, a general model for regulation of the mammalian globin loci could be formulated in which the transcriptional activation of the globin genes involves the physical and functional interactions between the multiple protein complexes formed at LCRs and the different globin promoters (5, 10, 12, 30). In particular, the LCR facilitates the loading of transcriptionally competent RNA polymerase II (RNAP II) molecules from the LCR to the active globin promoter(s) (22, 25, 44). This is in line with the observation that in embryonic and fetal erythroid cells, the mammalian β globin locus is colocalized with the RNAP II factories (37, 47).
Beside NF-E2 and GATA-1, another key transcription factor for mammalian globin switch is erythroid Krüppel-like factor (EKLF), first identified in mouse adult erythroleukemic (MEL) cells (34). EKLF is essential for the switch-on and activation of the adult β globin gene of either mice (36, 41) or humans (40, 58). In particular, EKLF-knockout mice died of anemia by embryonic day 16 due to failure to express the mouse βmaj globin gene (36, 41). Also, it appeared to be important for the maintenance of an active spatial organization of the β-LCR (11). The activation function of EKLF is carried out by its proline-rich transcription activation domain at the N terminus and the three C2H2-type zinc fingers at the C terminus (3). Through its zinc fingers, EKLF activates the mouse βmaj globin gene by binding to the CACCC promoter box (34). Indeed, ChIP analysis of the mouse adult and embryonic erythroid cell lines has demonstrated the in vivo specificity of binding of EKLF to the βmaj globin promoter (20, 48) but not to the ɛy or βh1 promoter, although those promoters also consist of CACCC boxes (48). In addition to the βmaj promoter results, EKLF binding was also detected at the HS2 site of β-LCR in these cells (20, 48). A model for EKLF activation of the adult β globin genes has been suggested in which the interaction between the zinc fingers of EKLF and the SWI/SNF complex targets the former to the β globin promoter, leading to the opening of the local chromatin (23).
Key questions still remain to be answered regarding the mechanisms of EKLF involvement in globin switching. Although EKLF appears to be mainly responsible for turning on the β globin gene expression at the adult stage, EKLF mRNA and protein are present throughout development in erythroid tissues, including the embryonic yolk sac as early as day E7, the fetal liver, and the adult erythroblasts (34, 48, 49). Evidence has been presented that EKLF is functional in the primitive erythroid lineage, but its activity with respect to turning on the adult β globin gene is greatly suppressed in the early embryonic erythroid cells for unknown reasons (see references 3 and 54 and references therein). Also, overexpression of EKLF leads to increased transcription of the adult β globin gene and an earlier switch from fetal γ to adult β globin expression (33, 53), while decreasing the concentration of EKLF results in lowered expression of β with a reciprocal increase in γ gene expression (58).
Thus, how the cellular concentration and activity of EKLF are negatively and positively regulated during development is an interesting puzzle. In relation to this, both the human and mouse β globin loci in the erythroid cells have been shown to be colocalized with the RNAP II factories (37, 47). In addition, the transcription factor p45/NF-E2 is clustered in the human β globin locus-containing promyelocytic leukemia (PML) protein oncogene domain nuclear body, POD (47). These two observations suggest that the local concentrations of RNAP II and NF-E2 might be important for transcriptional activation of the β globin locus. Is EKLF also locally concentrated in the vicinity of the globin loci? Second, previous overexpression studies of EKLF in COS, a monkey kidney fibroblast cell line, and K562, a human embryonic/fetal erythroid cell line with no EKLF, showed that efficient nuclear localization of EKLF required at least two nuclear localization signals (NLS), 275-296 and 293-376, respectively (39, 42). More recently, Zhou et al. have shown that hemagglutinin (HA)-tagged EKLF protein expressed from a homologously replaced mouse EKLF gene locus was located in the nucleus, as suggested by CHIP analysis of E10.5 yolk sacs and E14.5 fetal livers and by anti-HA immunostaining of adult bone marrow erythroblasts (59, 60). However, the subcellular locations of endogenous EKLF protein in the erythroblasts during development have remained unknown, mainly due to the lack of appropriate anti-EKLF antibodies. In the following, we provide the first evidence that expression of the mouse adult βmaj globin gene in the erythroid cells is closely associated with developmental stage- as well as differentiation state-specific processes of the nuclear import and nuclear positioning of the EKLF molecules.
MATERIALS AND METHODS
Mouse tissues and cell lines.
The blood islands of E9.5 yolk sacs, the peripheral blood cells of the AGM region of E10.5 embryos, and fetal livers of E14.5 fetuses from pregnant female B6 or BALB/c mice were isolated and disrupted by repeated pipetting in phosphate-buffered saline (PBS) (10 mM phosphate, 0.15 M NaCl [pH 7.4]). The murine adult erythroleukemia cell line MEL (35) and human 293T cells were both cultured in Dulbecco's modified Eagle medium containing 20% fetal bovine serum (Gibco), 50 units/ml of penicillin, and 50 μg/ml of streptomycin (Invitrogen). For induction, MEL cells at a density of 5 × 105 N/ml were supplemented with 2% dimethyl sulfoxide (DMSO; Merck) or 5 mM hexamethylene-bis-acetamide (HMBA; Sigma), and the culturing was continued for another 24 to 96 h. For blocking of the EKLF transport, MEL cells were treated with 1 μM of Ratjadone A (Calbiochem) for 2 h before the addition of 2% DMSO. The human embryonic/fetal erythroleukemia cell line K562 (ATCC accession no. CCL-234) was cultured in RPMI medium containing 10% fetal bovine serum (Gibco), 50 units/ml of penicillin, and 50 μg/ml of streptomycin (Invitrogen).
Plasmid construction and DNA transfection.
pGEM-T βmaj plasmid was constructed in the following way. βmaj globin cDNA (amino acids [aa] 1 to 148) was synthesized by reverse transcription and PCR using murine E14.5 fetal liver RNA as the template. It was cloned into pGEM-T Easy vector (Promega), resulting in pGEM-T βmaj. To construct plasmids expressing Flag-tagged EKLF (aa 1 to 376), Flag-tagged EKLF (aa 1 to 296), Flag-tagged 3F of EKLF (aa 282 to 376), and HA-tagged EKLF (aa 1 to 376), the corresponding cDNA regions were PCR amplified from the reverse transcription products of DMSO-induced MEL RNA and cloned into the vectors pCMV-Flag (Invitrogen) or pCMV-HA (Clontech), resulting in pFlag-EKLF, pFlag-EKLF (1-296), pFlag-3F, and pHA-EKLF, respectively. The inserts of all the plasmids were verified by DNA sequencing before use.
The MEL cells were transfected using Optifect reagent (Invitrogen). The total amount of transfected DNA was kept constant by the addition of the carrier DNA. A 0.1 μg to 1 μg volume of pFlag-EKLF, pFlag-EKLF (1-296), pFlag-3F, or pHA-EKLF was mixed with 9.9 μg to 9 μg of carrier DNA (salmon sperm DNA) and transfected into 4 × 105 N/ml cells. For induction, 2% DMSO was added to the medium at 24 h posttransfection. The K562 cells were transfected by electroporation. A 0.5 μg to 10 μg volume of pFlag-EKLF, pFlag-EKLF (1-296), pFlag-3F, or pHA-EKLF was mixed with 0 μg to 6 μg of carrier DNA (salmon sperm DNA) and transfected into 1 × 107 N/ml cells. The 293T cells were transfected using the calcium phosphate precipitation method.
Antibodies.
