Abstract
The WW domain-containing oxidoreductase (WWOX) spans the second most common fragile site of the human genome, FRA16D, located at 16q23, and its expression is altered in several types of human cancer. We have previously shown that restoration of WWOX expression in cancer cells suppresses tumorigenicity. To investigate WWOX tumor suppressor function in vivo, we generated mice carrying a targeted deletion of the Wwox gene and monitored incidence of tumor formation. Osteosarcomas in juvenile Wwox−/− and lung papillary carcinoma in adult Wwox+/− mice occurred spontaneously. In addition, Wwox+/− mice develop significantly more ethyl nitrosourea-induced lung tumors and lymphomas in comparison to wild-type littermate mice. Intriguingly, these tumors still express Wwox protein, suggesting haploinsuffiency of WWOX itself is cancer predisposing. These results indicate that WWOX is a bona fide tumor suppressor.
Keywords: common fragile site, FHIT, knockout, osteosarcoma, lung cancer
The WW domain-containing oxidoreductase (WWOX) encodes a 46-kDa protein that contains two WW domains and a short-chain dehydrogenase/reductase domain (1–3). The WWOX gene is altered by deletions or translocations in a large fraction of many cancer types including breast, prostate, esophageal, lung, stomach, and pancreatic carcinomas (2, 4–9). WWOX protein is lost or reduced in the majority of these cancers and in a large fraction of other cancer types (10, 11). WWOX spans the second most active common fragile sites, FRA16D (reviewed in ref. 12). Common fragile sites are large regions of profound genomic instability found in all individuals. Following partial inhibition of DNA synthesis, those regions show site-specific gaps or breaks on metaphase chromosomes. In addition, common fragile sites exhibit induction of sister chromatid exchange and show a high rate of translocations and deletions in somatic cell hybrids (13). Because FRA16D maps within regions of frequent loss of heterozygosity, and is associated with homozygous deletions in various adenocarcinomas and with chromosomal translocations in multiple myeloma (2), these rearrangements have been suggested to inactivate the WWOX gene. On the other hand, ectopic overexpression of WWOX in cancer cells lacking expression of endogenous WWOX results in significant growth inhibition and prevents the development of tumors in athymic nude mice (14, 15). In addition, we reported that restoration of WWOX expression in cancer cells results in caspase-mediated apoptosis (15). Thus, these data suggest that WWOX may act as a tumor suppressor
Biochemical and functional characterization of WWOX has shown that it interacts with proline-tyrosine rich motif-containing proteins. WWOX associates via its first WW domain with p73 and enhances p73-mediated apoptosis (16). WWOX also binds to the PPPY motif of AP2γ and ErbB4, established oncogenes in breast cancer (17, 18). Interestingly, WWOX suppresses the transcriptional ability of these target proteins by sequestering them in the cytoplasm (16–18). Taken together, accumulating evidence, both in cell culture and in nude mice, suggests that WWOX functions as a tumor suppressor, although no direct in vivo proof has yet been reported to verify WWOX as a bona fide tumor suppressor. To define the role of WWOX protein in cancer development, we generated mice carrying a targeted deletion of the Wwox gene. The murine Wwox locus is similar to its human homolog (19), spans the Fra8E1 common fragile site, and is highly conserved. Here, we demonstrate that the loss of both alleles of Wwox results in osteosarcomas in some early postnatal mice, whereas loss of one allele significantly increases the incidence of spontaneous and chemically induced tumors. Altogether, our results provide the first in vivo evidence of WWOX tumor suppressor function.
Results
Targeted Ablation of the Wwox Gene in Mice.
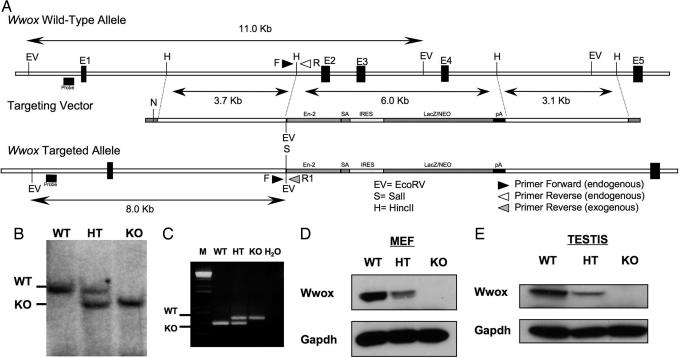
To investigate the role of the Wwox gene in vivo, we generated a targeting construct designed to disrupt mouse Wwox expression. The targeted allele replaced genomic sequences including exons 2, 3, and 4 of the Wwox gene (Fig. 1A). To identify homologous recombinants, electroporated embryonic stem (ES) clones were screened by Southern blot. Mice carrying the knockout (KO) allele (Wwox+/−) were established by blastocyst ES cell injection and germ-line transmission from chimeric founders. Progeny from Wwox+/− heterozygous (HET) intercrosses were genotyped by Southern blotting (Fig. 1B) and by PCR that generate a 590-bp product from the WT allele and a 665-bp product from the Wwox−/− allele (Fig. 1C). Genotype analysis of newborns obtained from a Wwox+/− intercross demonstrates the presence of all three genotypes with ratios consistent with the Mendelian distribution [Wwox−/− 131 (20%), n = 664]. By 4 weeks of age, 100% of the homozygous (KO) mice died, whereas HET pups were indistinguishable from wild-type (WT) littermate mice (data not shown).
