Abstract
Although indole-3-acetic acid (IAA), the predominant auxin in plants, plays a critical role in various plant growth and developmental processes, its biosynthesis and regulation have not been clearly elucidated. To investigate the molecular mechanisms of IAA synthesis in rice (Oryza sativa), we identified seven YUCCA-like genes (named OsYUCCA1-7) in the rice genome. Plants overexpressing OsYUCCA1 exhibited increased IAA levels and characteristic auxin overproduction phenotypes, whereas plants expressing antisense OsYUCCA1 cDNA displayed defects that are similar to those of rice auxin-insensitive mutants. OsYUCCA1 was expressed in almost all of the organs tested, but its expression was restricted to discrete areas, including the tips of leaves, roots, and vascular tissues, where it overlapped with expression of a β-glucuronidase reporter gene controlled by the auxin-responsive DR5 promoter. These observations are consistent with an important role for the rice enzyme OsYUCCA1 in IAA biosynthesis via the tryptophan-dependent pathway.
Indole-3-acetic acid (IAA), the predominant auxin in plants, plays a critical role in many plant growth and developmental processes, including cell division, differentiation, and elongation; flower and vascular development; and tropism (for review, see Teale et al., 2006). During the last decade, plant scientists have made significant advances in understanding the molecular mechanisms of IAA-mediated gene expression, signaling, and transport in the model dicot Arabidopsis (Arabidopsis thaliana; e.g. Leyser, 2002; Dharmasiri and Estelle, 2004; Dharmasiri et al., 2005; Kepinski and Leyser, 2005; Paponov et al., 2005). However, the biosynthesis and regulation of IAA are still not well understood.
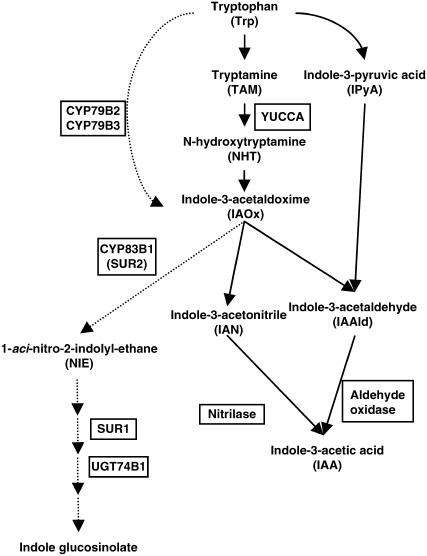
Two major pathways for IAA biosynthesis have been proposed: the Trp-dependent and Trp-independent pathways (Zazimalova and Napier, 2003). A large body of evidence indicates that plants convert Trp to IAA by several routes (Fig. 1); this evidence includes results from in vivo and in vitro labeling experiments using radiolabeled Trp and the identification of enzymes for each of the proposed steps. Furthermore, recent studies using gain-of-function approaches in Arabidopsis have revealed that two distinct routes lead from Trp to the intermediate indole-3-acetaldoxime (IAOx; Fig. 1). The YUCCA gene, which encodes a flavin monooxygenase-like enzyme, was identified by isolation of a dominant mutant with elevated levels of IAA (Zhao et al., 2001). Because YUCCA can convert tryptamine (TAM) to N-hydroxytryptamine (NHT) in vitro, Zhao et al. (2001) suggested that YUCCA catalyzes the N-oxygenation of TAM, a rate-limiting step in auxin biosynthesis in many plants. Involvement of YUCCA in IAA synthesis is also supported by molecular analysis of a petunia (Petunia hybrida) mutant, floozy (fzy; Tobena-Santamaria et al., 2002). The fzy mutant was first isolated as a flower mutant in which the formation of the outermost three floral whorl formations and one of the two bracts is blocked at an early stage. FZY encodes a flavin monooxygenase-like enzyme homologous to Arabidopsis YUCCA, and its overexpression results in increased IAA levels and in an auxin overproduction phenotype (Tobena-Santamaria et al., 2002). More recently, Cheng et al. (2006) reported that double, triple, and quadruple mutants of four Arabidopsis YUCCA genes display severe defects in floral patterning, vascular formation, and the other developmental processes. The defects in the Arabidopsis yuc mutants were rescued by expression of the bacterial auxin biosynthesis gene iaaM, indicating that these YUCCA genes are important for auxin biosynthesis in Arabidopsis (Cheng et al., 2006).