Anti-RNAP II rabbit antibody N-20, anti-PML goat antibody L-16, anti-NF-κB goat antibody C-19, anti-NF-κB p65 goat antibody, anti-GATA-1 goat antibody M-20, anti-p45/NF-E2 rabbit antibody C-19, anti-CBP rabbit antibody A-22, and anti-hnRNP A1 mouse monoclonal antibody 4B10 were purchased from Santa Cruz Inc. Antitubulin, anti-SC-35, and anti-Flag M2 mouse monoclonal antibodies were from Sigma. The mouse monoclonal anti-HA antibody used was from Roche. The generation of the polyclonal rabbit antibody AEK-1 recognizing mouse EKLF has been described previously (48). Following the approach of Hsu et al. (19), a polypeptide consisting of tandemly arranged, trimeric repeats of the sequence SEETQDLGPG (aa 51 to 60 of mouse EKLF) was purified from bacterial culture and used as the antigen for antibody generation in rabbits. The antibody was then purified from sera with use of an affinity column, concentrated, and stored at −20°C before use (see references 19 and 48 for more details).
Western blot analysis of extracts.
For Western blotting, a whole-cell extract of MEL was prepared by lysis of the cell pellets containing 5× 106 N PBS-washed cells in 300 μl of ice-cold M-PER reagent (PIERCE). For preparation of extracts from different mouse tissues, E14.5 B6 or BALB/c mice were sacrificed, and the tissues were dissected and homogenized in T-PER reagent (PIERCE) at a ratio of 0.1 g tissue per 200 μl of reagent.
The preparation of the nuclear and cytoplasmic extracts followed the procedures by Chen et al. (6) but with some modifications (47). MEL cells (5 × 106 N), E9.5 yolk sacs, E10.5 AGMs, and E14.5 fetal livers were washed with PBS and resuspended in 100 μl of ice-cold sucrose buffer I (0.32 M sucrose, 3 mM CaCl2, 2 mM magnesium acetate, 0.1 mM EDTA, 10 mM Tris-HCl [pH 8.0], 1 mM dithiothreitol, 0.5% NP-40) containing protease inhibitor cocktail (1 mM phenylmethylsulfonyl fluoride, 1 μg/ml of leupeptin, 1 μg/ml of pepstatin). The suspension was centrifuged at 500 × g for 10 min. The supernatant was mixed with 22 μl of 5× cytoplasmic extraction buffer (0.15 M HEPES-NaOH [pH 7.9], 0.7 M KCl, 0.015 M MgCl2), resulting in the cytoplasmic extract. The pellet was processed to prepare the nuclear extract as described previously (47).
For Western blotting assays, aliquots of the extracts (for whole-cell extracts, 40 μg per lane; for nuclear or cytoplasmic extracts, the equivalent from 2 × 104 N cells) were electrophoresed on a 10% sodium dodecyl sulfate-10% polyacrylamide gel and then electrotransferred to a nitrocellulose membrane (Amersham Biosciences). Staining with Ponceau S (Pierce) was used to estimate the loading and blotting efficiencies. The blots were hybridized with the primary antibodies, and then the secondary antibodies were prepared in Tris-buffered saline solution containing 5% nonfat dry milk. The hybridizing bands were identified by use of an enhanced chemiluminescence (ECL) detection system (Amersham Biosciences).
ChIP.
The mammalian ChIP procedures described previously by Daftari et al. (9) were used to analyze EKLF binding in the murine β-like globin locus of the E14.5 fetal liver. Fetal liver cells (5 × 106 N) were fixed at room temperature for 10 min in 60 ml of Dulbecco's modified Eagle medium containing 1% formaldehyde. After sonication, the protein-DNA complexes were immunoprecipitated with the anti-EKLF antibody AEK-1 (48). The precipitated chromatin DNAs were purified and amplified by PCRs using the appropriate primers as follows: 95°C for 7 min followed by 30 to 35 cycles at 94°C for 30 s, 58°C for 40 s, and 72°C for 40 s. The primers used for PCR amplification of the regions of HS2 (34 cycles), HS3 (32 cycles), the βmaj promoter (34 cycles), and actin and intergenic β (30 cycles) have been described previously (48). The primers for amplifications of HS1 (34 cycles), HS4 (34 cycles), and the ɛy (32 cycles) and βh1 (33 cycles) promoter regions and the lengths of the PCR fragments generated were as follows: for HS1 (336 bp), 5′-CTCTCCCATAACCCATACCTCTT-3′ and 5′-CCAATTTTCTCCCTCCATGTTCTTCT-3′; for HS4 (309 bp), 5′-TTGCCCTCTCTTACATCTAGCAGC-3′ and 5′-CACACACACCAAAACAAAACCCCA-3′; for the ɛy promoter (241 bp), 5′-GAGAGTTTTTGTTGAAGGAGGAGC-3′ and 5′-CAGGAGTGTCAGAAGCCAGTACGT3′ (45); and for the βh1 promoter (321 bp), 5′-TTGCAAATCTCCTGTTGGGGGAAG-3′ and 5′-ATGGTGTGAGGTCTAGAAGCTTGG-3′. The DNA amounts used for the individual PCRs were determined according to the intensities of the β-actin signals. The calibration of the PCR signals was done as described preciously (48). Presumably due to the differential effects of the qualities and quantities of DNA samples on the PCR amplifications of different genomic regions, the signals from the input DNA and preimmune serum (see lanes Input and Pre in the Fig. 1B legend) differed from region to region. Each histogram consists of the averages of data from two to three sets of PCR analyses of chromatin DNAs precipitated from two different fetal liver preparations. The severalfold differences are given as means ± standard errors of the means.
Northern blot analysis.
The Northern blotting procedures followed those of Liu et al. (32). The total RNAs were extracted by means of the use of commercial TRIzol reagent (Invitrogen), loaded on 1% agarose gels containing 6% formaldehyde, and transferred to a nylon membrane. The blots were analyzed by autoradiography after hybridization with radioactive DNA probes amplified by PCR.
Immunostaining.
The procedures followed essentially those described previously (47). The cells were fixed in 4% formaldehyde for 20 min, permeabilized with 0.1% Triton X-100-PBS, blocked in 10% normal donkey serum (Jackson ImmunoResearch), and incubated with the appropriate antibodies at 4°C overnight. Secondary antibodies conjugated to Alexa Fluor 488, Alexa Fluor 546, or Alexa Fluor 647 (Molecular Probes) were then added, and the incubation continued for another 2 h at 25°C. Staining of DNA was carried out using DAPI (4′,6′diamidino-2-phenylindole; Molecule Probes). MetaMorph software (Meta Imaging Series version 6.1; Universal Imaging Corporation) was used to quantify the relative levels of the signals of immunostaining in the nucleus and cytosol.
DNA FISH.
A fluorescence in situ hybridization (FISH) probe for the murine βmaj globin locus was generated by nick translation of the DNA insert of pGEM-T βmaj. The DNA probe was labeled with biotin by PCR. The protocols used for the FISH experiments were similar to those described previously (31, 47). After hybridization, the biotin-labeled probe was detected with an Alexa Fluor 488 (Molecular Probes)-conjugated antibody.
Immuno-DNA-FISH.
To check the relative locations of the murine β-like globin locus and different nuclear bodies, immuno-DNA-FISH (46) was carried out by a combination of the procedures of DNA FISH and immunofluorescent staining as described previously (47).
Image analysis.