Fig. 1.
Targeted disruption of the Wwox gene. (A) The Wwox genomic locus was altered by using a targeting vector that replaced ≈6 kb of genomic sequences that include exons 2, 3, and 4 (vertical dark boxes, E2, E3, E4) with a targeting cassette containing a chimeric sequence derived from the in-frame fusion of LacZ and Neo genes. A 5′ genomic probe (horizontal dark box) that recognized an ≈11-kb WT fragment and an ≈8-kb targeted EcoRV fragment was used for genotyping by Southern blotting. Three primers (arrows), a shared 5′ primer (F), distinct 3′ primers that recognized WT locus (R), and the other 3′ primers specific for LacZ-Neo sequence (R1) allowed PCR genotyping. (B) Southern blot analysis of genomic DNA extracted from mouse tails showing WT, HT, and homozygous (KO) mutant genotypes. (C) PCR genotype of DNA extracted from mouse tails showing the different genotypes. B6129F1-F4 mice were genotyped at age 14–21 days. M, marker. (D and E) Western blot analysis of mouse embryonic fibroblasts and testis with monoclonal anti-WWOX antibody indicating undetectable levels of Wwox in homozygous mutant mice. GAPDH levels were used for normalization.
To assess the impact of the mutant allele on Wwox protein levels, we isolated mouse embryonic fibroblast cells from E13.5 and carried out Western blot analysis using lysates from the three different genotypes and anti-WWOX antibody (16). As shown in Fig. 1D, we did not detect the endogenous 46-kDa Wwox protein in KO cells. To confirm that Wwox expression is absent in mouse tissues, we analyzed testis protein lysates by immunoblotting. As expected, no Wwox protein expression was detected in KO mice testis, whereas heterozygote (HET) mice displayed 50% or less of Wwox protein expression (Fig. 1E).
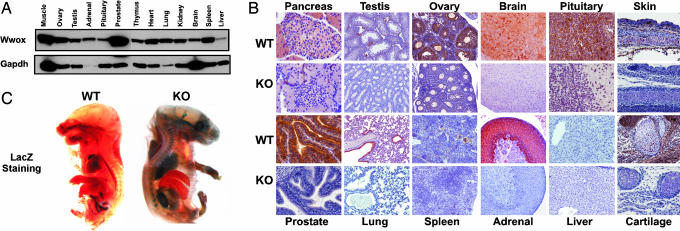
To gain insight into Wwox function, we characterized the tissue distribution patterns of Wwox expression. Analysis of Wwox protein revealed expression in most mouse tissues (Figs. 2 A and B), with particularly high levels in prostate, skeleton, lung, endocrine tissues, and brain. Wwox protein was predominantly cytoplasmic, as assessed by immunohistochemical staining with an anti-WWOX antibody (Fig. 2B). As expected, Wwox expression was absent in KO mice (Fig. 2B). Because our targeting vector included the LacZ gene (Fig. 1A), we monitored Wwox promoter activity in KO mice. Staining of 17.5-day whole embryo with X-Gal revealed that Wwox is ubiquitously expressed except in the liver (Fig. 2C).
Fig. 2.
Wwox expression pattern in mouse tissue. (A) Protein lysate from the indicated tissues was probed with monoclonal anti-WWOX antibody or GAPDH antibody as control. (B) Immunohistochemical staining of Wwox in main tissues from WT and KO mice with polyclonal anti-WWOX antibody, indicating high expression of Wwox in different epithelial tissues and in the endocrine system. Stained sections from pancreases, testis, ovary, pituitary, brain (brainstem), skin, prostate, lung, spleen, adrenal gland (cortex), liver, and cartilage primordium of the skull are shown. (C) LacZ staining of embryo from E17.5. E17.5 embryos were fixed in paraformaldehyde, stained with X-Gal, and analyzed by dissecting light microscope for gross morphology and LacZ expression (blue). Homozygous mutant embryo shows expression of LacZ in brain, lung, kidney, adrenal gland, stomach, and bones, whereas WT embryo is not stained.
Spontaneous Tumor Phenotype in Mutant Wwox Mice.
