Abstract
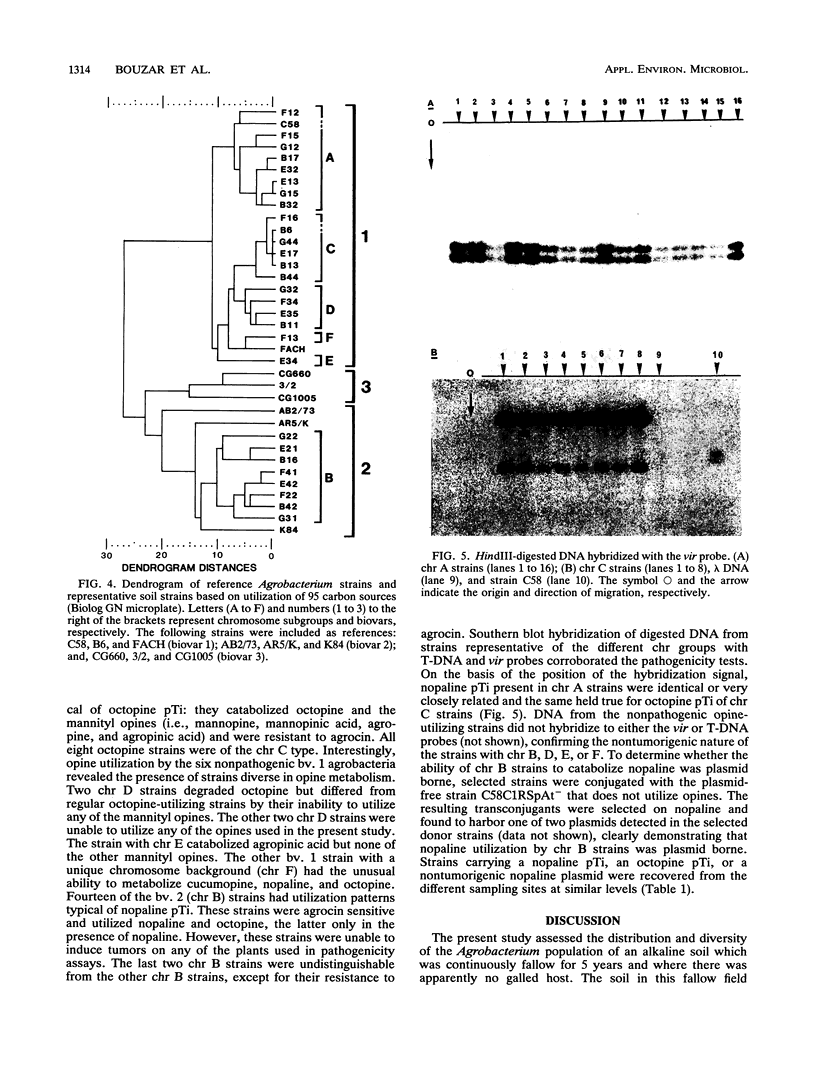
Soil samples collected from a fallow field which had not been cultivated for 5 years harbored a population of Agrobacterium spp. estimated at 3 × 107 CFU/g. Characterization of 72 strains selected from four different isolation media showed the presence of biovar 1 (56%) and bv. 2 (44%) strains. Pathogenicity assays on five different test plants revealed a high proportion (33%) of tumorigenic strains in the resident population. All tumorigenic strains belonged to bv. 1. Differentiation of the strains by restriction fragment length polymorphism analysis, sodium dodecyl sulfate-polyacrylamide gel electrophoresis of cellular proteins, and utilization patterns of 95 carbon substrates (Biolog GN microplate) revealed a diversified bv. 1 population, composed of five distinct chromosomal backgrounds (chr A, C, D, E, and F), and a homogeneous bv. 2 population (chr B). chr A, B, C, and D were detected at similar levels throughout the study site. According to opine metabolism, pathogenicity, and agrocin sensitivity, chr A strains carried a nopaline Ti plasmid (pTi), whereas chr C strains had an octopine pTi. In addition, four of six nontumorigenic bv. 1 strains (two chr D, one chr E, and one chr F) had distinct and unusual opine catabolism patterns. chr B (bv. 2) strains were nonpathogenic and catabolized nopaline. Although agrocin sensitivity is a pTi-borne trait, 14 chr B strains were sensitive to agrocin 84, apparently harboring a defective nopaline pTi similar to pAtK84b. The other two chr B strains were agrocin resistant. The present analysis of chromosomal and plasmid phenotypes suggests that in this Agrobacterium soil population, there is a preferential association between the resident plasmids and their bacterial host.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akiyoshi D. E., Klee H., Amasino R. M., Nester E. W., Gordon M. P. T-DNA of Agrobacterium tumefaciens encodes an enzyme of cytokinin biosynthesis. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5994–5998. doi: 10.1073/pnas.81.19.5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomhoff G., Klapwijk P. M., Kester H. C., Schilperoort R. A., Hernalsteens J. P., Schell J. Octopine and nopaline synthesis and breakdown genetically controlled by a plasmid of Agrobacterium tumefaciens. Mol Gen Genet. 1976 May 7;145(2):177–181. doi: 10.1007/BF00269591. [DOI] [PubMed] [Google Scholar]
- Bouzar H., Moore L. W. Isolation of different agrobacterium biovars from a natural oak savanna and tallgrass prairie. Appl Environ Microbiol. 1987 Apr;53(4):717–721. doi: 10.1128/aem.53.4.717-721.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilton M. D., Drummond M. H., Merio D. J., Sciaky D., Montoya A. L., Gordon M. P., Nester E. W. Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell. 1977 Jun;11(2):263–271. doi: 10.1016/0092-8674(77)90043-5. [DOI] [PubMed] [Google Scholar]