Image analyses of the fluorescent patterns from DNA FISH and immunostaining were reframed using a Zeiss LSM 510 Meta confocal microscope. Images were from single optical section(s). For each set of the analyses, three different pools of the cells were collected for immunostaining and/or DNA FISH. For each randomly chosen field, the confocal stacks of the single optimal sections of all the cells observed in the field were collected and analyzed. The distances between different signals on the three-dimensional reconstructed image stacks were measured using LSM 5 image analysis software. The association-colocalization of signals from DNA FISH and those from immunostaining using anti-AEK-1, anti-SC35, anti-PML, and anti-RNAP II were assessed by capturing the image stacks of the nuclei for three-dimensional image reconstruction. The thickness of the single optical sections ranged between 0.2 and 0.3 μm. The colocalized signals were then counted. For the “EKLF dots,” the PML bodies, the RNAP II factories, and the SC-35 speckles, only those with a diameter greater than 0.2 μm were counted and analyzed. For DNA FISH to locate the β-like globin locus, metaphase cells were first used to validate the signals. We usually detected one to three spots in MEL cells and one to two spots in the mouse yolk sac or fetal liver cells. The latter number range is similar to that reported by Trimborn et al. (54). However, the immunostaining signals found after DNA FISH were usually weaker due to the step of incubation at 65°C. This reduced the apparent numbers of the nuclear substructures, e.g., those of the RNAP II factories and the speckles, under the microscope.
For quantitation of the relative levels of the immunostaining signals in the nucleus and cytosol, MetaMorph software (Meta Imaging Series version 6.1; Universal Imaging Corporation) was used. We used DNA-staining DAPI as the nuclear marker and antitubulin staining for the cytosol. The differential interference contrast was also used to examine the nucleus and cytosol. The threshold for analysis of the immunostaining signals was based on the signals from the parallel use of the preimmune sera.
RESULTS
Binding in vivo of EKLF at the βmaj promoter and β-LCR in mouse E14.5 fetal liver cells.
Since the identification of EKLF in 1993 (see the introduction), it has been difficult to generate specific and sensitive antibodies against EKLF that are suitable for Western blotting, ChIP, and immunostaining experiments. Recently, we produced and purified a rabbit anti-mouse EKLF antibody, AEK-1, with use of the approach by Hsu et al. (also see Materials and Methods). This method takes advantage of the fact that tandem repeats of an epitope sequence often possess potent antigenicity (19, 48).
The specificity of AEK-1 against EKLF was initially demonstrated by Western blot analysis of total extracts prepared from a variety of cell lines and mouse tissues (48). As exemplified in the upper left panel of Fig. 1A, AEK-1 detected endogenous EKLF in the mouse adult erythroleukemia cell line MEL and an HA-tagged mouse EKLF transiently expressed in the human 293T cells. The specificity of AEK-1 was further demonstrated by the fact that it detected Flag-EKLF and Flag-EKLF (1-296), within both of which the epitope used to generate AEK-1 (aa 51 to 60 of EKLF) resided, but not Flag-3F (282-376), which lacked the epitope (Fig. 1A, upper right panel). AEK-1 also detected EKLF in Western blot analysis of E9.5 yolk sac, E14.5 fetal liver, and adult spleen tissues (Fig. 1A, lower panel, lanes 1, 3, and 5) but not in analysis of nonerythroid fetal and adult tissues (Fig. 1A, lower panel, lanes 2 and 4). These Western blotting data paralleled the RNA expression profiles of EKLF during mouse development (34).
AEK-1 was then used in ChIP assays for the analysis of EKLF binding in vivo in mouse E14.5 fetal liver samples. Fixed chromatin samples were prepared from the fetal liver cells as described in Materials and Methods and were immunoprecipitated (IP) with AEK-1 or preimmune serum. DNAs were purified from the precipitates and supernatants of the IP reactions and subjected to PCR analysis of the enrichment of different regions of the murine β globin locus in the DNA samples pulled down by AEK-1.
As shown in Fig. 1B, EKLF binding was detected not only at the βmaj promoter but also at HS1, HS2, and HS4 of the β-LCR. Interestingly, we also detected EKLF binding, albeit relatively weaker, at the embryonic ɛy globin promoter region (Fig. 1B). The ChIP data shown in Fig. 1B indicate that EKLF indeed bound in vivo at the regulatory elements of the murine β globin locus in erythroid tissue actively transcribing the βmaj globin gene.
Developmental stage-specific localization of EKLF in the nuclei of mouse erythroid tissues.
The subcellular location of the endogenous EKLF had been unknown before. Based on the DNA binding (34) and chromatin binding (Fig. 1) (20, 48, 59) properties of EKLF, it was expected to be located mainly in the nucleus of the erythroid cells. To check the subcellular locations of EKLF in erythroid cells at different developmental stages, the affinity-purified antibody AEK-1 was again used to immunostain cells isolated from blood islands of E9.5 yolk sac samples, peripheral blood in the AGM of E10.5 embryos, and E14.5 fetal livers. As shown in Fig. 2, while nonerythroid 293T cells exhibited no obvious staining of EKLF (exemplified in the first row shown in Fig. 2), EKLF proteins in the EKLF-positive cells in AGM of E10.5 embryos and in E14.5 fetal livers, both of which expressed the βmaj globin gene, appeared to be distributed mainly (63% and 81%, respectively) in the nucleus, with a dotted pattern (exemplified in the third and fourth rows of Fig. 2). The numbers of these EKLF “dots” ranged from 5 to 28 per nucleus, and their diameters ranged from 0.2 μm to 0.5 μm. In great contrast, the majority (95%; n = 42) of the EKLF-positive cells in the blood islands of E9.5 yolk sacs, which expressed embryonic ɛy and βh1 but not adult βmaj, contained EKLF proteins mainly in their cytoplasm (exemplified in the second row of Fig. 2). Western blotting of the nuclear and cytoplasmic extracts has confirmed the immunostaining result (Fig. 2, lower panels). Thus, there appeared to be a tight correlation between the switches of the βmaj globin gene transcription and the subcellular locations of EKLF during development. As shown later, this correlation was also observed for the inducible activation of βmaj in a mouse adult erythroid cell line.
FIG. 2.
Subcellular locations of EKLF. 293T cells (top row), blood islands of E9.5 mouse yolk sacs (second row), peripheral blood cells of E10.5 AGM (third row), and E14.5 fetal liver cells (bottom row) were stained with AEK-1 and examined with the confocal microscope. The broken lines in the first column indicate the boundaries of the nuclei, as defined by the DAPI staining. Lower-magnification pictures are shown in the last column. The percentage values represent the fractions of EKLF-positive cells containing the majority of EKLF (more than 90%) in the nucleus. n values represent the numbers of cells analyzed for each set of the experiments. Each set consists of three or more independent analyses. Western blot analysis results of the nuclear (Nu) and cytoplasmic (Cyto) extracts prepared from the blood island cells of E9.5 yolk sacs, peripheral blood cells of the E10.5 AGM, and E14.5 fetal liver cells are shown in the lower panel, with tubulin as the cytoplasmic marker and hnRNP A1 as the nuclear marker.
Colocalization of the β globin locus with POD/RNAP II factory containing concentrated EKLF and SC35 in mouse fetal liver cells.
Previously, it had been shown that in mouse or human erythroid cells, the β globin locus is colocalized with nuclear substructures, including the RNAP II factory and the nuclear bodies POD and speckles (see references 4, 37, and 47; see also the latter sections of Results and Discussion). To investigate whether the nuclear substructure (the dots) formed by EKLF in the fetal liver was also colocalized with the β globin locus, we carried out combined DNA-FISH immunostaining analysis as described before (47). Among the 38 fetal liver nuclei analyzed that both were EKLF-positive and exhibited two β globin alleles, 36 (95%) of them had at least one of their β globin alleles colocalized with an EKLF dot (Fig. 3A). Of these 36 nuclei, 31 (86%) of them had only one of the β globin alleles colocalized with an EKLF dot, as exemplified in Fig. 3A. In the rest of the nuclei examined, 8% (3/36) exhibited two β globin alleles each colocalized with an EKLF dot and 6% (2/36) had both alleles colocalized with the same EKLF dot (data not shown).
FIG. 3.