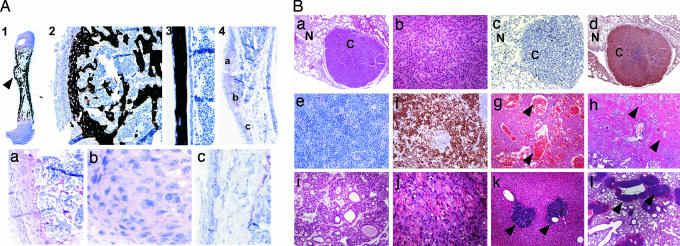
To study the tumor suppressor activity of Wwox in vivo, we examined both KO and HET mutant mice for a tumor-susceptibility phenotype. In the Wwox−/− mouse, we observed four mice from a total of 13 examined histologically having focal lesions along the diaphysis that appeared neoplastic at age day 3 and day 5 and two mice at age 2.5 weeks (Fig. 3A). The periosteal origin of the lesions and the very irregular immature trabeculae suggest a periosteal osteosarcoma (Fig. 3 A1 and A2 vs. A3). Although human periosteal osteosarcomas may contain cartilage tissue (20), the appearance of a cartilage cap underlying the periosteal proliferative cells in two tumors and a developing growth plate in two other lesions indicate the lesion may be a chondroid osteosarcoma (Fig. 3A4). Along the diaphysis of the KO mouse limbs containing the localized tumor, increased numbers of osteoblasts are observed, suggesting proliferation of the progenitors arising from the periosteum. Enlarged cells in cartilage matrix with multiple nucleoli suggest a transformed phenotype (Fig. 3A4, b). These tumors, formed in young postnatal mice in the absence of any carcinogenic treatment, indicate the importance of WWOX as a tumor suppressor in the bone.
Fig. 3.
Tumor phenotype in Wwox mutant mice. (A) Formation of osteosarcomas in Wwox null mice. Typical histological appearance is shown for one tumor (A1–A3, toluidine blue with von Kossa stain). Calcified chondroid tissue forms the diaphysis (A1, arrowhead) with immature trabeculae in the osteosarcoma (A2). A section of diaphysis of WT mouse from the same litter showing a single layer of osteoblasts constituting the endosteal and periosteal surfaces of cortical bone (A3). Decalcified femur bone, age 17 days (A4), reveals a cartilage cap under the periosteum with enlarged hypertrophic cells forming a growth plate (Aa). Proliferating cells adjacent to this lesion appear to be transformed (Ab). (Ac) Hyperproliferative periosteal cells along the diaphysis. (B) Histopathology of tumors in Wwox+/− mice. (Ba) Lung (H&E staining) nodule composed of epithelial proliferation with glandular and papillary growth pattern (bronchiolo-alveolar carcinoma). N (normal) versus C (carcinoma). (Magnification: × 200.) (Bb) Higher magnification (H&E staining) of the lung nodule shown in Ba, indicating marked nuclear and cellular atypia with rare atypical mitoses. (Magnification: ×400.) (Bc) Immunohistochemistry for Ki67 showing low proliferation rate in tumor cells in a lung nodule. (Magnification: ×200.) (Bd) Immunohistochemistry for WWOX showing positive expression in tumor cells in a lung nodule. (Magnification: ×100.) (Be) Spleen (H&E staining) monomorphic lymphopid population expanding the red pulp: diffuse large B cell lymphoma. (Magnification: ×400.) (Bf) Immunohistochemistry for Ki67 showing intense positivity in the lymphoblastic population in spleen. (Magnification: ×100.) (Bg) Hemangioma in liver (H&E staining). Arrows show large vascular channels. (Magnification: ×100.) (Bh) Large mass of malignant chondroblasts surrounded by chondroid material (arrowheads) in lung (H&E staining): chondrosarcoma. (Magnification: ×100.) (Bi) Fibroadenoma in mammary gland (H&E staining). (Magnification: ×100.) (Bj) Large mass of malignant squamous cells with incomplete keratinization in stomach (H&E staining): squamous cell carcinoma. (Magnification: ×400.) (Bk) Atypical B cell infiltrates (arrowheads) in liver (H&E staining). (Magnification: ×100.) (Bl) Perivascular atypical B cell infiltrates (arrowheads) in lung (H&E staining). (Magnification: ×40.)
