Abstract
Cyclin E is a critical G1-S cell cycle regulator aberrantly expressed in bronchial premalignancy and lung cancer. Cyclin E expression negatively affects lung cancer prognosis. Its role in lung carcinogenesis was explored. Retroviral cyclin E transduction promoted pulmonary epithelial cell growth, and small interfering RNA targeting of cyclin E repressed this growth. Murine transgenic lines were engineered to mimic aberrant cyclin E expression in the lung. Wild-type and proteasome degradation-resistant human cyclin E transgenic lines were independently driven by the human surfactant C (SP-C) promoter. Chromosome instability (CIN), pulmonary dysplasia, sonic hedgehog (Shh) pathway activation, adenocarcinomas, and metastases occurred. Notably, high expression of degradation-resistant cyclin E frequently caused dysplasia and multiple lung adenocarcinomas. Thus, recapitulation of aberrant cyclin E expression as seen in human premalignant and malignant lung lesions reproduces in the mouse frequent features of lung carcinogenesis, including CIN, Shh pathway activation, dysplasia, single or multiple lung cancers, or presence of metastases. This article reports unique mouse lung cancer models that replicate many carcinogenic changes found in patients. These models provide insights into the carcinogenesis process and implicate cyclin E as a therapeutic target in the lung.
Keywords: lung carcinogenesis, lung cancer, sonic hedgehog
Cyclin E binds to and activates cyclin-dependent kinase 2 (Cdk2) and promotes G1 cell cycle transition (1, 2). Cyclin E overexpression shortens the G1 cell cycle, alters S-phase progression, and causes chromosomal instability (CIN) (3, 4). Cyclin E has oncogenic potential. Transgenic cyclin E expression in the mammary gland causes hyperplasia and carcinoma (5). Aberrant cyclin E expression occurs in premalignant lung lesions (6). Cyclin E expression has a negative prognostic impact in lung cancers (7–9). Tobacco carcinogens can transform immortalized human bronchial epithelial (HBE) cells and augment cyclin E expression (10). All-trans-retinoic acid (RA) chemoprevention represses cyclin E and associated Cdk2 kinase activity, causing G1 arrest (10). This would permit repair of genomic DNA damage by carcinogens and was proposed as a chemoprevention mechanism (10, 11).
Regulation of cyclin E is critical for cell cycle progression. Cyclin E accumulates late in G1 and declines through S phase (1, 2). Cyclin E is regulated by the ubiquitin–proteasome pathway (12). The ubiquitin ligase Cullin 3 promotes ubiquitination of free cyclin E not bound to Cdk2 (13). Ubiquitination of Cdk2-bound cyclin E depends on phosphorylation of threonines Thr-62 and -380 as well as Ser-372 and -384 (14–16). Phosphorylation of these residues allows cyclin E to be recognized by Fbw7 (hCdc4) (15, 17, 18), a phosphoepitope-specific substrate recognition component of the Skp1-Cullin1 F-box protein (SCF) ubiquitin ligase. Fbw7 mutations occur in malignancies and contribute to cell cycle deregulation (15, 18–21). Mutations of residues 62 and/or 380 stabilize cyclin E (14, 15, 22). Effects of wild-type (WT) and degradation-resistant (T62A/T380A) transgenic cyclin E expression were examined in this study. Notably, transgenic cyclin E recapitulates in the mouse many features of human lung carcinogenesis including CIN, sonic hedgehog (Shh) pathway activation, pulmonary dysplasia, single or multiple lung adenocarcinomas, and presence of metastases. The therapeutic implications of these findings are discussed.
Results
siRNA Targeting of Cyclin E.
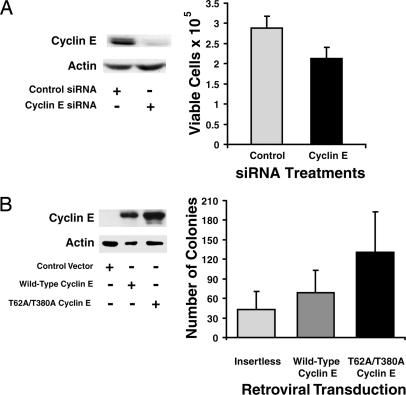
Targeting cyclin E with siRNAs repressed HBE cell growth. This targeting reduced cyclin E protein expression by 75% as compared with controls (Fig. 1A) and caused significant growth suppression (P < 0.0001; Fig. 1A). A second siRNA targeting a different cyclin E domain confirmed these results, and mock transfection did not appreciably affect cyclin E expression (data not shown).
Fig. 1.
Cyclin E effects on growth of pulmonary epithelial cells. (A) BEAS-2B cells were independently transiently transfected with cyclin E targeting or control siRNAs. (Left) Cyclin E protein expression was examined by immunoblot analyses 96 h after siRNA transfection. Actin expression served as a loading control. (Right) Viable cell numbers 96 h after transfection. Mean values and SDs from three independent experiments are shown. Significant difference was observed between control and cyclin E targeting siRNAs (P < 0.0001). (B) C10 cells were independently transduced with WT cyclin E, degradation-resistant cyclin E or an insertless control vector. (Left) Exogenous cyclin E expression was examined by immunoblot analysis. The degradation-resistant cyclin E species was stabilized. Actin expression served as a loading control. (Right) Anchorage-independent colonies ≥75 μm in diameter were scored at 2 weeks.