DNA-FISH immunostaining of mouse fetal liver and yolk sac cells. Combined DNA-FISH immunostaining of cells from murine E14.5 fetal livers and blood islands of E9.5 yolk sacs was carried out using DNA probe specific for the mouse β globin locus and different antibodies. Note that the immunostaining signals after DNA FISH were usually weaker due to the step of incubation at 65°C. This was already mentioned previously in reference 47. (A) Colocalization of EKLF with the β globin locus in the E14.5 fetal liver but not the E9.5 yolk sac. The arrows point to the two β globin alleles in the cells shown. The diagram shows the locations of the EKLF dots (red) and the β globin alleles (green). The percentage values represent the fractions of EKLF-positive and two allele-exhibiting cells that have at least one of their β globin alleles colocalized with an EKLF dot. (B) Colocalization of the β globin locus with EKLF, POD, RNAP II factory, and SC35 speckles in the fetal liver cells. DNA FISH was combined with double immunostaining and use of the antibody pairs AEK-1/anti-PML (top row), AEK-1/N-20 (middle row), and AEK-1/anti-SC35 (bottom row). The fractions (%) of cells in which at least one β globin allele was colocalized with the pair(s) of nuclear bodies and/or RNAP II factory are indicated. n represents the numbers of cells analyzed for each set of the experiments.
To further examine whether the EKLF dot coinciding with the β globin locus also colocalized with other nuclear substructures or nuclear bodies in the fetal liver cells, we carried out DNA FISH in combination with immunostaining using AEK-1 as well as antibodies against RNAP II, the POD signature protein PML, and the splicing factor SC35 (Fig. 3B). Interestingly, there was a high percentage of colocalization of the EKLF dots with the β globin locus, the POD, the RNAP II factories, and the SC35-staining speckles. As exemplified in Fig. 3B, in approximately 76% of the EKLF-positive fetal liver cells, at least one of their β globin alleles was colocalized with an EKLF dot and a POD; in 83% of them, the EKLF-β complexes were colocalized with the RNAP II factories. Relatively fewer of the EKLF-β complexes were colocalized with the speckles (Fig. 3B).
These DNA-FISH immunostaining data indicated that EKLF proteins were concentrated in the PODs and/or the RNAP II factories previously shown to be colocalized with the β globin loci in erythroid cells in humans (47) and mice (37). The data further suggested that not only the nuclear localization of EKLF but also the concentration in specific nuclear substructures colocalizing with the β globin locus is crucial for the adult β globin switch.
Nuclear import of EKLF accompanied with induced βmaj globin gene activation of MEL cells.
MEL is a mouse adult erythroleukemia cell line. Upon treatment with different reagents such as DMSO, HMBA, etc., the MEL cells differentiate and a number of erythroid-specific genes, in particular the adult α and βmaj globins, become transcriptionally active (35). It was thus interesting to see whether nuclear import of EKLF is involved in turning on the βmaj promoter during MEL differentiation, as suggested by the different subcellular locations of EKLF in the mouse E14.5 fetal livers and blood islands of E9.5 yolk sacs.
As shown in Fig. 4A, both immunostaining and Western blotting experiments demonstrated that the EKLF proteins were indeed located in the cytosol of most (80% or so) of the MEL cells prior to induction. Upon induction with either DMSO or HMBA, a major portion of the EKLF proteins became located in the nucleus. Furthermore, a dotted pattern of nuclear distribution of EKLF was also observed in the induced MEL cells (Fig. 4A). In fact, as shown by Northern blotting, the nuclear import of EKLF upon induction of MEL by DMSO paralleled well with the induced transcription of the βmaj globin gene (Fig. 4B). These data indicated that there was an active nuclear import process of EKLF during erythroid differentiation of the MEL cells and that the nuclear import of EKLF might be required a priori for the induced activation of the βmaj transcription by DMSO.
FIG. 4.
Subcellular location of EKLF in MEL cells. (A) Subcellular location of the endogenous EKLF in MEL cells. The locations of EKLF in MEL cells before (top row) and after DMSO (middle row) or HMBA (bottom row) induction were examined by immunostaining with AEK-1 (first column). The nuclei were identified by DAPI staining. The fractions (%) of cells with the majority (more than 90%) of EKLF present in the nucleus are indicated. The subcellular distributions of EKLF in MEL cells were further analyzed by Western blotting of the whole-cell extract (Total), nuclear extract (Nu), and cytoplasmic extract (Cyto). Note the absence of the control tubulin bands in the “Nu” lanes. Also, most of the EKLF of MEL (+) DMSO and MEL (+) HMBA cells were in the nuclear fraction. (B) Changes of the expression levels of the βmaj globin gene and the subcellular location of EKLF during MEL differentiation. The expression levels of βmaj and G3PDH genes in MEL cells at different days after induction with DMSO were analyzed by Northern blotting. The fractions of cells containing the majority (more than 90%) of EKLF in their nuclei were also scored by immunostaining, as indicated below the Northern panel. Note the apparently concomitant appearance of βmaj RNA and the abrupt increase in the fraction of cells containing nuclear EKLF on day 2 of the induction.
The nuclear positioning of the β globin locus relative to the distributions of PODs, RNAP II factories, and speckles in DMSO-induced MEL cells has also been analyzed by combined DNA-FISH immunostaining. In similarity to the E14.5 fetal liver results, there was also a high frequency of colocalization of the β globin locus with these three classes of nuclear substructure (Fig. 5).
FIG. 5.
Combined DNA-FISH immunostaining of MEL cells. MEL cells without (top row) and with DMSO induction for 3 days were analyzed by combined DNA-FISH immunostaining as described for Fig. 3 for the E14.5 fetal liver cells. Again, the fractions (%) of cells containing at least one β globin allele colocalized with the nuclear body and/or RNAP II factory are indicated. Note that the immunostaining signals after DNA FISH were usually weaker due to the step of incubation at 65 °C. This was also evident in the results reported in reference 47.
Repression of βmaj globin gene transcription in DMSO-induced MEL cells by blockage of the nuclear import of EKLF.
In order to examine whether the nuclear import of EKLF was one of the essential steps for activation of the βmaj globin gene during DMSO induction of MEL cells, we have attempted to use several inhibitors of nuclear import or export to block DMSO-induced EKLF import into the nuclei of MEL cells. Due to the intrinsic properties of these drugs and the relatively long time of DMSO induction, most of the drugs were not suitable for our purpose (data not shown). However, a known distinct inhibitor of the nuclear export factor CRM1 (chromosome region maintenance 1), Ratjadone A (28), was found to effectively block the nuclear import of EKLF. As shown in the immunostaining patterns of Fig. 6A, inclusion of Ratjadone A during DMSO treatment of MEL kept EKLF (exemplified in the second row of Fig. 6A), but not GATA-1, CBP, and p45/NF-E2 (exemplified in the third, fourth, and bottom rows, respectively, of Fig. 6A), from being imported into the nuclei. As a control, a high portion of the p65 subunit of NF-κB was retained in the nucleus upon Ratjadone A treatment, as shown by Western blotting (Fig. 6B) and by immunostaining (data not shown).
FIG. 6.