As mentioned before, Wwox−/− mice die by 4 weeks postnatally precluding adult tumor analysis. Therefore, we monitored WT mice for spontaneous tumors. One hundred eighteen age-matched mice (60 Wwox+/+ and 58 Wwox+/−; average age 15 ± 1.5 months) were killed and autopsied. Tissues were then examined by gross pathology and histology. The number of tumor-bearing animals among Wwox+/− mice was 5-fold higher (P = 0.03) than in WT mice (Table 1). The incidence of tumors per mouse in Wwox+/− mice was 5-fold higher (P = 0.003) than WT littermates. Sixteen percent (9/58) of WT mice develop lung papillary carcinomas, whereas only 3.0% (2/60) of WT mice were diagnosed with lung tumors (P = 0.028). Multiplicity of total spontaneous tumors in HET versus WT mice was significant (P = 0.04), however multiplicity of lung tumors only was not significant (P = 0.09, Table 1). Lung tumor nodules were grossly visible, localized at the periphery of the pulmonary parenchyma, and composed of gland-like structures lined by tall columnar atypical epithelium actively proliferating and forming multiple branching intraluminal papillae, with rare atypical mitoses (Fig. 3 Ba and Bb). Immunohistochemistry for Ki67 (proliferation antigen) showed low positivity for all tumors (Fig. 3Bc), as expected. To examine the status of Wwox protein expression in those tumors, we performed immunohistochemistry with anti-WWOX antibody. We found that tumors from both WT and HET mice were positively stained for Wwox (Fig. 3Bd), suggesting that loss of one allele of Wwox is probably enough to predispose normal cells to malignant transformation.
Table 1.
Incidence of spontaneous and induced tumors in Wwox+/+ and Wwox+/+ mice
| Characteristic | Heterozygous | Wild type | P value |
|---|---|---|---|
| Spontaneous tumors | |||
| Number | 58 | 60 | |
| Total tumors | 19 | 4 | 0.04 |
| Tumors incidence | 10/58 (17.2%) | 2/60 (3.3%) | 0.03 |
| Tumor/mouse | 19/58 (0.33) | 4/60 (0.067) | 0.003 |
| Lung cancer incidence | 9/58 (15.5%) | 2/60 (3.3%) | 0.028 |
| Lung cancer multiplicity | 16 | 4 | 0.09 |
| ENU-induced tumors | |||
| Number | 46 | 42 | |
| Total tumors | 119 | 45 | <0.0001 |
| Tumor incidence | 37/46 (80%) | 20/42 (48%) | 0.002 |
| Lung cancer incidence | 33/46 (72%) | 15/42 (36%) | 0.001 |
| Lymphoma incidence | 27/46 (59%) | 13/42 (31%) | 0.01 |
| Others incidence | 9/46 (20%) | 1/42 (2.4%) | 0.015 |
| Lung tumor multiplicity* | 83 | 31 | 0.0004 |
| Lymphoma aggressiveness† | 63 | 32 | 0.03 |
To evaluate spontaneous tumors, 118 B6129F1-F4 mice (9–18 months of matched age) were sacrificed and autopsies were performed after CO2 asphyxiation. Tissues were fixed in 10% phosphate-buffered formalin and examined histologically after H&E staining for the presence of hyperplasia, adenomas, and carcinomas. Tumor incidence was analyzed by two-tailed Fisher's test, and lung tumor multiplicity was analyzed by one-way ANOVA. To evaluate ENU-induced tumors, 118 B6129F1-F4 mice (6–8 weeks of age) were given an intraperitoneal dose of ENU (20 mg/kg body weight). All mice were killed 40 weeks after ENU dose, and tissues were treated as above. Fourteen (23%) HET and 16 (28%) WT mice died during the course of the experiment.
*Lung multiplicity represents number of lung tumor nodules in each group.
†Lymphoma aggressiveness was calculated based on intensity of Ki67 staining. Tumor incidence was analyzed by two-tailed Fisher's test, and multiplicity and aggressiveness of tumors were analyzed by one-way ANOVA.
Of note, WT mice also spontaneously developed lymphomas. We found approximately the same incidence of large cell lymphoma in HET and WT mice and therefore did not tabulate them in our comparison. Interestingly, HET mice develop not only large cell lymphoma but also centroblastic lymphoma (10%) and Burkitts cell lymphoma (5%). A number of other tumors were found only in HET mice such as liver hemangioma and lymphoma infiltrations into internal organs such as livers and lungs (Fig. 3B).
Ethyl Nitrosourea (ENU)-Induced Tumors in Wwox+/− Mice.
To confirm the tumor suppressor activity of WWOX and learn about the tumor spectrum in Wwox-mutant mice under stress conditions, we investigated the possibility that loss of one Wwox allele may increase ENU-induced tumor development. ENU is a powerful chemical mutagen that permits the analysis of a spectrum of tumors in a given genetic background (21). One hundred eighteen mice (60 Wwox+/− and 58 Wwox+/+) at 6–8 weeks of age were injected intraperitoneally with a single dose of ENU (20 mg/kg). After 40 weeks, all mice were killed and their organs examined macroscopically and histologically. As shown in Table 1, the incidence of tumors, including lung cancer and lymphoblastic lymphomas in the HET mice, was significantly higher than in WT mice. In particular, 72% (33/46) of mice developed lung papillary carcinomas, whereas only 36% (15/42) of WT mice did (P = 0.0012). Furthermore, the number of lung tumors per mouse in HET mice was significantly higher than in WT mice (Table 1, P = 0.0004). Histology of lung tumors was very similar to those developed by the spontaneous tumor group, although tumors were larger in size (data not shown).