Cyclin E Transduction.
Exogenous cyclin E expression promotes HBE cell growth (10). To learn whether murine cells were similarly affected, C10 cells were transduced with a retrovirus encoding WT or degradation-resistant human cyclin E (hcyclin E) or an insertless control vector. Expression of degradation-resistant cyclin E was stabilized relative to WT cyclin E (Fig. 1B). An anti-cyclin E antibody was used for immunoblot analyses to identify exogenous human, but not endogenous murine, cyclin E. WT cyclin E transduced C10 cells increased anchorage independent growth relative to controls. An even greater increase followed transduction of degradation-resistant cyclin E (P = 0.001; Fig. 1B). These findings established a role for cyclin E in murine pulmonary epithelial cell growth.
Transgenic Cyclin E Expression.
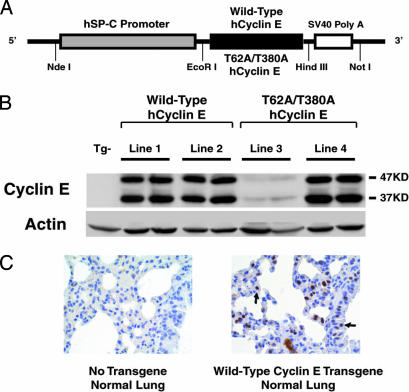
To explore cyclin E effects, murine transgenic lines were engineered to express in the lung WT or degradation-resistant hcyclin E (T62A/T380A). Transgenes were each driven by the human surfactant C (SP-C) promoter (Fig. 2A), which conferred expression in alveolar type II and bronchioalveolar cells (23). Two independent WT cyclin E and two independent degradation-resistant cyclin E founder mice were identified by Southern blot analyses (data not shown). Transgenic hcyclin E protein was detected by using an immunoblot assay with an antibody recognizing hcyclin E protein in all transgenic lines, but not in nontransgenic (Tg−) control mice (Fig. 2B). As expected from prior work (24), transgenic proteins migrated at 47Kd and 37Kd (Fig. 2B). These species were also present in human lung cancer cell lines A549 and H520 and other lines (data not shown).
Fig. 2.
Generation of human SP-C (hSP-C) driven WT cyclin E or degradation-resistant (T62A/T380A) hcyclin E transgenic lines. (A) The cyclin E transgenic constructs. Restriction endonuclease sites used for cloning these species are shown. (B) Exogenous hcyclin E immunoblot expression profiles in normal lung tissues of transgenic mice. Actin expression served as a loading control. (C) Immunohistochemical detection of hcyclin E in lung tissues of a representative WT (line 2) transgenic cyclin E mouse and a nontransgenic (Tg−) control mouse that did not stain. The left and right arrows in Right indicate hcyclin E nuclear staining in representative pneumocyte and bronchiole epithelial cells, respectively.
Transgenic cyclin E protein expression was comparable in WT cyclin E lines 1 and 2 and degradation-resistant cyclin E line 4 (Fig. 2B). The degradation-resistant cyclin E transgenic line 3 had low expression of transgenic protein (Fig. 2B). Immunohistochemical analyses revealed that hcyclin E expression was detected in nuclei of bronchiole epithelial cells and pneumocytes of WT cyclin E lines and degradation-resistant cyclin E transgenic line 4 (data not shown) but was undetected in the Tg− mouse lung (Fig. 2C). Nuclear localization of exogenous hcyclin E protein was confirmed (Fig. 2C). Cyclin E immunostaining was detected in line 3, but fewer cells stained than in other lines (data not shown).
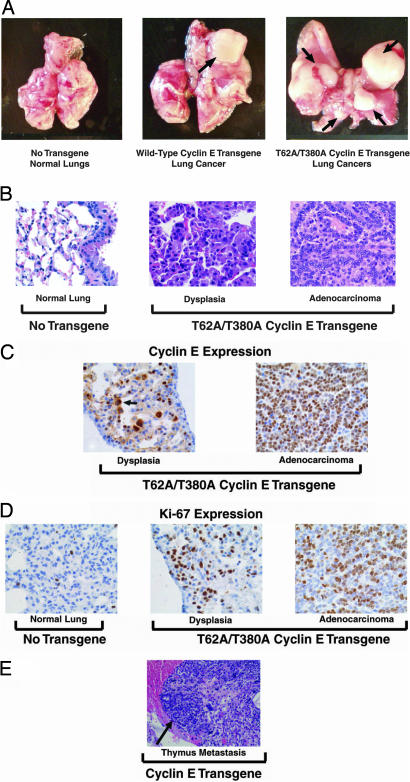
WT and proteasome-degradation-resistant cyclin E transgenic mice developed pulmonary premalignancy and malignancy (Fig. 3 A and B). Histopathological analyses revealed dysplasia alone or with lung adenocarcinoma in affected mice (Fig. 3B). Histopathologic features of dysplasia were similar to those in clinical lesions (6). Lung adenocarcinomas in these mice (Fig. 2B) resembled lesions found in clinical lung cancers (25).
Fig. 3.