Repression of βmaj globin gene transcription by Ratjadone A-mediated blockage of the nuclear import of EKLF. (A) The subcellular locations of EKLF in MEL cells subjected to 3 days of DMSO induction with (bottom four rows) or without (first row) pretreatment with Ratjadone A were analyzed by immunostaining. Note that the Ratjadone A treatment prevented the EKLF from entering into the nucleus during DMSO induction (second row). As controls, anti-GATA-1, anti-CBP, and anti-p45/NF-E2 antibodies were also used to immunostain the (+) Ratjadone A/(+) DMSO cells (bottom three rows). The fractions (%) of cells with the majority (more than 90%) of the signals in their nuclei are indicated. (B) The subcellular locations of EKLF in MEL cells subjected to 3 days of DMSO induction with or without pretreatment with Ratjadone A were analyzed by Western blotting. Nu, nucleus; Cyto, cytosol. (C) Northern blotting of RNAs isolated from uninduced MEL cells (lane 1), MEL cells induced subjected to 3 days of DMSO induction (lane 2) or HMBA induction (lane 3), MEL cells treated with Ratjadone A for 2 h (lane 4), and MEL cells pretreated with Ratjadone A for 2 h and then induced with DMSO for 3 days (lane 5). Note the significantly lowered RNA levels of βmaj globin and ALAS-E in DMSO-induced cells pretreated with Ratjadone A in comparison to those of DMSO-MEL cells without the Ratjadone A treatment (compare lane 5 to lane 2).
Significantly, this blockage of EKLF import by Ratjadone A was accompanied by transcriptional repression (by more than 80%) of βmaj as well as of the erythroid-specific δ-aminolaevulinate synthase gene (ALAS-E), another erythroid gene known to be activated by EKLF (18, 51) (compare lane 5 to lane 2 in Fig. 6C). On the other hand, DMSO-induced transcription of the other erythroid genes tested, such as α globin and porphobilinogen deaminase (PBGD), was not affected (Fig. 6C). Thus, the process of nuclear import of EKLF appeared to be specifically required for transcriptional activation of EKLF-regulated genes, including the βmaj globin, during MEL differentiation as induced by DMSO.
Mapping of EKLF domain(s) required for its induced nuclear import in DMSO-treated MEL cells.
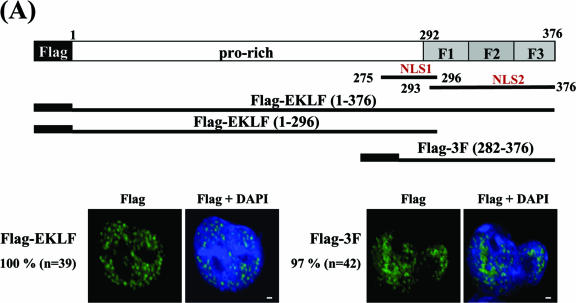
It would be helpful to map the domain(s) of EKLF required for its DMSO-induced nuclear import in MEL cells. Previously, two nuclear localization signals of murine EKLF, NLS1 (aa 275 to 296) and NLS2 (aa 293 to 376), have been mapped by immunostaining of exogenously expressed, green fluorescent protein-tagged EKLF in the human erythroid cell line K562 and in nonerythroid monkey CV1 cells (42). Of the two NLSs, NLS1 covers a basic region while NLS2 encompasses the three zinc fingers of EKLF (top diagram in Fig. 7A) (42).
FIG. 7.
Mapping of EKLF domains required for DMSO-induced nuclear import of EKLF in MEL cells. (A) The subcellular locations of transiently expressed Flag-EKLF and Flag-3F in K562 cells. Top, schematic representation of the domain organization of EKLF. The proline-rich (pro-rich) region and the three-zinc-finger region (aa 292 to 376) are indicated. Shown below the EKLF map are the inserts of plasmid constructs pFlag-EKLF, pFlag-EKLF (1-296), and pFlag-3F used to transfect K562 and MEL cells. The subcellular locations of Flag-EKLF and Flag-3F in the transfected K562 cells were detected by immunostaining with anti-Flag. In either case, nearly all of the proteins were located in the nucleus. (B) The subcellular locations of transiently expressed Flag, Flag-EKLF, Flag-EKLF (1-296), and Flag-3F in MEL cells. Again, the locations of the exogenous proteins in the transfected MEL cells, before and after DMSO induction, were identified by immunostaining using anti-Flag as the probe. The nuclei were identified by DAPI staining. MetaMorph software (Meta Imaging Series version 6.1; Universal Imaging Corporation) was used to quantify the relative amounts of transiently expressed, Flag-tagged proteins in the nucleus and cytosol. For each experimental set, the percentage value (%) indicates the average fraction per cell of the corresponding exogenous protein located in the nucleus. The comparisons of the amounts of transiently expressed proteins in MEL cells before (white bars) and after (black bars) DMSO induction are shown schematically in the histogram for Flag, Flag-EKLF, Flag-EKLF (1-296), and Flag-3F. Note that transfected HA-EKLF behaved similarly to Flag-EKLF in MEL cells during DMSO induction (Shyu and Shen, unpublished results).
To see whether either or both of these NLSs mediated DMSO-induced nuclear import of EKLF in MEL cells, we have made constructs capable of expressing EKLF, aa 1 to 296 of EKLF, and aa 282 to 376 of EKLF, each tagged with a Flag epitope. These constructs were transfected into different cell lines, including MEL, and the immunostaining patterns as stained by anti-Flag were examined (Fig. 7). In similarity to the results of previous studies (39, 42), the transfected Flag-EKLF was almost exclusively localized in the nuclei of transfected K562 (Fig. 7A). However, in similarity to the results for endogenous EKLF shown in Fig. 4A, few (approximately 7%) of the Flag-EKLF proteins were located in the nuclei of uninduced MEL cells, but the nuclear content of Flag-EKLF greatly increased (to approximately 82%) upon DMSO induction (second row of Fig. 7B). Flag-EKLF lacking the NLS1 region of aa 275 to 296 behaved similarly to the full-length Flag-EKLF (Y.-C. Shyu and C.-K. J. Shen, unpublished results). In contrast, there was no regulation of the nuclear import of transfected Flag-EKLF (1-296) or Flag-3F (282-376). Flag-EKLF (1-296) was present in both the cytosol and nuclei of transfected MEL cells with or without DMSO induction (third row of Fig. 7B). On the other hand, Flag-3F (282-376), while being exclusively localized in the nuclei of transfected K562 cells (Fig. 7A and reference 42), was mainly present in the cytoplasm of transfected MEL cells before (77%) and after (61%) DMSO induction (fourth row of Fig. 7B). The data of Fig. 7 indicated that unlike the results seen for the other cell types tested, NLS2 encompassing the zinc finger region of EKLF was the main element responsible for keeping EKLF in the cytoplasm of MEL cells. Furthermore, it appeared that the full-sequence context of EKLF was required for its regulated nuclear import process in DMSO-induced MEL cells and, by inference, during early erythroid development.
DISCUSSION
It has been clear that mammalian EKLF is an essential transcription factor for the activation of the adult β globin gene in erythroid cells of definitive lineage. It is also known that this function of EKLF requires its direct binding at the CACCC box in the adult β globin promoter (see the introduction for references). In this study, we demonstrated that the mouse β globin locus is physically associated with the nuclear body POD concentrated with EKLF, RNAP II, and the splicing factor SC35. These data, together with those of our previous analysis of K562 cells (47), indicate that POD is indeed the nuclear domain within which transcription of the β globin locus is regulated. Remarkably, our study has revealed that activation of the mouse adult βmaj globin gene involves regulation of the nuclear transport processes of EKLF in erythroid cells that are both developmental stage and differentiation state specific.
We first showed, by ChIP assays using a high-specificity antibody (AEK-1) against EKLF (Fig. 1A) (48), that in the E14.5 mouse fetal liver in which 99% of the cells are definitive erythroid cells (54) expressing the adult βmaj globin gene, EKLF bound at the βmaj promoter and HS1, HS2, and HS4 sites of β-LCR and, to a relatively low extent, the embryonic ɛy promoter (Fig. 1B). The in vivo binding pattern of EKLF in the E14.5 fetal liver is in interesting similarity to those in the DMSO-induced MEL cells (48) and in the E14.5 fetal liver and E10.5 yolk sac cells from mice with a knocked-in, HA-tagged EKLF gene (59). Our failure to detect EKLF binding at HS3 might be due to the masking in vivo of the epitope(s) recognized by AEK-1. The binding of EKLF at both the βmaj promoter and the HS sites suggested that EKLF could also recognize and bind at the CACCC boxes within those latter-named regulatory elements. Alternatively, the coappearance of EKLF might reflect the long-range interaction between the β-LCR holocomplex and the EKLF-βmaj promoter complex. In either scenario, the ChIP analysis results supported the notion that EKLF is an integral component of the transcription complex activating the βmaj globin promoter in the definitive erythroid cells.