In addition, 59% (27/46) of ENU-treated WT mice presented visibly enlarged spleens, compared with 31% (13/42) of ENU-treated WT mice (Table 1, P = 0.01). Microscopically, the red pulp was greatly expanded by an atypical lymphoblastoid proliferation, positive for Ki67, gradually replacing the white pulp and, in some cases, completely replacing lymphoid follicles (Fig. 3 Be and Bf). Lymphoma aggressiveness was higher in WT mice where two thirds of these cases, compared with 30% in WT mice, featured effacement of the overall histological spleen architecture, with marked atypia of the malignant lymphoblasts, both hallmarks of an aggressive form of lymphoma (Table 1, P = 0.03). Flow cytometry analysis showed that the lymphoproliferation in spleen was mainly composed of B220+/IgM+ B cells [supporting information (SI) Fig. 4].
A number of other tumors were found only in Wwox+/− mice (Fig. 3 Bg–Bl), including liver hemangiomas (5/46, 11%), chondrosarcomas (1/46, 2%), fibroadenoma (1/46, 2%), squamous cell carcinomas (2/46, 4%), and lymphoma infiltrations into internal organs, mainly livers and lungs (4/46, 9%).
Discussion
Several reports have suggested that WWOX behaves as a tumor suppressor (4, 14). Recently, we have shown that ectopic expression of WWOX in lung cancer cells lacking expression of endogenous WWOX dramatically suppresses tumorigenicity in athymic nude mice (15). To further address the role of WWOX in carcinogenesis, we generated a mouse lacking expression of Wwox and examined whether Wwox mutant mice develop more tumors compared with WT mice. Targeted deletion of Wwox led to growth retardation and eventually to postnatal lethality. Blood chemistry analysis of homozygous mice showed marked alterations in serum levels of proteins, carbohydrates, and lipids (data not shown). Because no significant histological lesions in the main organs were observed, our analysis indicate that KO pups most likely suffered from severe metabolic defect (data not shown).
Given the phenotype of the KO, we also analyzed HET mice for tumor formation. Here, we provide the first in vivo evidence that Wwox functions as a tumor suppressor gene. The occurrence of osteosarcomas in bone of newborn KO mice and the increased incidence of spontaneous and induced tumors in ENU-treated Wwox+/− mice provide direct evidence that WWOX is a bona fide tumor suppressor. WWOX spans the second most common fragile site in the human genome, FRA16D. Fragile sites are genomic regions that are more prone to chromosomal alterations following treatment with specific chemicals or cultured under stress conditions. It has been suggested that loss of WWOX expression in several cancer types is due mainly to its location within FRA16D and might be a secondary event not associated with tumorigenesis. In our study, we clearly demonstrate that targeted deletion of Wwox in mice leads to tumor phenotype. This finding strongly suggests that loss of WWOX expression in human tumors may be a primary event selected by transformed cancer cells for growth advantage. The first tumor suppressor fragile gene that has been cloned was the FHIT gene spanning FRA3B (22). WWOX and FHIT share a number of similarities, and decrease or loss of their expression was reported in a number of common cancers (reviewed in ref. 12). Moreover, mice carrying one or two inactivated Fhit alleles (Fhit+/− or Fhit−/−) developed spontaneous tumors and showed increased incidence of induced tumors compared with WT mice (23). This suggests that mechanism of WWOX and FHIT inactivation could be similarly (e.g., carcinogen damage) contributing to cancer development.
High incidence of lung tumors formed either spontaneously or following stress conditions in Wwox+/− mice suggests that WWOX plays a pivotal role in lung carcinogenesis. Alteration of fragile genes, such as FHIT, has been linked to tobacco smoke carcinogens (24), therefore we may speculate that WWOX may also be prone to direct DNA damage due to carcinogens implicated in lung cancer. Several lines of evidence support the alteration of WWOX in human lung cancer. First, a high incidence of loss of heterozygosity was observed in nonsmall-cell lung carcinoma (36%) (7). Second, altered expression of WWOX in lung cancer can be due to epigenetic modifications, such as promoter hypermethylation (62%) (25). Third, WWOX protein expression is significantly reduced or absent in the vast majority of non-small-cell lung carcinoma (85%) (26). Fourth, restoration of WWOX expression in lung cancer cells induces apoptosis and prevents tumor growth in nude mice (15). Finally, we show here that Wwox+/− mice develop lung tumors more frequently than WT mice. These results suggest that WWOX plays an important role in human lung cancer etiology and might be a promising candidate for their gene therapy.