Figure 1.
Proposed Trp-dependent pathway for IAA biosynthesis. Genes or mutants that have been identified with particular enzymatic steps are specified boxes. The steps indicated by dotted arrows are shared with the glucosinolate pathway, which is restricted to a few plant families but not in rice.
On the other hand, Zhao et al. (2002) reported evidence that the multifunctional cytochrome P450 enzymes CYP79B2 and CYP79B3, which were previously identified as enzymes that catalyze conversion of Trp to IAOx in vitro (Hull et al., 2000; Mikkelsen et al., 2000), are critical enzymes in auxin biosynthesis in vivo. Arabidopsis plants overproducing CYP79B2 contained increased levels of free auxin and exhibited auxin overproduction phenotypes, and double knockout of the CYP79B2 and CYP79B3 genes caused a reduction in IAA levels and induced growth defects related to partial auxin deficiency. These findings concerning YUCCA and CYP79B2/CYP79B3 strongly suggest that a Trp-dependent pathway occurring via an IAOx intermediate is a major source of IAA in Arabidopsis.
Although recent molecular genetic studies have successfully identified a few genes involved in the Trp-dependent IAA biosynthetic pathway in Arabidopsis, auxin biosynthesis mechanisms in monocots have not been defined (Zhao et al., 2002). As a model monocot, rice (Oryza sativa) has been extensively studied. The entire rice genome sequence is known, and full-length cDNAs, transformation systems, and many mutant collections are available. Using these tools, we have systematically screened and characterized the genes involved in biosynthesis of gibberellin and brassinosteroid (e.g. Sakamoto et al., 2004; Hong et al., 2005). As a result, we have concluded that the mechanisms of gibberellin and brassinosteroid biosynthesis differ somewhat in rice and Arabidopsis.
By analogy to our previous work, and because of the lack of obvious CYP79B2/CYP79B3 genes in rice, we have postulated that the mechanism of IAA synthesis in rice may be different from that in Arabidopsis. In this study, we identified and characterized a YUCCA-like gene in rice, OsYUCCA1. Plants overexpressing OsYUCCA1 exhibited increased IAA levels and auxin overproduction phenotypes, whereas plants suppressing antisense OsYUCCA1 cDNA showed abnormal phenotypes similar to auxin-insensitive rice. Moreover, OsYUCCA1 is not expressed ubiquitously; rather, its expression is restricted to discrete places such as the tips of leaves and roots and parenchyma cells surrounding large vascular stem tissues. Based on these observations, we propose a molecular mechanism for IAA synthesis in rice.
RESULT
Cloning of Rice YUCCA Homologs and Analysis of Their Expression Patterns
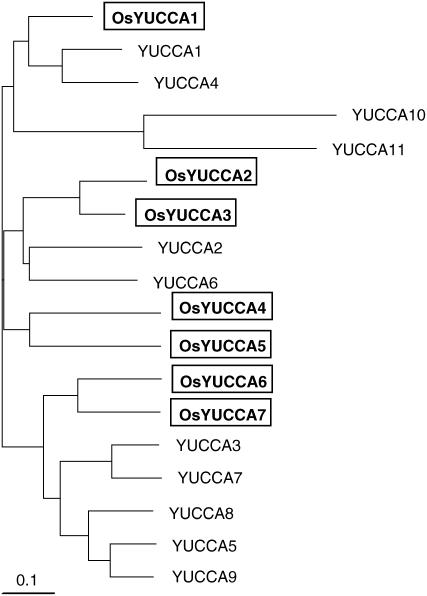
To identify rice homologs of the Arabidopsis YUCCA genes, we performed a tBLASTn search against all available rice DNA databases using the known Arabidopsis YUCCA1 protein as a query sequence (Zhao et al., 2001). This search yielded seven rice homologs (OsYUCCA1-7), each of which encodes a protein containing the conserved binding motifs for FAD and NADPH (Supplemental Fig. S1). We also analyzed the phylogenic relationships among these homologs based on alignments of their full-length amino acid sequences (Fig. 2). As previously reported (Zhao et al., 2001; Woodward et al., 2005; Cheng et al., 2006), the rice and Arabidopsis YUCCA proteins cluster into three groups. One group includes YUCCA1, YUCCA4, YUCCA10, YUCCA11, and one of the OsYUCCA proteins, which we named OsYUCCA1. The second group contains YUCCA2, YUCCA6, and four of the OsYUCCA proteins, which we named OsYUCCA2 to 5. The third group contains YUCCA3, YUCCA5, YUCCA7, YUCCA8, YUCCA9, and two rice homologs, which we named OsYUCCA6 and OsYUCCA7.