- Clare B. G., Kerr A., Jones D. A. Characteristics of the nopaline catabolic plasmid in Agrobacterium strains K84 and K1026 used for biological control of crown gall disease. Plasmid. 1990 Mar;23(2):126–137. doi: 10.1016/0147-619x(90)90031-7. [DOI] [PubMed] [Google Scholar]
- Dessaux Y., Tempé J., Farrand S. K. Genetic analysis of mannityl opine catabolism in octopine-type Agrobacterium tumefaciens strain 15955. Mol Gen Genet. 1987 Jun;208(1-2):301–308. doi: 10.1007/BF00330457. [DOI] [PubMed] [Google Scholar]
- Dhaese P., De Greve H., Decraemer H., Schell J., Van Montagu M. Rapid mapping of transposon insertion and deletion mutations in the large Ti-plasmids of Agrobacterium tumefaciens. Nucleic Acids Res. 1979 Dec 11;7(7):1837–1849. doi: 10.1093/nar/7.7.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dughri M. H., Bottomley P. J. Effect of Acidity on the Composition of an Indigenous Soil Population of Rhizobium trifolii Found in Nodules of Trifolium subterraneum L. Appl Environ Microbiol. 1983 Nov;46(5):1207–1213. doi: 10.1128/aem.46.5.1207-1213.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt T. A rapid method for the identification of plasmid desoxyribonucleic acid in bacteria. Plasmid. 1978 Sep;1(4):584–588. doi: 10.1016/0147-619x(78)90016-1. [DOI] [PubMed] [Google Scholar]
- KING E. O., WARD M. K., RANEY D. E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954 Aug;44(2):301–307. [PubMed] [Google Scholar]
- Kerr A., Manigault P., Tempé J. Transfer of virulence in vivo and in vitro in Agrobacterium. Nature. 1977 Feb 10;265(5594):560–561. doi: 10.1038/265560a0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Michel M. F., Brasileiro A. C., Depierreux C., Otten L., Delmotte F., Jouanin L. Identification of different agrobacterium strains isolated from the same forest nursery. Appl Environ Microbiol. 1990 Nov;56(11):3537–3545. doi: 10.1128/aem.56.11.3537-3545.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesme X., Michel M. F., Digat B. Population Heterogeneity of Agrobacterium tumefaciens in Galls of Populus L. from a Single Nursery. Appl Environ Microbiol. 1987 Apr;53(4):655–659. doi: 10.1128/aem.53.4.655-659.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STONIER T. Agrobacterium tumefaciens Conn. II. Production of an antibiotic substance. J Bacteriol. 1960 Jun;79:889–898. doi: 10.1128/jb.79.6.889-898.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder G., Waffenschmidt S., Weiler E. W., Schröder J. The T-region of Ti plasmids codes for an enzyme synthesizing indole-3-acetic acid. Eur J Biochem. 1984 Jan 16;138(2):387–391. doi: 10.1111/j.1432-1033.1984.tb07927.x. [DOI] [PubMed] [Google Scholar]
- Stachel S. E., Nester E. W. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 1986 Jul;5(7):1445–1454. doi: 10.1002/j.1460-2075.1986.tb04381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Larebeke N., Genetello C., Schell J., Schilperoort R. A., Hermans A. K., Van Montagu M., Hernalsteens J. P. Acquisition of tumour-inducing ability by non-oncogenic agrobacteria as a result of plasmid transfer. Nature. 1975 Jun 26;255(5511):742–743. doi: 10.1038/255742a0. [DOI] [PubMed] [Google Scholar]