Following treatment with ENU, Wwox+/− mice develop tumors earlier than WT mice, and a statistically significant greater number of tumors per mouse is formed. Both lung tumor multiplicity and lymphoma incidence and aggressiveness were higher in the ENU-treated group compared with the spontaneous group (Table 1). In addition, a wide variety of tumors of epithelial origin were observed (Fig. 3B). Taken together, these results suggest that beyond loss of one Wwox allele, additional mutations are necessary for tumor formation and lead to more aggressive tumors. In this study, we also observed that a number of tumors in Wwox+/− mice showed Wwox protein expression (Fig. 3Bd). These results may suggest that WWOX may be a haploinsufficient tumor suppressor. It is thus possible that our tumor phenotype observed in HET mice represents a condition that frequently occurs because of the fragility of the WWOX locus and the high incidence of loss of heterozygosity observed in WWOX gene in many human cancer types. Therefore, Wwox+/− mice represent a good model system for the study of putative carcinogens and chemoprevention studies.
Our analyses of the Wwox-KO mouse phenotype demonstrated the occurrence of bone tumors. WWOX expression in the skeleton is detected in embryonic stages and by staining for LacZ (β-Gal activity) in embryo sections identified Wwox expression in the connective tissue tendon, bone and cartilage of limbs, vertebrae and calvaria (Fig. 2C and data not shown). Thirty-one percent of KO mice develop very early osteosarcomas. Although it is difficult to draw comparisons between the various types of human ostesarcomas and the mouse tumors (20), the KO mouse tumors have characteristics of chondroid osteosarcoma for which there is no mouse model (27). Nevertheless, our data clearly indicate that Wwox deletion predisposes to the development of osteosarcoma.
In conclusion, our findings demonstrate a direct evidence of WWOX tumor suppressor function and indicate further haploinsufficient characteristics.
Materials and Methods
Generation of the Wwox-Deficient Mice.
A 403-nt probe generated by PCR amplification from a 129/Svj mouse ES cells genomic DNA (primer pair: 5′-TCCTgTCACAgACTggggA-3′ and 5′-CTCATTAATCTCTgggTCCTg-3′) was used to screen a 129/Svj mouse genomic λ FIX-II phage library (Stratagene, La Jolla, CA). A phage containing an ≈15-kb insert was isolated and sequenced. Sequence analysis showed that this insert contained exons 2, 3, and 4 of Wwox. The Wwox gene targeting construct was generated by subcloning a 3.5-kb HincII/HincII fragment as the 5′ arm upstream of exon 2 and a 3.1-kb HincII/HincII fragment as the 3′ arm downstream of exon 4 into a pBluescript SK-vector (Stratagene). The resulting vector replaced nearly a 6.0-kb HincII/HincII fragment containing exons 2, 3, and 4 of Wwox with an ≈7.0-kb β-Geo cassette (28). The β-Geo cassette consists of mouse intronic sequences (En-2), a splice acceptor site, an internal ribosome entry site (IRES), lacZ fused in-frame with NeoR, and a poly(A) sequence (Fig. 1A). Thus, the resulting transcript driven by the Wwox promoter consists only of lacZ and Neo. The targeting vector was linearized with NotI and transfected by electroporation into 129/Svj ES cells. Selection with G418 yielded 65 clones. Genomic DNA was extracted from each clone, and 5 μg of DNA was digested with EcoRV and separated on a 0.8% agarose gel. Southern blot screening identified three homologous recombination clones. All three clones were injected into C57BL/6J blastocysts and implanted into foster mothers. Male chimeras derived from all three ES clones were selected by agouti color and were mated to C57BL/6J females to identify mice with germ-line transmission.
Genotyping.
ES cells, mouse tails, and tissues were genotyped by Southern blot or PCR-based methods. We generated a 190-bp probe by PCR amplification of an intronic sequence. Primers used for the production of this probe are the follows: (i) forward primer: 5′-ACCCTCCCAAATTCTggAgTC-3′; and (ii) reverse primer: 5′-gCATTTATTAgTATCCCgggCC-3′. This probe recognizes an ≈11-kb genomic fragment and an ≈8-kb fragment from the targeted allele after digestion with EcoRV. For PCR genotyping, three primers were used: a shared 5′ primer of the WT Wwox allele (WF, 5′-gCAgAATgTCTTgCTAgAgCTTTg-3′), a 3′ primer in sequence deleted in the targeted allele (WR, 5′-ATACTgACATCTgCCTCTAC-3′), and a 3′ primer internal to the β-Geo cassette (β-GeoR1, 5′-CAAAAgggTCTTTgAgCACCAgAg-3′) (Fig. 1A).
Histology and Immunohistochemistry.