Figure 2.
Comparison of the amino acid sequences of rice and Arabidopsis YUCCA proteins. Phylogenic analysis of rice and Arabidopsis YUCCA and YUCCA-like proteins. The tree was constructed by the UPGMA method based on the alignment in the Supplemental Figure S1.
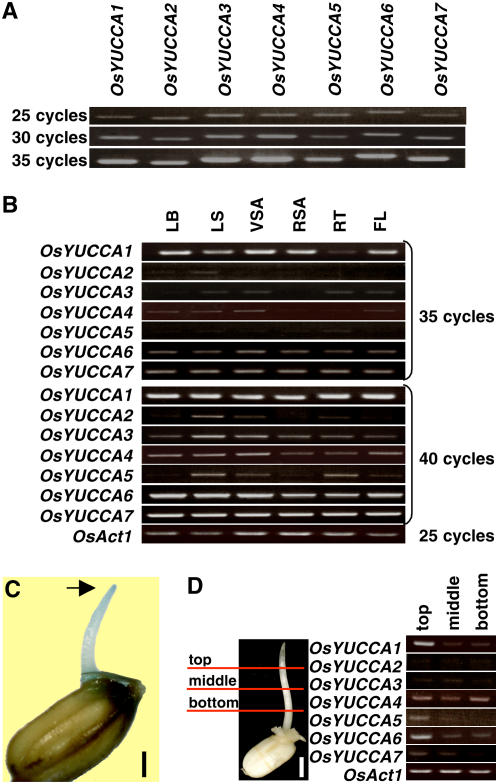
To determine whether the OsYUCCA genes are actually expressed in rice organs, we used semiquantitative reverse transcription-PCR (qRT-PCR) analysis with primers that specifically amplified the 3′-noncoding regions of OsYUCCA1-7. This approach was necessary, because the levels of OsYUCCA mRNA were too low to detect using RNA gel-blot analysis (data not shown). Before performing the qRT-PCR analysis, we tested the efficiencies of the primer sets by using them in PCR reactions with the rice genomic DNA as a template. The amounts of the resulting PCR products increased similarly as a function of cycle number (Fig. 3A), confirming that the primer sets were of similar efficiencies.
Figure 3.
Analysis of OsYUCCA genes expression. A, To compare the efficiency of the PCR primer sets for each OsYUCCA gene, they were used in PCR reactions with the rice genomic DNA as a template. The primer sets amplified their respective OsYUCCA genes with similar efficiency. Product amounts are compared at 25, 30, and 35 cycles. B, qRT-PCR analysis was conducted for detection of OsYUCCA gene expression using OsYUCCA cDNAs produced from total RNA. Total RNA was isolated from leaf blade (LB), leaf sheath (LS), vegetative shoot apical (VSA), reproductive shoot apical (RSA), root (RT), and flower (FL). The amount of products was compared at 35 and 40 cycles. OsAct1 was used as control (25 cycles). C, GUS staining for 24 h in 3-d-old coleoptile of a DR5-GUS transgenic plant grown in the dark. Bar = 3 mm. D, qRT-PCR analysis of OsYUCCA gene expression in the top, middle, and bottom of the coleoptiles. Bar = 5 mm. OsAct1 was used as a control. The OsYUCCA genes and the OsAct1 gene were amplified for 40 and 25 cycles, respectively.