Pathology and immunohistochemical expression of hcyclin E in transgenic lines. (A) Lungs from representative nontransgenic (Tg−) control (Left), WT cyclin E (line 2, Center), and degradation-resistant cyclin E (line 4, Right) transgenic mice are shown. Independent lung tumors are indicated by arrows. (B) Hematoxylin/eosin staining of lung tissue sections. Histopathologically normal lung tissue (Left) from a representative Tg− control mouse and dysplasia (Center) and adenocarcinoma (Right) from a representative degradation-resistant transgenic cyclin E line 4 mouse are shown. (C) Immunohistochemical detection of hcyclin E in dysplastic and malignant lung lesions from the same degradation-resistant cyclin E transgenic mouse in B. Arrow indicates hcyclin E nuclear staining in a cell from a dysplastic lesion. (D) Immunohistochemical detection of Ki-67 in a representative Tg− control mouse and the same lung lesions from the cyclin E line in B. (E) A representative metastasis (arrow depicts thymic metastasis) present in WT cyclin E transgenic line 2 is shown.
Incidences of dysplasia and lung cancer were each examined in cyclin E transgenic lines and control Tg− mice (Table 1). Significant sex differences in these incidences were not observed (P < 0.1). Tumors were observed in WT and degradation-resistant cyclin E transgenic lines as early as 5 months of age. Most cyclin E transgenic mice were euthanized at 12–14 months of age for analyses of dysplasia and lung cancer incidence. Mice with advanced tumors were euthanized for symptoms of labored breathing and decreased body weight.
Table 1.
Incidences of lung cancer, dysplasia, and multiple lung cancers in nontransgenic control and transgenic mice
| Line | N | Cancers |
Dysplasia |
Multiple cancers |
|||
|---|---|---|---|---|---|---|---|
| N (%) | P | N (%) | P | N (%) | P | ||
| Tg− | 48 | 4 (8.3) | — | 2 (4.2) | — | 0 (0) | — |
| Line 1 | 40 | 9 (22.5) | 0.067 | 6 (15) | 0.082 | 1 (2.5) | 0.37 |
| Line 2 | 16 | 9 (56.3) | 0.0002 | 1 (6.3) | 0.76 | 0 (0) | 1.0 |
| Line 3 | 42 | 7 (16.7) | 0.198 | 4 (9.5) | 0.27 | 1 (2.4) | 0.35 |
| Line 4 | 34 | 17 (50) | <0.0001 | 11 (32.3) | 0.001 | 7 (20.6) | 0.0026 |
Summary of incidences (number, N, and percentage) of lung cancer, dysplasia, and multiple lung cancers in nontransgenic (Tg−) control mice, WT cyclin E transgenic lines 1 and 2, and degradation-resistant cyclin E transgenic lines 3 and 4.
To confirm pulmonary dysplasia and tumors were increased by transgenic cyclin E expression in the murine FVB/N line, cohorts of age-matched WT (Tg−) control mice were evaluated for pulmonary dysplasia and lung tumor formation. Of 48 control mice, only 4 had lung cancers, and 2 others developed pulmonary dysplasia without cancer. In marked contrast, 18 of 56 transgenic WT cyclin E mice of lines 1 and 2 in this age range had lung adenocarcinomas. Six of them developed both pulmonary dysplasia and adenocarcinoma, and one mouse had dysplasia without adenocarcinoma. Seventeen of 34 degradation-resistant cyclin E transgenic line 4 mice developed lung adenocarcinomas, and 8 of these mice developed both pulmonary dysplasia and adenocarcinoma. Three other mice had pulmonary dysplasia without lung adenocarcinoma. Seven of 42 degradation-resistant cyclin E transgenic line 3 mice developed lung adenocarcinoma. WT cyclin E transgenic lines 1 and 2 had increased incidence of dysplasia as compared with control mice, but this was not statistically significant (Table 1). Of WT cyclin E transgenic mice, 22.5% (line 1) and 56.3% (line 2) had lung cancers as compared with 8.3% of Tg− control mice (Table 1). The degradation-resistant cyclin E transgenic line 4 displayed a substantially higher incidence of dysplasia than control mice (32.3% vs. 4.2%). This difference was significant (P = 0.001; Table 1).
Half of degradation-resistant cyclin E transgenic line 4 mice developed lung cancers (Table 1). Cancer incidence between line 4 and control mice was significantly different (P < 0.0001). Degradation-resistant cyclin E line 3 had much lower transgenic cyclin E expression relative to line 4 (Fig. 2B). Incidence of dysplasia in line 3 was higher than in control mice (9.5% vs. 4.2%) but was not statistically significant (Table 1). Similar results were observed for incidence of lung cancers (16.7% vs. 8.3%; Table 1). Dysplasia and lung cancer incidences for cyclin E lines 3 and 4 were statistically different (P = 0.013 and 0.001, respectively). Dysplasia incidence was highest in degradation-resistant cyclin E line 4 (32.3%) and was significantly different compared with control mice (P = 0.001). All of the cyclin E transgenic lines had increased dysplasia and lung cancer compared with Tg− controls.
Multiple lung cancer incidences were examined. All Tg− control mice with lung cancer had a single tumor (Table 1); only 2.5% (line 1) WT cyclin E transgenic mice and 2.4% (line 3) degradation-resistant cyclin E transgenic mice developed two lung cancers (Table 1). Strikingly, 20.6% of degradation-resistant cyclin E transgenic line 4 mice developed at least two lung adenocarcinomas (Table 1). Line 4 displayed a significant increase in incidence of multiple lung cancers relative to Tg− mice (P = 0.0026). Metastases to pleura, pulmonary lymph nodes or thymus were detected in lines 1 (one mouse), 2 (one mouse), and 4 (three mice). An example of a thymic metastasis appears in Fig. 3E. Metastases were not detected in Tg− mice.