Tissue from different organs were processed, embedded, and sectioned (4 μm) according to standard methods. Antibodies used for immunohistochemical staining were polyclonal anti-WWOX antibody (a gift from Kay Huebner, Ohio State University; dilution 1:8,000); and staining was performed as described in ref. 8; rat anti-mouse Ki67 antibody (Dako, Carpinteria, CA; dilution 1:500) was used as a marker for proliferation. The detection system used was Vectastain Elite (Vector Laboratories, Burlingame, CA). Detailed protocols are available upon request. For LacZ staining of whole mouse embryos, 17.5-day embryos were fixed in 2% paraformaldehyde and 0.2% gluteraldehyde-containing buffer for 2 h, washed with PBS, and stained with X-Gal (0.8 mg/ml). Bones were dissected for fixation in 4% paraformaldehyde for 24 h and either demineralized in 18% EDTA for paraffin embedding or embedded in methyl methacrylate for examination of sections of mineralized tissues. Sections were stained for mineral by using the von Kossa 3% silver nitrate stain, toluidine blue to distinguish cartilage and bone (Sigma, St. Louis, MO).
Immunoblot Analysis.
Cells were lysed by using Nonidet P-40 lysis buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 10% glycerol, 0.5% Nonidet P-40, and protease inhibitors. Lysates were resolved on SDS/PAGE. Antibodies used were mouse monoclonal anti-WWOX (16) and mouse monoclonal anti-GAPDH (Calbiochem, San Diego, CA).
Carcinogenesis Study.
One hundred eighteen 6- to 8-week-old mice (60 HET and 58 WT) were injected intraperitoneally with a single dose of ENU (Sigma), 20 mg/kg body weight. All of the mice were killed 40 weeks after ENU injection and examined for end-point tumor incidence. At autopsy, main organs including liver, spleen, stomach, intestine, colon, lung, heart, thymus, testis, ovary, and lymph nodes were examined for tumors. Tissues were fixed in 10% buffered formalin and examined histologically after H&E staining for the presence of dysplasia, adenomas, and carcinomas.
Statistical Analysis.
Tumor incidence differences were analyzed by two-tailed Fisher's exact test, and tumor multiplicity was analyzed by one-way ANOVA.
Supplementary Material
Acknowledgments
We thank Dr. Joanna Groden for suggestions and critical reading of the manuscript; and Lisa Rawahneh, Susie Jones, and Christina McKeegan (Histology Laboratory, Ohio State University) for technical assistance. This work was supported by Kimmel Scholar Award (to R.I.A.), National Cancer Institute grants (to C.M.C.), and National Institutes of Health Grants P01 CA082834 and P01 AR048818 (to G.S.S. and J.B.L.).
Abbreviations
- KO
knockout
- HET
heterozygous
- ENU
ethyl nitrsourea
- WWOX
WW domain-containing oxidoreductase
- ES
embryonic stem.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609783104/DC1.
References
- 1.Bednarek AK, Laflin KJ, Daniel RL, Liao Q, Hawkins KA, Aldaz CM. Cancer Res. 2000;60:2140–2145. [PubMed] [Google Scholar]
- 2.Ried K, Finnis M, Hobson L, Mangelsdorf M, Dayan S, Nancarrow JK, Woollatt E, Kremmidiotis G, Gardner A, Venter D, Baker E, Richards RI. Hum Mol Genet. 2000;9:1651–1663. doi: 10.1093/hmg/9.11.1651. [DOI] [PubMed] [Google Scholar]
- 3.Chang NS, Pratt N, Heath J, Schultz L, Sleve D, Carey GB, Zevotek N. J Biol Chem. 2001;276:3361–3370. doi: 10.1074/jbc.M007140200. [DOI] [PubMed] [Google Scholar]
- 4.Paige AJ, Taylor KJ, Taylor C, Hillier SG, Farrington S, Scott D, Porteous DJ, Smyth JF, Gabra H, Watson JE. Proc Natl Acad Sci USA. 2001;98:11417–11422. doi: 10.1073/pnas.191175898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuroki T, Trapasso F, Shiraishi T, Alder H, Mimori K, Mori M, Croce CM. Cancer Res. 2002;62:2258–2260. [PubMed] [Google Scholar]
- 6.Driouch K, Prydz H, Monese R, Johansen H, Lidereau R, Frengen E. Oncogene. 2002;21:1832–1840. doi: 10.1038/sj.onc.1205273. [DOI] [PubMed] [Google Scholar]