Using the same experimental conditions as for the genomic template, we performed qRT-PCR with template cDNA produced from the total RNA isolated from various organs. At 35 cycles, strong bands corresponding to OsYUCCA1 were amplified from cDNAs from leaf blade, vegetative shoot apex, reproductive shoot apex, and flower, bands of intermediate intensity were amplified from leaf sheath cDNA, and faint bands were amplified from root cDNA (Fig. 3B, 35 cycles). In contrast, only intermediate or faint intensity bands of ambiguous specificity were observed for OsYUCCA6 or 7, and very faint or no bands were observed for OsYUCCA2 through 5 from the same cDNAs under the same conditions (Fig. 3B, 35 cycles). At 40 cycles, OsYUCCA2 through 5 produced intermediate or faint bands (Fig. 3B, 40 cycles). Products corresponding to OsYUCCA3 and 4 were observed in all organs tested, with stronger bands from leaf sheath and vegetative shoot apex, whereas the products corresponding to OsYUCCA2 and 5 were preferentially produced from leaf sheath, vegetative shoot apex, and root. When we used DNA samples skipped the reverse transcriptase step, no bands were produced. These results indicate that all of the OsYUCCA genes are expressed in rice, but OsYUCCA1 expression predominates in almost all organs.
To help elucidate which of the OsYUCCA genes are associated with IAA biosynthesis, we examined their spatial expression patterns in coleoptiles because of the localization of auxin in the tip of grass coleoptiles (Fig. 3C). If the OsYUCCA gene products are involved in IAA production, the OsYUCCA mRNAs may be preferentially expressed at the coleoptile tip. As we expected, OsYUCCAs 1, 5, and 6 were preferentially expressed in the top of rice coleoptiles in comparison to the middle or bottom, whereas the other OsYUCCAs did not exhibit a preferential pattern (Fig. 3D). From these observations, we expected that OsYUCCA1 may dominantly play a role in IAA production in rice and therefore focused on this gene in further study.
Phenotypes of Plants Overexpressing OsYUCCA1 mRNA
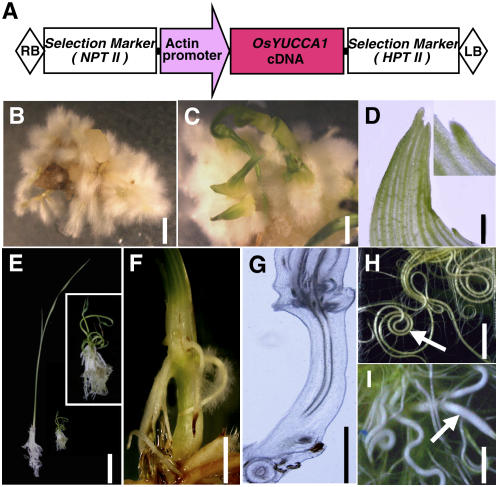
Next, we produced transgenic rice plants overexpressing OsYUCCA1 cDNA. The rice ACT1 gene promoter (McElroy et al., 1990) was used to induce constitutive, high-level expression of OsYUCCA1 in rice calli (Fig. 4A). The growth rate and regeneration frequency of calli transformed with pAct-OsYUCCA1 were greatly decreased, as compared to those of calli transformed with a control vector, probably because increased auxin production is inhibitory. Because addition of 1.5× kinetin to the growth and regeneration media partially improved the frequency of overexpressing calli (data not shown), we used 1.5× kinetin in subsequent transformations of several hundred rice calli with pAct-OsYUCCA1. Twenty-one independent reentrants were obtained.
Figure 4.