The binding of EKLF to β-LCR and the βmaj promoter in the fetal liver, as revealed by the ChIP assay results, was accompanied by our observation in the DNA FISH-immunostaining experiments that EKLF and the splicing factor SC35 were concentrated in the POD-RNAP II factory within which the β globin locus resided (Fig. 3). Previously, we had shown that in the human embryonic-fetal erythroid K562 cell line, the β globin locus is also colocalized with POD containing concentrated RNAP II and SC35 as well as sumoylated p45/NF-E2 (47). The data from our DNA-FISH immunostaining analyses and the RNA-FISH studies by others (4, 37) together point to a model in which the multiple protein-DNA complexes of the mammalian β globin locus in actively transcribing erythroid cells are formed and/or maintained within or near the nuclear body POD containing concentrated transcription factors, including both EKLF and p45/NF-E2 as well as RNAP II. The α globin locus is organized in POD in a similar fashion (Shyu and Shen, unpublished data). Presumably, the local concentrations of RNAP II as well as of different factors, including EKLF in the β locus-associated POD/RNAP II factory, facilitate the assembly of the transcription initiation and the propagation of the transcription as well as splicing of the primary globin transcripts.
Most interestingly, we have identified development stage- and differentiation state-specific processes of nuclear transport of EKLF in erythroid cells actively transcribing the adult βmaj globin gene. In particular, the data of Fig. 2 indicated that during the early developmental stage there was a switch of the subcellular localization of EKLF in the erythroid cells from the cytosol to the nucleus. This switch coincided with the timing of the switch-on of βmaj globin gene transcription (13, 26, 38, 54, 57). The nuclear localization of EKLF in the E10.5 yolk sac and E14.5 fetal liver tissues (Fig. 2) was consistent with the ChIP analysis of the same two tissues (Fig. 1B) (59) and with the anti-HA immunostaining pattern of the adult bone marrow erythroblasts from the genetically engineered mice (60). Thus, there appeared to be a developmental stage-specific switch of the subcellular compartments in which the majority of EKLF proteins were present in the erythroblasts.
Our observation of a developmental coswitch of the subcellular locations of EKLF and the transcription status of the adult βmaj globin gene in the erythroblasts during the E9.5 to E10.5 transition paralleled the results of the study of EKLF in the MEL cells (Fig. 4). In differentiated MEL as induced by DMSO, EKLF joins p18/NF-E2 to undergo active, subcellular transport. While p18/NF-E2 migrated from the heterochromatin to euchromatin regions during DMSO induction of MEL (14), EKLF was imported from the cytosol to the nuclei (Fig. 4), some becoming concentrated in the β globin locus-associated POD/RNAP II factory (Fig. 5). Interestingly, treatment of MEL cells with Ratjadone A, the inhibitor of nuclear export, blocked the DMSO-induced import of EKLF (Fig. 6A and 6B), and it specifically blocked the activation of the βmaj globin gene as well (Fig. 6C). Notably, several transcription factors known to be activators of βmaj globin gene transcription still remained in the nucleus after Ratjadone A treatment (Fig. 6A). Also, this inhibition of βmaj activation by Ratjadone A did not appear to be the result of blockage of MEL differentiation per se, since PBGD, one of the marker genes of MEL differentiation, was activated in DMSO-treated MEL cells whether or not Ratjadone A was present (Fig. 6C). In relation to this, the transcription of PBGD is known not to require EKLF (18). On the other hand, ALAS-E, another erythroid gene known to be activated by EKLF (18, 51), was also repressed in the presence of Ratjadone A (Fig. 6C). Ratjadone A blocks the nuclear export of several proteins (28, 50; Shyu and Shen, data not shown), and it does so by blocking the exportin protein CRM1 (28). The mechanism(s) utilized by Ratjadone A to block the nuclear import of EKLF is unknown at the moment. It might block the export of a certain vehicle protein(s) required for the import of EKLF. Alternatively, the suppression of the βmaj globin gene by Ratjadone A could be accomplished through direct transcriptional repression. In any case, the analysis of MEL cells (Fig. 4 to 6) further indicated the importance of nuclear import of EKLF in the transcriptional activation of βmaj in the fetal and adult erythroid cells.
The molecular and cellular basis of the regulated process of nuclear import of EKLF during erythroid development (Fig. 2) and differentiation (Fig. 4) is unclear at the moment. Previous studies of other systems have shown that regulation of the nuclear import processes could be mediated through protein-protein interaction, posttranslational modification, and sometimes even conformational changes of the protein (7, 8, 27). For example, the nuclear translocation of the NF-κB/RelA proteins is controlled by release of NF-κB from IκB upon phosphorylation and consequent degradation of the latter (7). N-terminal phosphorylation signals the nuclear retention of p53 by blocking its interaction with MDM2, thus inhibiting the ubiquitination and subsequent nuclear export of p53 (16). Also, sumoylation of RanGAP1 promotes its interaction with the Ran-GTP binding protein RanBP2 at the cytoplasmic face of the nuclear pore complex (43, 52), and sumoylation is required for the calcineurin-mediated dephosphorylation and nuclear import of the transcription factor NFAT1 (52).
In the case of EKLF, its regulated nuclear import in MEL cells during DMSO induction appeared to require the full-sequence context of the factor (Fig. 7). In particular, the function of efficient nuclear import-nuclear localization of NLS2 spanning the zinc finger region of EKLF, as observed in the nonerythroid cells (39, 42; Shyu and Shen, unpublished results) or erythroid K562 cells (42) (Fig. 7A), was lost in MEL cells with or without DMSO induction (Fig. 7B). Similarly, there was no regulation of the nuclear import of the EKLF (1-296) fragment during DMSO induction of the transfected MEL cells (Fig. 7B). While identification of the mechanisms and pathways regulating the EKLF import awaits future investigation, we speculate, among the different possibilities, on the following scenario of regulation. In particular, it is suggested that although the zinc finger region/NLS2 is required for nuclear localization of EKLF, this element is also responsible for the retention of EKLF in uninduced MEL cells, as reflected by the nuclear fractions of Flag-EKLF and Flag-3F being relatively lower than that of Flag-EKLF (1-296) (Fig. 7B). The retention is likely mediated through the physical interaction between NLS2 and a certain cytosolic factor(s). Upon DMSO induction, this interaction is disrupted, possibly due to an induced covalent modification of EKLF, e.g., phosphorylation or sumoylation as observed in the other systems mentioned above, somewhere within the region (1-296) carboxyl to the zinc finger domain. The originally cytosolic EKLF molecules are consequently released and transported into the nucleus. This scenario, which exists in MEL but not in K562 cells, would partially explain the observations shown in Fig. 7.
In summary, this study revealed a novel pathway for EKLF to activate the adult βmaj globin gene during mouse erythroid development and during differentiation of the erythroid cells in culture. The existence of this pathway, i.e., a regulated process of nuclear import of EKLF, provides a logical explanation for the silencing of the adult βmaj globin gene in embryos before the stage of E9.5, despite the presence of EKLF in the erythroblasts during early development. Whether a specific protein modification(s) and/or protein-protein interaction(s) is involved in the initiation of this developmental stage- and differentiation state-specific import process is currently under investigation.