- 7.Yendamuri S, Kuroki T, Trapasso F, Henry AC, Dumon KR, Huebner K, Williams NN, Kaiser LR, Croce CM. Cancer Res. 2003;63:878–881. [PubMed] [Google Scholar]
- 8.Aqeilan RI, Kuroki T, Pekarsky Y, Albagha O, Trapasso F, Baffa R, Huebner K, Edmonds P, Croce CM. Clin Cancer Res. 2004;10:3053–3058. doi: 10.1158/1078-0432.ccr-03-0594. [DOI] [PubMed] [Google Scholar]
- 9.Kuroki T, Yendamuri S, Trapasso F, Matsuyama A, Aqeilan RI, Alder H, Rattan S, Cesari R, Nolli ML, Williams NN, et al. Clin Cancer Res. 2004;10:2459–2465. doi: 10.1158/1078-0432.ccr-03-0096. [DOI] [PubMed] [Google Scholar]
- 10.Park SW, Ludes-Meyers J, Zimonjic DB, Durkin ME, Popescu NC, Aldaz CM. Br J Cancer. 2004;91:753–759. doi: 10.1038/sj.bjc.6602023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pimenta FJ, Gomes DA, Perdigao PF, Barbosa AA, Romano-Silva MA, Gomez MV, Aldaz CM, De Marco L, Gomez RS. Int J Cancer. 2006;118:1154–1158. doi: 10.1002/ijc.21446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Keefe LV, Richards RI. Cancer Lett. 2006;232:37–47. doi: 10.1016/j.canlet.2005.07.041. [DOI] [PubMed] [Google Scholar]
- 13.Glover TW. Cancer Lett. 2006;232:4–12. doi: 10.1016/j.canlet.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 14.Bednarek AK, Keck-Waggoner CL, Daniel RL, Laflin KJ, Bergsagel PL, Kiguchi K, Brenner AJ, Aldaz CM. Cancer Res. 2001;61:8068–8073. [PubMed] [Google Scholar]
- 15.Fabbri M, Iliopoulos D, Trapasso F, Aqeilan RI, Cimmino A, Zanesi N, Yendamuri S, Han SY, Amadori D, Huebner K, Croce CM. Proc Natl Acad Sci USA. 2005;102:15611–15616. doi: 10.1073/pnas.0505485102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aqeilan RI, Pekarsky Y, Herrero JJ, Palamarchuk A, Letofsky J, Druck T, Trapasso F, Han SY, Melino G, Huebner K, Croce CM. Proc Natl Acad Sci USA. 2004;101:4401–4406. doi: 10.1073/pnas.0400805101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aqeilan RI, Palamarchuk A, Weigel RJ, Herrero JJ, Pekarsky Y, Croce CM. Cancer Res. 2004;64:8256–8261. doi: 10.1158/0008-5472.CAN-04-2055. [DOI] [PubMed] [Google Scholar]
- 18.Aqeilan RI, Donati V, Palamarchuk A, Trapasso F, Kaou M, Pekarsky Y, Sudol M, Croce CM. Cancer Res. 2005;65:6764–6772. doi: 10.1158/0008-5472.CAN-05-1150. [DOI] [PubMed] [Google Scholar]
- 19.Krummel KA, Denison SR, Calhoun E, Phillips LA, Smith DI. Genes Chromosomes Cancer. 2002;34:154–167. doi: 10.1002/gcc.10047. [DOI] [PubMed] [Google Scholar]
- 20.Murphey MD, Jelinek JS, Temple HT, Flemming DJ, Gannon FH. Radiology. 2004;233:129–138. doi: 10.1148/radiol.2331030326. [DOI] [PubMed] [Google Scholar]
- 21.Weber JS, Salinger A, Justice MJ. Genesis. 2000;26:230–233. [PubMed] [Google Scholar]
- 22.Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, et al. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 23.Zanesi N, Fidanza V, Fong LY, Mancini R, Druck T, Valtieri M, Rudiger T, McCue PA, Croce CM, Huebner K. Proc Natl Acad Sci USA. 2001;98:10250–10255. doi: 10.1073/pnas.191345898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sozzi G, Sard L, De Gregorio L, Marchetti A, Musso K, Buttitta F, Tornielli S, Pellegrini S, Veronese ML, Manenti G, et al. Cancer Res. 1997;57:2121–2123. [PubMed] [Google Scholar]
- 25.Iliopoulos D, Guler G, Han SY, Johnston D, Druck T, McCorkell KA, Palazzo J, McCue PA, Baffa R, Huebner K. Oncogene. 2005;24:1625–1633. doi: 10.1038/sj.onc.1208398. [DOI] [PubMed] [Google Scholar]
- 26.Donati V, Fontanini G, Dell'Omodarme M, Prati MC, Nuti S, Lucchi M, Mussi A, Fabbri M, Basolo F, Croce CM, et al. Clin Cancer Res. 2007;13:884–891. doi: 10.1158/1078-0432.CCR-06-2016. [DOI] [PubMed] [Google Scholar]
- 27.Ek ET, Dass CR, Choong PF. Crit Rev Oncol Hematol. 2006;60:1–8. doi: 10.1016/j.critrevonc.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 28.Wallace MJ, Batt J, Fladd CA, Henderson JT, Skarnes W, Rotin D. Nat Genet. 1999;21:334–338. doi: 10.1038/6866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.