Phenotypes of transgenic rice plants overexpressing OsYUCCA1. A, Schematic representation of the pAct-OsYUCCA1 construct. The full-length OsYUCCA1 cDNA is expressed under the control of the rice actin1 promoter. B and C, Typical phenotypes of transgenic calli 2 weeks after regeneration. Bar = 5 mm. Callus produced crown roots with abundant hairy roots (B) and often also produced several leaflets (C). D, Aberrant morphology of leaflet protruded from transgenic callus. Bar = 4 mm. Vascular tissues often stopped at the leaf margin (superimposed photo at top right). E, Gross morphology of regenerated plants carrying the control vector (left) or pAct-OsYUCCA1 with mild phenotype (right) at 3 weeks after regeneration. The pAct-OsYUCCA1 plant is enlarged at top right. F, Abnormal elongation of internode and ectopic development of crown roots from elongated node in pAct-OsYUCCA1 plants. Bar = 5 mm. G, Longitudinal section of the elongated internode of F. Bar = 5 mm. H and I, Root morphology of control (H) and pAct-OsYUCCA1 (I) plants. Roots of pAct-OsYUCCA1 plants were often thicker than those of the control plants and grew in random directions (indicated by arrows). Bar = 5 mm.
In many cases, the transformed calli did not produce plantlets; instead, they produced extensive hairy roots that developed from adventitious roots directly formed from calli (Fig. 4B). Sometimes, small leaflets sporadically protruded from calli enveloped with hairy roots (Fig. 4C). The leaflet morphology was aberrant in that the vascular tissues often stopped at the leaf margin (Fig. 4D). Several regenerated plantlets produced several or more leaves with a more normal structure (Fig. 4E), but these plantlets with the mildest phenotypes still exhibited dwarfism (Fig. 4E, right).
In addition to inhibiting leaf growth, overexpression of OsYUCCA1 also severely inhibited root elongation in the mild phenotype plants, whereas formation of crown root (adventitious root, the major component of the rice root system) was promoted (Fig. 4E). Interestingly, some plantlets with a mild phenotype initiated internode elongation at an early stage (Fig. 4, F and G); initiation of internode elongation does not occur in wild-type plants under normal conditions until the vegetative-to-reproductive phase changes. Active crown root formation still occurred at the upper node after internode elongation (Fig. 4F). The increased formation of crown root corresponded well to the high frequency of adventitious root formation from calli, demonstrating that increased IAA promotes adventitious root formation from both callus and stem. When the crown roots of regenerated plantlets carrying the control vector reached the bottom of the plant box, almost all of the roots coiled at the bottom (Fig. 4H), whereas the roots of plants overexpressing OsYUCCA1 did not coil, and their root diameters were greater than those of wild-type plants (Fig. 4I). The lack of root coiling in the overexpressing plants may be related to their abnormal gravitropism, as discussed below.
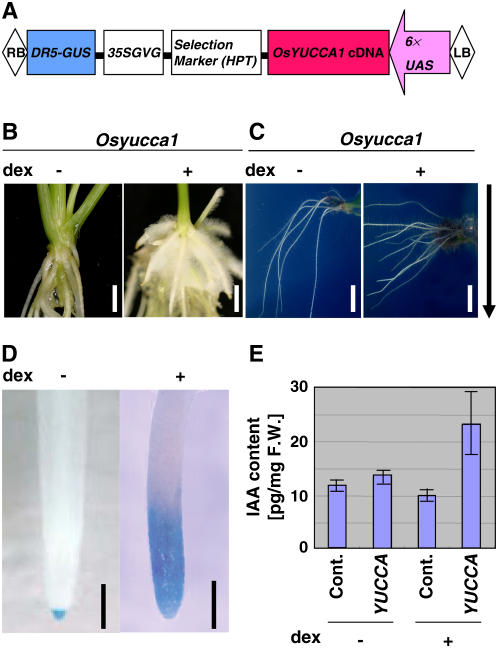
All of the above observations indicate that overexpression of OsYUCCA1 causes abnormal leaf, root, and stem development. These phenotypes are so severe, however, that the possibility that indirect effects of OsYUCCA1 overexpression had contributed to the observed phenotypes could not be discounted. Therefore, to more directly observe the effects of increased OsYUCCA1 function, we used a steroid-hormone-inducible system for OsYUCCA1 expression (Aoyama and Chua, 1997). We also constructed a plasmid containing the β-glucuronidase (GUS) reporter gene under the control of the DR5 promoter (Fig. 5A). The DR5 promoter, which is often used as an artificial auxin-responsive promoter in Arabidopsis, was used to create a marker for visualization of the in vivo distribution of auxin in rice (Ulmasov et al., 1997; Sabatini et al., 1999; Scarpella et al., 2003).