The presence of increased transgenic cyclin E immunohistochemical expression in dysplastic and malignant lung relative to normal lung tissues implicated cyclin E involvement in murine lung carcinogenesis (Fig. 3C and data not shown). The Ki-67 immunohistochemical proliferation marker was readily detected in dysplasia and lung cancers from degradation-resistant cyclin E transgenic line 4 vs. normal lungs of Tg− control mice (Fig. 3D). Similar histopathology, hcyclin E and Ki-67 immunohistochemical expression profiles were detected in dysplastic and malignant lung lesions from WT cyclin E transgenic mice (data not shown).
Cyclin E expression causes genomic instability as does cyclin E stabilization through an alanine transversion of residue 380 (T380A) (4). This increased associated kinase activity and CIN (4). The degradation-resistant transgenic cyclin E species studied here is reported as more stable than cyclin E T380A (15). CIN was examined in lung cancers of WT and degradation-resistant cyclin E transgenic mice by FISH analyses using chromosome 4 and 6 probes. Aneuploidy of these chromosomes was frequently observed in lung adenocarcinoma lines (26). Aneuploid cells for each of these chromosome markers were prominent in lung cancers from transgenic cyclin E mice relative to adjacent normal lung tissues. This degree of aneuploidy was not seen in normal or malignant lung tissues from Tg− control mice (Table 2).
Table 2.
FISH analyses on paired lung cancers and normal lung tissues from nontransgenic and transgenic lines
| Line | Tissue | Chromosome 4 |
Chromosome 6 |
||
|---|---|---|---|---|---|
| N | >2, % | N | >2, % | ||
| Tg−-1 | Cancer | 100 | 0 | 25 | 0 |
| Normal | 157 | 0 | 75 | 0 | |
| Tg−-2 | Cancer | 50 | 0 | 50 | 2 |
| Normal | 160 | 0 | 150 | 0 | |
| Line 2 | Cancer | 85 | 100 | 143 | 88.8 |
| Normal | 100 | 0 | 50 | 0 | |
| Line 2 | Cancer | 100 | 27 | 176 | 91.5 |
| Normal | 250 | 0 | 55 | 9.1 | |
| Line 4 | Cancer | 188 | 86.2 | 91 | 78 |
| Normal | 100 | 1 | 50 | 0 | |
| Line 4 | Cancer | 150 | 100 | 100 | 100 |
| Normal | 241 | 0 | 125 | 0 | |
The FISH analyses independently performed by using chromosome 4 and 6 probes on paired lung cancers (cancer) and normal lung (normal) tissues from two different nontransgenic (Tg−-1 and Tg−-2) mice and different transgenic lines (line 2, WT cyclin E and line 4, degradation-resistant cyclin E). The number (N) and percentage (%) are displayed. Findings reveal that CIN prominently occurs in transgenic cyclin E lung cancers but not in normal lung tissues of transgenic and Tg− mice or in lung cancers arising in Tg− mice.
Hedgehog (Hh) Pathway Activation.
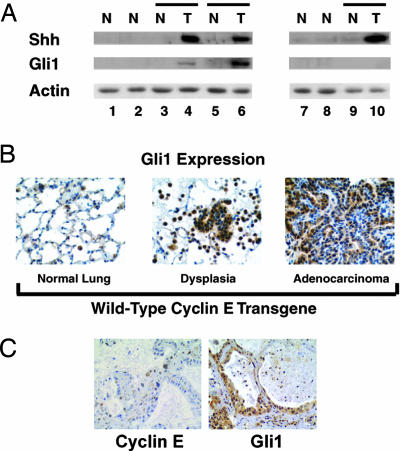
Not all cyclin E transgenic mice developed pulmonary dysplasia or malignancy. This indicated that cyclin E likely cooperated with other genetic alterations in lung carcinogenesis. Aberrant Hh signaling occurs in many cancers, including lung cancers (27–29). Whether this pathway was activated in dysplasia or lung cancer from cyclin E transgenic mice was examined by immunoblot analyses (Fig. 4A). Shh was overexpressed in seven of eight examined lung cancers from WT cyclin E transgenic mice (data not shown) and five of six lung tumors from degradation-resistant transgenic cyclin E mice, as compared with normal lung tissues from the same transgenic or control mice (Fig. 4A). Gli1, an Hh pathway transcriptional target (27), was overexpressed in four of eight examined WT transgenic cyclin E lung cancers and four of six lung cancers from degradation-resistant transgenic cyclin E mice (Fig. 4A and data not shown), relative to normal lung tissues from the same transgenic mice. Normal lungs from age- and sex-matched nontransgenic mice, WT cyclin E, and degradation-resistant cyclin E transgenic mice without evidence of lung adenocarcinomas were found to have low or undetected Shh and Gli1 protein expression (Fig. 4A and data not shown). Gli1 and Shh were often coexpressed in these tumors (Fig. 4A and data not shown). Of five examined cancers from degradation-resistant cyclin E transgenic mice, four had both Gli1 and Shh overexpression, and one had only Shh overexpression. These findings were confirmed by an immunohistochemical assay (Fig. 4B) establishing that activation of Gli1 expression occurs within dysplastic and malignant lung lesions of transgenic cyclin E mice.
Fig. 4.