Acknowledgments
We thank our laboratory colleagues Guang-Yuh Chiou, Ruei-Lin Chen, Kuen-Jer Tsai, and Bose Jayarama Krishnan for various reagents and help. Chi-Hung Lin's help with the DNA FISH technique at the beginning stage of this work is appreciated. Sue-Ping Lee from the imaging core of the Institute of Molecular Biology of Academia Sinica helped on the use of confocal microscope. We also thank Mark Groudine and his colleagues for their critical comments.
This research was supported by a grant from the National Health Research Institute (NHRI), and by the Academia Sinica, Taipei, Taiwan, Republic of China.
Footnotes
Published ahead of print on 22 January 2007.
REFERENCES
- 1.Anguita, E., J. Hughes, C. Heyworth, G. A. Blobel, W. G. Wood, and D. R. Higgs. 2004. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 23:2841-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron, M. H. 2003. Embryonic origins of mammalian hematopoiesis. Exp. Hematol. 31:1160-1169. [DOI] [PubMed] [Google Scholar]
- 3.Bieker, J. J. 2005. Probing the onset and regulation of erythroid cell-specific gene expression. Mt. Sinai J. Med. 72:333-338. [PubMed] [Google Scholar]
- 4.Brown, J. M., J. Leach, J. E. Reittie, A. Atzberger, J. Lee-Prudhoe, W. G. Wood, D. R. Higgs, F. J. Iborra, and V. J. Buckle. 2006. Coregulated human globin genes are frequently in spatial proximity when active. J. Cell Biol. 172:177-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bulger, M., and M. Groudine. 1999. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 13:2465-2477. [DOI] [PubMed] [Google Scholar]
- 6.Chen, H. J., C. L. Hwong, C. H. Wang, and J. Hwang. 2000. Degradation of DNA topoisomerase I by a novel trypsin-like serine protease in proliferating human T lymphocytes. J. Biol. Chem. 275:13109-13117. [DOI] [PubMed] [Google Scholar]
- 7.Chen, L. F., and W. C. Greene. 2004. Shaping the nuclear action of NF-κB. Nat. Rev. Mol. Cell Biol. 5:392-401. [DOI] [PubMed] [Google Scholar]
- 8.Crabtree, G. R., and E. N. Olson. 2002. NFAT signaling: choreographing the social lives of cells. Cell 109(Suppl.):S67-S79. [DOI] [PubMed] [Google Scholar]
- 9.Daftari, P., N. R. Gavva, and C. K. Shen. 1999. Distinction between AP1 and NF-E2 factor-binding at specific chromatin regions in mammalian cells. Oncogene 18:5482-5486. [DOI] [PubMed] [Google Scholar]
- 10.de Laat, W., and F. Grosveld. 2003. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 11:447-459. [DOI] [PubMed] [Google Scholar]
- 11.Drissen, R., R. J. Palstra, N. Gillemans, E. Splinter, F. Grosveld, S. Philipsen, and W. de Laat. 2004. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 18:2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engel, J. D., and K. Tanimoto. 2000. Looping, linking, and chromatin activity: new insights into beta-globin locus regulation. Cell 100:499-502. [DOI] [PubMed] [Google Scholar]
- 13.Fantoni, A., A. Bank, and P. A. Marks. 1967. Globin composition and synthesis of hemoglobins in developing fetal mice erythroid cells. Science 157:1327-1329. [DOI] [PubMed] [Google Scholar]
- 14.Francastel, C., W. Magis, and M. Groudine. 2001. Nuclear relocation of a transactivator subunit precedes target gene activation. Proc. Natl. Acad. Sci. USA 98:12120-12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser, P., and F. Grosveld. 1998. Locus control regions, chromatin activation and transcription. Curr. Opin. Cell Biol. 10:361-365. [DOI] [PubMed] [Google Scholar]
- 16.Geyer, R. K., Z. K. Yu, and C. G. Maki. 2000. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat. Cell Biol. 2:569-573. [DOI] [PubMed] [Google Scholar]
- 17.Haar, J. L., and G. A. Ackerman. 1971. A phase and electron microscopic study of vasculogenesis and erythropoiesis in the yolk sac of the mouse. Anat. Rec. 170:199-223. [DOI] [PubMed] [Google Scholar]
- 18.Hodge, D., E. Coghill, J. Keys, T. Maguire, B. Hartmann, A. McDowall, M. Weiss, S. Grimmond, and A. Perkins. 2006. A global role for EKLF in definitive and primitive erythropoiesis. Blood 107:3359-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu, C. T., C. Y. Ting, C. J. Ting, T. Y. Chen, C. P. Lin, J. Whang-Peng, and J. Hwang. 2000. Vaccination against gonadotropin-releasing hormone (GnRH) using toxin receptor-binding domain-conjugated GnRH repeats. Cancer Res 60:3701-3705. [PubMed] [Google Scholar]
- 20.Im, H., J. A. Grass, K. D. Johnson, S. I. Kim, M. E. Boyer, A. N. Imbalzano, J. J. Bieker, and E. H. Bresnick. 2005. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc. Natl. Acad. Sci. USA 102:17065-17070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jahn, C. L., C. A. Hutchison III, S. J. Phillips, S. Weaver, N. L. Haigwood, C. F. Voliva, and M. H. Edgell. 1980. DNA sequence organization of the beta-globin complex in the BALB/c mouse. Cell 21:159-168. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, K. D., H. M. Christensen, B. Zhao, and E. H. Bresnick. 2001. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell 8:465-471. [DOI] [PubMed] [Google Scholar]
- 23.Kadam, S., G. S. McAlpine, M. L. Phelan, R. E. Kingston, K. A. Jones, and B. M. Emerson. 2000. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 14:2441-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiekhaefer, C. M., J. A. Grass, K. D. Johnson, M. E. Boyer, and E. H. Bresnick. 2002. Hematopoietic-specific activators establish an overlapping pattern of histone acetylation and methylation within a mammalian chromatin domain. Proc. Natl. Acad. Sci. USA 99:14309-14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, A., and A. Dean. 2004. Developmental stage differences in chromatin subdomains of the beta-globin locus. Proc. Natl. Acad. Sci. USA 101:7028-7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kingsley, P. D., J. Malik, R. L. Emerson, T. P. Bushnell, K. E. McGrath, L. A. Bloedorn, M. Bulger, and J. Palis. 2006. “Maturational” globin switching in primary primitive erythroid cells. Blood 107:1665-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kochel, I., and L. Strzadala. 2004. [FK506-binding proteins in the regulation of transcription factors activity in T cells]. Postepy Hig Med. Dosw. 58:118-127. (In Polish.) (Online.) [PubMed] [Google Scholar]
- 28.Köster, M., S. Lykke-Andersen, Y. A. Elnakady, K. Gerth, P. Washausen, G. Hofle, F. Sasse, J. Kjems, and H. Hauser. 2003. Ratjadones inhibit nuclear export by blocking CRM1/exportin 1. Exp. Cell Res. 286:321-331. [DOI] [PubMed] [Google Scholar]
- 29.Leder, A., H. I. Miller, D. H. Hamer, J. G. Seidman, B. Norman, M. Sullivan, and P. Leder. 1978. Comparison of cloned mouse alpha- and beta-globin genes: conservation of intervening sequence locations and extragenic homology. Proc. Natl. Acad. Sci. USA 75:6187-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, Q., K. R. Peterson, X. Fang, and G. Stamatoyannopoulos. 2002. Locus control regions. Blood 100:3077-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, Y. C., C. Lee, D. Sanoudou, T. H. Hseu, S. Y. Li, and C. C. Lin. 2000. Interstitial colocalization of two cervid satellite DNAs involved in the genesis of the Indian muntjac karyotype. Chromosome Res. 8:363-373. [DOI] [PubMed] [Google Scholar]