Figure 5.
Phenotypes of transgenic rice plants expressing OsYUCCA1 under the control of a steroid hormone-inducible system. A, Schematic representation of a construct for expression of OsYUCCA1 cDNA under the control of the steroid hormone-inducible system regulated by DEX (Aoyama and Chua, 1997). The plasmid also contains a DR5-GUS construct for monitoring auxin expression. B, DEX-dependent root-hair induction. The OsYUCCA1 plant was treated with (+) or without (−) 100 μm DEX for 1 week. Bar = 5 mm. C, DEX-dependent agravitropic response. The OsYUCCA1 plant was treated as in B. Bar = 5 mm. D, DEX-dependent enhancement of GUS activity in root. The OsYUCCA1 plant was treated as in B. The GUS reporter gene was controlled by the DR5 promoter. Bar = 1 mm. E, DEX-dependent increase in IAA. Control and OsYUCCA1 plants were treated as in B.
Ten independent transgenic plants carrying the DR5-GUS transgene were obtained. From these plants, we selected three lines that expressed GUS at their root tips and did not express detectable OsYUCCA1 mRNA in the absence of the inducer dexamethasone (DEX; data not shown). In the absence of DEX treatment, these transgenic plants exhibited normal phenotypes, as did plants carrying the control vector (Fig. 5B, left). In contrast, when the plants were treated with DEX for 1 week, the newly developed crown roots produced abundant hairy roots, as was also observed for the constitutively overexpressing plants (compare Fig. 4C and 5B, right). In DEX-containing agar medium, the roots of the transgenic plants did not respond normally to gravity, but their gravity response was normal in a medium without DEX (Fig. 5C). These results indicate that induction of OsYUCCA1 expression by DEX treatment disrupted gravitropic responses of the transgenic plants. DEX treatment appeared to directly induce auxin production. A non-DEX-treated plant exhibited only faint GUS staining in the root tip, whereas DEX-treated plants displayed stronger and more broadly localized GUS staining around the root tip (Fig. 5D). Furthermore, OsYUCCA1-overexpressing seedlings without DEX treatment contained about 14 pg IAA/mg fresh weight, similar to that of plants carrying the control vector with or without DEX treatment, whereas OsYUCCA1-overexpressing plants treated with DEX contained about 24 pg IAA/mg fresh weight (Fig. 5E). Thus, OsYUCCA1 overexpression induced by DEX treatment caused an increase in the level of IAA, disturbed gravitropism, and induced hairy root production, indicating that OsYUCCA1 functions as an enzyme for IAA production in rice.