Activation of the Shh pathway in cyclin E transgenic mice and comparison with human adenomatous hyperplasia. (A) Shh, Gli1, and actin immunoblot expression profiles in lung tissues from Tg− (control), WT transgenic cyclin E (data not shown), and degradation-resistant transgenic cyclin E mice. Two representative age- and sex-matched groups are shown. Lanes 1 and 7, normal (N) lung tissues from Tg− control mice; lanes 2 and 8, normal (N) lung tissues from transgenic cyclin E line 4 mice; lanes 3 and 4, a pair (depicted by line) of normal (N) lung and malignant (T) lung tissues of a representative transgenic cyclin E line 4 mouse. Comparisons appear for paired specimens from other transgenic cyclin E line 4 mice in lanes 5 and 6 and in lanes 9 and 10. Shh and Gli1 immunoblot expression was observed in malignant lung tissues from WT transgenic cyclin E lines (data not shown). Actin expression served as a loading control. (B) Gli1 findings in B were independently confirmed by a Gli1 immunohistochemical assay on tissues from WT and degradation-resistant (data not shown) cyclin E transgenic lines. Increased Gli1 expression (relative to normal lung) appears in dysplasia and lung adenocarcinoma from the depicted cyclin E transgenic line 2. (C) Immunohistochemical detection of cyclin E (Left) and Gli1 (Right) in a human adenomatous hyperplasia lesion.
Whether cyclin E overexpression activated the Shh pathway in C10 cells was examined. RT-PCR assays were used to detect Shh pathway members Smoothened (Smo), Gli1, and Patched 1 (Ptch1) in C10 cells transduced with WT or degradation-resistant hcyclin E. Findings were compared with insertless vector-transduced C10 cells. An increase in mRNA expression for Smo, Gli1, and Ptch1 was observed after exogenous cyclin E expression. Smo activates the Hh pathway through the Gli1 transcription factor (27). Ptch1, a transmembrane receptor, when bound to Hh ligands, blocks Smo function (27). Both Ptch1 and Gli1 are transcriptional targets of Hh signaling (27). Expression for Smo, Gli1, and Ptch1 increased in C10 cells transduced with WT or degradation-resistant hcyclin E, relative to controls (see supporting information (SI) Fig. 5). Shh mRNA was undetected in these cells.
Whether results from these transgenic models predicted findings in clinical lung carcinogenesis was examined. A series of 101 lung cancer cases (including 45 adenocarcinomas) revealed that >90% of these cases had Gli1 immunostaining (29). Nine of these lung cancers and adjacent normal lung were immunostained for hcyclin E with the same antibody used in analyses of these transgenic lines. Each case overexpressed cyclin E relative to adjacent normal lung tissues (data not shown). Studies were extended to the premalignant lesion, adenomatous hyperplasia. A representative lesion was independently immunostained for cyclin E and Gli1; both species were detected in Fig. 4C.
Discussion
Cyclin E, an important cell cycle regulator, is deregulated in pulmonary dysplasia and malignancy; it confers a poor prognosis in lung cancer (1–4, 6–9). Findings reported here reveal retroviral transduction of cyclin E promoted growth of pulmonary epithelial cells; targeting cyclin E expression had the opposite effect (Fig. 1). Murine transgenic lines driven by the SP-C promoter expressed WT or proteasome-degradation-resistant cyclin E species. These lines exhibited CIN, pulmonary dysplasia, single or multiple lung adenocarcinomas, Shh pathway activation, and presence of metastases (Tables 1 and 2 and Figs. 3 and 4). Incidences of dysplasia and multiple lung cancers were highest in degradation-resistant cyclin E line 4, as shown in Figs. 3 and 4 and Tables 1 and 2. Aberrant expression for both cyclin E and the Shh target gene Gli1 were found in dysplastic and malignant transgenic lung tissues. This provided a basis to search for and find similar expression patterns in human premalignant (Fig. 4C) and malignant lung tissues (data not shown).
Lung cancer is the leading cause of cancer mortality for men and women (30). There is a need to develop transgenic lines that mimic key features of clinical lung carcinogenesis to better understand lung cancer biology and molecular therapeutics. This study was undertaken to reproduce in the mouse lung aberrant cyclin E expression. This is frequently found in human premalignant and malignant lung lesions; this study confirmed and extended prior work (6). This carcinogenesis program occurred without exposure of mice to a tobacco carcinogen. Multiple genetic changes exist in human lung cancers (30). CIN was hypothesized to confer other carcinogenic events such as Shh pathway activation as in lung cancers (27–29).
Findings observed in engineered lung cancer models provide a rationale to search for similar changes in human lung carcinogenesis. This was the case for these new transgenic cyclin E lines that exhibited CIN as well as Gli1 activation in dysplastic and malignant lung. Single and multiple lung cancers arose with kinetics consistent to that seen in the clinic. These transgenic lung lesions also had histopathology similar to those found in patients (6). Other genetically engineered mouse lung cancer models exist, involving p53, ras, c-raf1, RON receptor tyrosine kinase, and other alterations, as reviewed in ref. 31. The transgenic lines described in this study are distinct from prior models because these have reproduced in the mouse a frequent single genetic change found in human premalignant and malignant lung lesions, aberrant cyclin E expression. This causes CIN, Shh pathway activation, dysplasia, and individual or multiple lung cancers. Metastasis is not routinely reported in transgenic mice (31) but occurs in human lung cancers. This was detected in some transgenic cyclin E lines. In contrast to prior transgenic models, these transgenic cyclin E lines reproduce many features of human lung carcinogenesis. These lines thus are useful tools to explore the biology of lung cancer.