- 32.Liu, J. J., S. C. Hou, and C. K. Shen. 2003. Erythroid gene suppression by NF-κB. J. Biol. Chem. 278:19534-19540. [DOI] [PubMed] [Google Scholar]
- 33.McMorrow, T., A. van den Wijngaard, A. Wollenschlaeger, M. van de Corput, K. Monkhorst, T. Trimborn, P. Fraser, M. van Lohuizen, T. Jenuwein, M. Djabali, S. Philipsen, F. Grosveld, and E. Milot. 2000. Activation of the beta globin locus by transcription factors and chromatin modifiers. EMBO J. 19:4986-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, I. J., and J. J. Bieker. 1993. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Krüppel family of nuclear proteins. Mol. Cell. Biol. 13:2776-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nudel, U., J. E. Salmon, M. Terada, A. Bank, R. A. Rifkind, and P. A. Marks. 1977. Differential effects of chemical inducers on expression of beta globin genes in murine erythroleukemia cells. Proc. Natl. Acad. Sci. USA 74:1100-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nuez, B., D. Michalovich, A. Bygrave, R. Ploemacher, and F. Grosveld. 1995. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 375:316-318. [DOI] [PubMed] [Google Scholar]
- 37.Osborne, C. S., L. Chakalova, K. E. Brown, D. Carter, A. Horton, E. Debrand, B. Goyenechea, J. A. Mitchell, S. Lopes, W. Reik, and P. Fraser. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36:1065-1071. [DOI] [PubMed] [Google Scholar]
- 38.Palis, J., S. Robertson, M. Kennedy, C. Wall, and G. Keller. 1999. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 126:5073-5084. [DOI] [PubMed] [Google Scholar]
- 39.Pandya, K., and T. M. Townes. 2002. Basic residues within the Kruppel zinc finger DNA binding domains are the critical nuclear localization determinants of EKLF/KLF-1. J. Biol. Chem. 277:16304-16312. [DOI] [PubMed] [Google Scholar]
- 40.Perkins, A. C., K. M. Gaensler, and S. H. Orkin. 1996. Silencing of human fetal globin expression is impaired in the absence of the adult beta-globin gene activator protein EKLF. Proc. Natl. Acad. Sci. USA 93:12267-12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perkins, A. C., A. H. Sharpe, and S. H. Orkin. 1995. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 375:318-322. [DOI] [PubMed] [Google Scholar]
- 42.Quadrini, K. J., and J. J. Bieker. 2002. Kruppel-like zinc fingers bind to nuclear import proteins and are required for efficient nuclear localization of erythroid Kruppel-like factor. J. Biol. Chem. 277:32243-32252. [DOI] [PubMed] [Google Scholar]
- 43.Saitoh, H., R. Pu, M. Cavenagh, and M. Dasso. 1997. RanBP2 associates with Ubc9p and a modified form of RanGAP1. Proc. Natl. Acad. Sci. USA 94:3736-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sawado, T., J. Halow, M. A. Bender, and M. Groudine. 2003. The beta-globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev. 17:1009-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sawado, T., K. Igarashi, and M. Groudine. 2001. Activation of beta-major globin gene transcription is associated with recruitment of NF-E2 to the beta-globin LCR and gene promoter. Proc. Natl. Acad. Sci. USA 98:10226-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shiels, C., S. A. Islam, R. Vatcheva, P. Sasieni, M. J. Sternberg, P. S. Freemont, and D. Sheer. 2001. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J. Cell Sci. 114:3705-3716. [DOI] [PubMed] [Google Scholar]
- 47.Shyu, Y.-C., T.-L. Lee, C.-Y. Ting, S.-C. Wen, L.-J. Hsieh, Y.-C. Li, J.-L. Hwang, C.-C. Lin, and C.-K. J. Shen. 2005. Sumoylation of p45/NF-E2: nuclear positioning and transcriptional activation of the mammalian beta-like globin gene locus. Mol. Cell. Biol. 25:10365-10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shyu, Y. C., S. C. Wen, T. L. Lee, X. Chen, C. T. Hsu, H. Chen, R. L. Chen, J. L. Hwang, and C. K. Shen. 2006. Chromatin-binding in vivo of the erythroid kruppel-like factor, EKLF, in the murine globin loci. Cell Res. 16:347-355. [DOI] [PubMed] [Google Scholar]
- 49.Southwood, C. M., K. M. Downs, and J. J. Bieker. 1996. Erythroid Kruppel-like factor exhibits an early and sequentially localized pattern of expression during mammalian erythroid ontogeny. Dev. Dyn. 206:248-259. [DOI] [PubMed] [Google Scholar]
- 50.Strunze, S., L. C. Trotman, K. Boucke, and U. F. Greber. 2005. Nuclear targeting of adenovirus type 2 requires CRM1-mediated nuclear export. Mol. Biol. Cell 16:2999-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Surinya, K. H., T. C. Cox, and B. K. May. 1997. Transcriptional regulation of the human erythroid 5-aminolevulinate synthase gene. Identification of promoter elements and role of regulatory proteins. J. Biol. Chem. 272:26585-26594. [DOI] [PubMed] [Google Scholar]
- 52.Terui, Y., N. Saad, S. Jia, F. McKeon, and J. Yuan. 2004. Dual role of sumoylation in the nuclear localization and transcriptional activation of NFAT1. J. Biol. Chem. 279:28257-28265. [DOI] [PubMed] [Google Scholar]
- 53.Tewari, R., N. Gillemans, M. Wijgerde, B. Nuez, M. von Lindern, F. Grosveld, and S. Philipsen. 1998. Erythroid Kruppel-like factor (EKLF) is active in primitive and definitive erythroid cells and is required for the function of 5′HS3 of the beta-globin locus control region. EMBO J. 17:2334-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trimborn, T., J. Gribnau, F. Grosveld, and P. Fraser. 1999. Mechanisms of developmental control of transcription in the murine alpha- and beta-globin loci. Genes Dev. 13:112-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vieira, K. F., P. P. Levings, M. A. Hill, V. J. Crusselle, S. H. Kang, J. D. Engel, and J. Bungert. 2004. Recruitment of transcription complexes to the beta-globin gene locus in vivo and in vitro. J. Biol. Chem. 279:50350-50357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitelaw, E., S. F. Tsai, P. Hogben, and S. H. Orkin. 1990. Regulated expression of globin chains and the erythroid transcription factor GATA-1 during erythropoiesis in the developing mouse. Mol. Cell. Biol. 10:6596-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whitney, J. B., III. 1977. Differential control of the synthesis of two hemoglobin beta chains in normal mice. Cell 12:863-871. [DOI] [PubMed] [Google Scholar]
- 58.Wijgerde, M., J. Gribnau, T. Trimborn, B. Nuez, S. Philipsen, F. Grosveld, and P. Fraser. 1996. The role of EKLF in human beta-globin gene competition. Genes Dev. 10:2894-2902. [DOI] [PubMed] [Google Scholar]
- 59.Zhou, D., K. M. Pawlik, J. Ren, C. W. Sun, and T. M. Townes. 2006. Differential binding of erythroid Krupple-like factor to embryonic/fetal globin gene promoters during development. J. Biol. Chem. 281:16052-16057. [DOI] [PubMed] [Google Scholar]
- 60.Zhou, D., J. X. Ren, T. M. Ryan, N. P. Higgins, and T. M. Townes. 2004. Rapid tagging of endogenous mouse genes by recombineering and ES cell complementation of tetraploid blastocysts. Nucleic Acids Res. 32:e128. [DOI] [PMC free article] [PubMed] [Google Scholar]