Phenotype of Plants Expressing the OsYUCCA1 Antisense
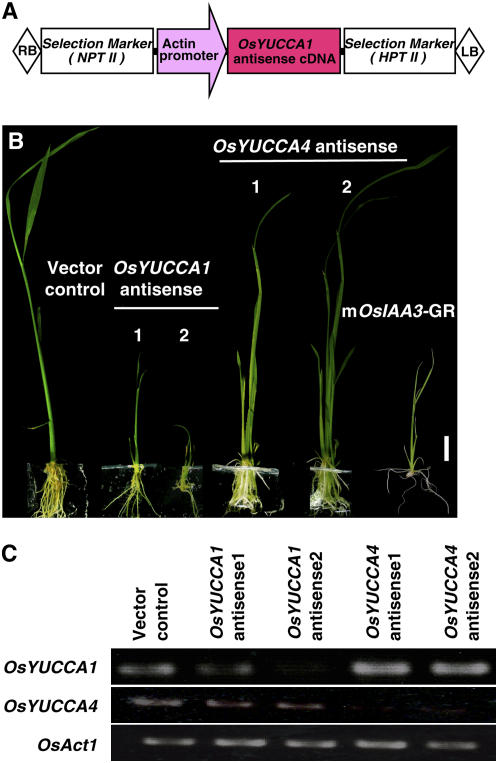
To confirm the involvement of OsYUCCA1 in auxin synthesis, we produced transgenic rice plants expressing the antisense cDNA of OsYUCCA1 under the control of the ACT1 promoter (Fig. 6A). We also produced transgenic plants carrying the antisense cDNA of OsYUCCA4, because OsYUCCA4 is classified into a different group from other OsYUCCAs and Arabidopsis YUCCAs, except OsYUCCA5 (Fig. 2), and the level of its expression was relatively stronger than that of OsYUCCA2-3 in various organs, including coleoptiles (Fig. 3, B and D). The growth rate and regeneration frequency of calli transformed with the pAct-OsYUCCA1 antisense were reduced extremely in comparison with those of calli transformed with a control vector, probably because decreased auxin production is inhibitory for growth and regeneration of rice calli (data not shown). We produced more than several hundred rice calli carrying the pAct-OsYUCCA1 antisense construct and finally obtained five independent regenerants. These transgenic plants showed severe dwarfism of shoot elongation and inhibited root formation and elongation (Fig. 6B), which are very similar to those observed in the auxin-insensitive rice plants carrying the constitutive active IAA proteins (Nakamura et al., 2006). The degree of the developmental defects correlated well with the suppression of the OsYUCCA1 expression levels (Fig. 6C), indicating that these abnormal phenotypes are caused by the knockdown of the OsYUCCA1 function. In contrast, pAct-OsYUCCA4 antisense plants showed no abnormal phenotype (Fig. 6B), although its expression was almost completely suppressed in these antisense plants (Fig. 6C). These observations strongly support that OsYUCCA1 is important for growth and development of shoot and root in rice.
Figure 6.
Phenotypes of transgenic rice plants expressing the OsYUCCA1 antisense. A, Schematic representation of the pAct-OsYUCCA1 antisense construct. The antisense OsYUCCA1 or OsYUCCA4 cDNA is expressed under the control of the rice actin1 promoter. B, Typical phenotypes of the OsYUCCA1 and OsYUCCA4 antisense plants 2 weeks after regeneration. Bar = 1 cm. The OsYUCCA1 antisense plants show dwarf shoot and inhibited development and elongation of root, while the OsYUCCA4 antisense plants show no abnormal phenotype. The mOsIAA3-GR transgenic plant, which carries the constitutive active IAA protein, was also shown. C, qRT-PCR analysis of OsYUCCA1 and OsYUCCA4 gene expression in their antisense plants. Total RNAs were isolated from leaf blades of the antisense plants presented in B. The amount of products was compared at 35 cycles for OsYUCCA1 and 40 cycles for OsYUCCA4.
Sites of OsYUCCA1 Expression in Rice Plants
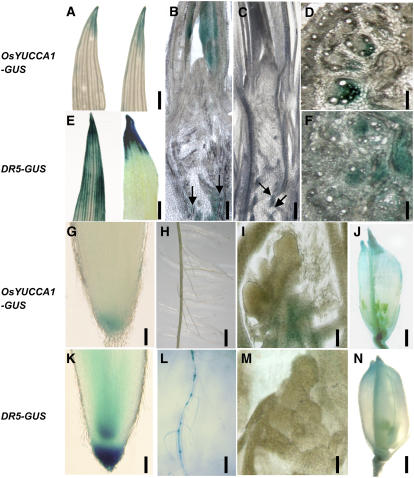
From the above observations, we concluded that OsYUCCA1 is an important enzyme for auxin biosynthesis in rice. Reasoning that the specific localization of OsYUCCA1 expression might indicate the site of auxin biosynthesis, we next examined OsYUCCA1 expression by comparing the expression levels of GUS reporter genes under control of either the OsYUCCA1 promoter or the DR5 promoter (Fig. 7). In shoots, GUS staining controlled by the OsYUCCA1 promoter was observed at the tip of young leaves (Fig. 7A, left). Expression of DR5-GUS was observed in a broader area around the leaf tip (Fig. 7E, left), and it was localized into the leaf tip after treatment with N-(1-naphthyl) phthalamic acid (NPA), an inhibitor of IAA transport (Fig. 7E, right). On the other hand, NPA treatment did not affect localization of GUS controlled by the OsYUCCA1 promoter (Fig. 7A, right). These observations are consistent with localized synthesis of IAA at the leaf tip and its rapid transport from the tip to the base of the leaf, as previously discussed (Jones et al., 1998).