These transgenic findings implicate cyclin E as an important regulator of lung carcinogenesis. What needs to be learned is whether cyclin E is a molecular therapeutic target in the lung. Future work will determine whether these models are useful to assess efficacy of antineoplastic agents affecting cyclin E or other oncogenic targets in lung cancer. That work will be aided by preliminary findings showing that MRI can detect transgenic cyclin E tumors (data not shown). Preclinical studies would help prioritize clinical testing of antineoplastic agents. Treatment of these transgenic lines with cyclin E targeting agents would establish whether persistent cyclin E expression or activity is required for maintenance or progression of dysplastic or malignant lung lesions. Future studies could uncover stem cell progenitors for these transgenic lung tumors and pathways in addition to the Hh pathway that cooperate with cyclin E to form lung cancers. Given the high incidence of dysplasia in transgenic line 4 mice, this line would be a useful model for future chemoprevention studies.
In summary, findings reported here establish a role for cyclin E in lung carcinogenesis. Cyclin E transgenic lines reproduce in the mouse many features of human lung carcinogenesis, including CIN, Hh pathway activation, dysplasia, single or multiple lung adenocarcinomas, and even metastases. Inhibition of cyclin E proteasome degradation enhanced neoplastic changes in transgenic lines. The precise mechanism responsible for cyclin E recapitulating these aspects of lung carcinogenesis needs to be discerned. This insight would provide a basis for targeting cyclin E to confer therapeutic or chemopreventive effects in the lung.
Materials and Methods
Cell Culture and Recombinant Plasmids.
BEAS-2B immortalized HBE cells were cultured in serum-free LHC-9 medium (Biofluids, Rockville, MD) as described (32). Nontumorigenic murine C10 alveolar type II epithelial cells (33) were cultured in CMRL 1066 medium (Life Technologies, Grand Island, NY) supplemented with 10% FBS, 2 mM l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. The GP2-293 packaging cell line was purchased from BD Bioscience Clontech (Palo Alto, CA) and cultured in DMEM (BD Bioscience Clontech). Cell lines were cultured at 37°C in a humidified incubator with 5% CO2. The pVSV-G expression vector was purchased from BD Bioscience Clontech. Full-length cDNAs for myc-tagged human WT and degradation-resistant (T62A/T380A) cyclin E were each engineered by DraI/EcoRV restriction endonuclease digestions of respective cyclin E plasmids (11) and independently ligated into the HpaI restriction site of the pMSCVhyg retroviral vector (BD Bioscience Clontech).
Plasmid Transfection and Retrovirus Infection.
The pMSCVhyg, pMSCVhyg-myc-cyclin E, and pMSCVhyg-myc-T62A/T380A cyclin E retroviral vectors were independently cotransfected with pVSV-G into GP2–293 cells using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) and manufacturer's techniques. Two days after transfection, medium were independently used for C10 cell viral transductions. Transductants were selected with hygromycin B (Sigma, St. Louis, MO).
siRNA Experiments.
An siRNA was synthesized (Qiagen, Valencia, CA) and designed to target the hcyclin E coding region with the sequence 5′-AAGTGCTACTGCCGCAGTATC-3′. The GL2 siRNA (Dharmacon, Lafayette, CO) served as a control. Transfection was performed as described (32). Viable BEAS-2B cells were measured by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (34). Three independent triplicate experiments were performed. A second cyclin E siRNA targeted a different cyclin E coding region: 5′-CAGTGGTGCGACATAGAGAAC-3′.
Anchorage-Independent Clonal Growth Assay.
Anchorage-independent clonal growth assays were performed as described (35) with 5 × 104 C10 cells plated onto each well of a six-well plate. Colonies ≥75 μm in size were scored in three independent triplicate experiments, with results pooled.
Cyclin E Transgenic Lines.
Human SP-C driven untagged WT and degradation-resistant hcyclin E (T62A/T380A) cDNAs were independently engineered by EcoRI and HindIII restriction endonuclease digestions of respective cyclin E plasmids (11) before insertion into the SP-C-3.7-SV40 vector. For microinjection, cyclin E constructs were each digested by NedI/NotI restriction endonucleases and agarose gel purified. Transgenic lines were engineered in FVB/N mice (The Jackson Laboratory, Bar Harbor, ME) using established techniques (36). Transgenic integration was confirmed by Southern blot analyses (37) of genomic DNA from tails of individual mice. Mice were screened for the transgene by using a PCR assay (data not shown) (32).
Histopathology and Immunohistochemistry.
Lung tissues were formalin-fixed, sectioned, and processed for immunostaining by using standard techniques (25). Tissues were incubated with a murine monoclonal anti-cyclin E antibody recognizing hcyclin E protein (HE-12; Neomarkers, Fremont, CA), a rabbit monoclonal Ki-67 antibody (Vector Laboratories, Burlingame, CA), or a rabbit polyclonal anti-Gli1 antibody (Abcam, Cambridge, MA). Hematoxylin counterstaining was used. Control tissues stained appropriately. Analyses were performed by a pathologist who was unaware of whether tissues were transgenic.
Immunoblot Analyses.
Protein extracts were isolated for immunoblot analyses by using established techniques (32). Independent primary antibodies included a murine monoclonal antibody recognizing hcyclin E (HE-12; Santa Cruz Biotechnology, Santa Cruz, CA), goat polyclonal antibody recognizing actin (C11; Santa Cruz Biotechnology), rabbit polyclonal antibodies for Shh (H-160; Santa Cruz Biotechnology), and Gli1 (Abcam). Anti-murine and anti-rabbit antisera were purchased from Amersham Biosciences (Piscataway, NJ), and anti-goat antisera were purchased from Santa Cruz Biotechnology.
FISH.
FISH analyses were performed by using paraffin-embedded tissue sections from transgenic cyclin E and control mice according to the manufacturer's methods (Cambio, Cambridge, U.K.). Murine chromosome 4 and 6 specific probes were purchased (Cambio) and independently analyzed.
Statistical Analyses.
Retroviral transduction and siRNA growth assays were expressed as mean ± SD. A two-sided test on proportion for group comparison (38) was used for analyses of dysplasia and single or multiple lung tumors with the S-Plus 6.1 statistical package (Insightful Inc., Seattle, WA). Statistical significance was considered for P < 0.05.
RT-PCR Assays.
Total cellular RNA was isolated by using the RNeasy Protect Mini Kit (Qiagen) or TRI Reagent (Molecular Research Center, Cincinnati, OH). Contaminating DNA was removed by using a DNA-free kit (Ambion, Austin, TX). RT-PCR assays were performed by using established methods (32) with the following primers: Smo forward, 5′-AGATTGTTTGCCGAGCAGAT-3′, and reverse, 5′-GTGAGGACAAAGGGGAGTGA-3′; Gli1 forward, 5′-CCTGGTGGCTTTCATCAACT-3′, and reverse, 5′-GCTAGACATGTCCCCTTCCA-3′; Ptch1 forward, 5′-TACGTGGAGGTGGTTCATCA-3′, and reverse, 5′-CCTGAGTTGTCGCAGCATTA-3′; GAPDH forward, 5′-AACTTTGGCATTGTGGAAGG-3′, and reverse, 5′-ACACATTGGGGGTAGGAACA-3′; and Shh forward, 5′-TTAAATGCCTTGGCCATCTC-3′, and reverse, 5′-CCACGGAGTTCTCTGCTTTC-3′.
Supplementary Material
Acknowledgments
We thank Dr. Jeffrey A Whitsett (Cincinnati Children's Hospital Medical Center, Cincinnati, OH) for providing the SP-C promoter; the Transgenic Core Facility of Dartmouth Medical School as well as Ms. Jennifer Fields, Ms. Laurie Horne, and Ms. Sandra L. Warner for technical assistance; and Ms. Maudine D. Waterman (Dartmouth–Hitchcock Medical Center) for technical assistance. This work was supported by National Institutes of Health and National Cancer Institute Grants R01-CA87546 (to E. Dmitrovsky) and R01-CA111422 (to E. Dmitrovsky), a Samuel Waxman Cancer Research Foundation Award (to E. Dmitrovsky), and a Lance Armstrong Foundation Award (to S.J.F).
Abbreviations
- Cdk2
cyclin-dependent kinase 2
- HBE
human bronchial epithelial
- RA
all-trans-retinoic acid
- SCF
Skp1-Cullin1 F-box protein
- SP-C
surfactant C
- hcyclin E
human cyclin E
- Hh
hedgehog
- Shh
sonic Hh
- Ptch1
Patched 1
- CIN
chromosomal instability
- Smo
smoothened.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0606537104/DC1.
References
- 1.Dulic V, Lees E, Reed SI. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- 2.Koff A, Giordano A, Desai D, Yamashita K, Harper JW, Elledge S, Nishimoto T, Morgan DO, Franza BR, Roberts JM. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- 3.Ohtsubo M, Theodoras AM, Schumacher J, Roberts JM, Pagano M. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spruck CH, Won KA, Reed SI. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 5.Bortner DM, Rosenberg MP. Mol Cell Biol. 1997;17:453–459. doi: 10.1128/mcb.17.1.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lonardo F, Rusch V, Langenfeld J, Dmitrovsky E, Klimstra DS. Cancer Res. 1999;59:2470–2476. [PubMed] [Google Scholar]
- 7.Fukuse T, Hirata T, Naiki H, Hitomi S, Wada H. Cancer Res. 2000;60:242–244. [PubMed] [Google Scholar]
- 8.Dosaka-Akita H, Hommura F, Mishina T, Ogura S, Shimizu M, Katoh H, Kawakami Y. Cancer Res. 2001;61:2500–2504. [PubMed] [Google Scholar]
- 9.Muller-Tidow C, Metzger R, Kugler K, Diederichs S, Idos G, Thomas M, Dockhorn-Dworniczak B, Schneider PM, Koeffler HP, Berdel WE, et al. Cancer Res. 2001;61:647–653. [PubMed] [Google Scholar]
- 10.Langenfeld J, Lonardo F, Kiyokawa H, Passalaris T, Ahn MJ, Rusch V, Dmitrovsky E. Oncogene. 1996;13:1983–1990. [PubMed] [Google Scholar]
- 11.Dragnev KH, Pitha-Rowe I, Ma Y, Petty WJ, Sekula D, Murphy B, Rendi M, Suh N, Desai NB, Sporn MB, et al. Clin Cancer Res. 2004;10:2570–2577. doi: 10.1158/1078-0432.ccr-03-0271. [DOI] [PubMed] [Google Scholar]
- 12.Clurman BE, Sheaff RJ, Thress K, Groudine M, Roberts JM. Genes Dev. 1996;10:1979–1990. doi: 10.1101/gad.10.16.1979. [DOI] [PubMed] [Google Scholar]
- 13.Singer JD, Gurian-West M, Clurman B, Roberts JM. Genes Dev. 1999;13:2375–2387. doi: 10.1101/gad.13.18.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Won KA, Reed SI. EMBO J. 1996;15:4182–4193. [PMC free article] [PubMed] [Google Scholar]
- 15.Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 16.Welcker M, Singer J, Loeb KR, Grim J, Bloecher A, Gurien-West M, Clurman BE, Roberts JM. Mol Cell. 2003;12:381–392. doi: 10.1016/s1097-2765(03)00287-9. [DOI] [PubMed] [Google Scholar]
- 17.Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 18.Moberg KH, Bell DW, Wahrer DC, Haber DA, Hariharan IK. Nature. 2001;413:311–316. doi: 10.1038/35095068. [DOI] [PubMed] [Google Scholar]
- 19.Spruck CH, Strohmaier H, Sangfelt O, Muller HM, Hubalek M, Muller-Holzner E, Marth C, Widschwendter M, Reed SI. Cancer Res. 2002;62:4535–4539. [PubMed] [Google Scholar]
- 20.Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- 21.Ekholm-Reed S, Spruck CH, Sangfelt O, van Drogen F, Mueller-Holzner E, Widschwendter M, Zetterberg A, Reed SI. Cancer Res. 2004;64:795–800. doi: 10.1158/0008-5472.can-03-3417. [DOI] [PubMed] [Google Scholar]
- 22.Loeb KR, Kostner H, Firpo E, Norwood T, D Tsuchiya K, Clurman BE, Roberts JM. Cancer Cell. 2005;8:35–47. doi: 10.1016/j.ccr.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Glasser SW, Korfhagen TR, Wert SE, Bruno MD, McWilliams KM, Vorbroker DK, Whitsett JA. Am J Physiol. 1991;261:L349–L356. doi: 10.1152/ajplung.1991.261.4.L349. [DOI] [PubMed] [Google Scholar]
- 24.Karsunky H, Geisen C, Schmidt T, Haas K, Zevnik B, Gau E, Moroy T. Oncogene. 1999;18:7816–7824. doi: 10.1038/sj.onc.1203205. [DOI] [PubMed] [Google Scholar]
- 25.Rusch V, Klimstra D, Venkatraman E, Pisters PW, Langenfeld J, Dmitrovsky E. Clin Cancer Res. 1997;3:515–522. [PubMed] [Google Scholar]
- 26.Sargent LM, Senft JR, Lowry DT, Jefferson AM, Tyson FL, Malkinson AM, Coleman AE, Reynolds SH. Cancer Res. 2002;62:1152–1157. [PubMed] [Google Scholar]
- 27.Pasca di Magliano M, Hebrok M. Nat Rev Cancer. 2003;3:903–911. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- 28.Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 29.Yuan Z, Goetz JA, Singh S, Ogden SA, Petty WJ, Black CC, Memoli VA, Dmitrovsky E, Robbins DJ. Oncogene. 2006 Aug 14; doi: 10.1038/sj.onc.1209860. [DOI] [PubMed] [Google Scholar]
- 30.Minna JD, Roth JA, Gazdar AF. Cancer Cell. 2002;1:49–52. doi: 10.1016/s1535-6108(02)00027-2. [DOI] [PubMed] [Google Scholar]
- 31.Dutt A, Wong KK. Clin Cancer Res. 2006;12(14 Suppl):4396s–4402s. doi: 10.1158/1078-0432.CCR-06-0414. [DOI] [PubMed] [Google Scholar]
- 32.Ma Y, Feng Q, Sekula D, Diehl JA, Freemantle SJ, Dmitrovsky E. Cancer Res. 2005;65:6476–6483. doi: 10.1158/0008-5472.CAN-05-0370. [DOI] [PubMed] [Google Scholar]
- 33.Malkinson AM, Dwyer-Nield LD, Rice PL, Dinsdale D. Toxicology. 1997;123:53–100. doi: 10.1016/s0300-483x(97)00108-x. [DOI] [PubMed] [Google Scholar]
- 34.Nason-Burchenal K, Allopenna J, Begue A, Stehelin D, Dmitrovsky E, Martin P. Blood. 1998;92:1758–1767. [PubMed] [Google Scholar]
- 35.Dmitrovsky E, Moy D, Miller WH, Jr, Li A, Masui H. Oncogene Res. 1990;5:233–239. [PubMed] [Google Scholar]
- 36.Early E, Moore MA, Kakizuka A, Nason-Burchenal K, Martin P, Evans RM, Dmitrovsky E. Proc Natl Acad Sci USA. 1996;93:7900–7904. doi: 10.1073/pnas.93.15.7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bender MA, Mehaffey MG, Telling A, Hug B, Ley TJ, Groudine M, Fiering S. Blood. 2000;95:3600–3604. [PubMed] [Google Scholar]
- 38.Rice JA. Mathematical Statistics and Data Analysis. 2nd Ed. Belmont, CA: Duxbury; 1995. pp. 389–390. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.