Figure 7.
Histochemical localization of GUS activity in transgenic rice carrying OsYUCCA1-GUS (A, B, D, and G–J) or DR5-GUS (C, E, F, and K–N). Plants were stained for GUS activity for 2 h (E, F, K, L, and N), 24 h (A–C, G, I, and J), or 48 h (D, H, and M). A and E, Tip area of leaf blade. Right and left leaves in each segment were treated without or with 10 μm NPA. Bar = 2 mm. B and C, Longitudinal sections around the shoot apex region at the vegetative stage. Bar = 100 mm. D and F, Cross sections of stem. Bar = 1 mm. G and K, Root tip region. Bar = 0.2 mm. H and L, Developing roots at the elongation zone. Bar = 5 mm. I and M, Longitudinal sections at the floral shoot meristem. Bar = 0.1 mm. J and N, Developing flowers. Bar = 2 mm.
OsYUCCA1-GUS expression was broadly seen in young leaf primordia around the vegetative shoot apical meristem (SAM) but was virtually absent in the SAM itself (Fig. 7B). Almost no GUS staining was observed in the DR5-GUS plants, and it was absent from the leaf primordia (Fig. 7C). Thus, IAA may undergo rapid transport after synthesis. OsYUCCA1-GUS expression was also seen in parenchymal cells surrounding large vascular bundles of stem (Fig. 7B, arrows), and a similar staining pattern was observed in transgenic plants carrying DR5-GUS with more faint activity (Fig. 7C, arrows). Stem cross sections of OsYUCCA1-GUS plants clearly showed localized GUS staining in cells surrounding large vascular bundles (Fig. 7D), as well as in a broader area of the peripheral vascular cylinder in DR5-GUS plants (Fig. 7F).
In root, OsYUCCA1-GUS expression was faint and was localized at the tip (Fig. 7G), whereas that of DR5-GUS was stronger and also occurred in the root apical meristem (Fig. 7K). OsYUCCA1-GUS expression was detected at the root tip and not in other root portions (Fig. 7H), whereas DR5-GUS expression was also observed strongly in stele, especially at the base of lateral root (Fig. 7L). OsYUCCA1-GUS expression was not detected in the floral SAM itself, but it was observed in cells surrounding the vascular bundle (Fig. 7I), whereas no DR5-GUS expression was detected in the same region (Fig. 7M). In developing flower, GUS expression was observed at the flower tip and vascular tissues for both promoters (Fig. 7, J and N). The near-overlapping expression of GUS controlled by the OsYUCCA1 and DR5 promoters in various rice organs supports the conclusion that OsYUCCA1 functions as an important enzyme in IAA synthesis.
DISCUSSION
A Possible Biosynthetic Pathway for IAA in Rice
Although the IAA biosynthetic pathway in plants has not yet been determined with certainty, a large body of evidence has accumulated showing that plants convert Trp to IAA via several parallel pathways (Cohen et al., 2003; Zazimalova and Napier, 2003). As a result of recent gain-of-function studies in Arabidopsis, two enzymes involved in these parallel Trp-dependent pathways are the subjects of much research interest. These enzymes, YUCCA and CYP79B2/CYP79B3, catalyze the conversion of TAM to NHT and the conversion of Trp to IAOx, respectively (Fig. 1; Hull et al., 2000; Mikkelsen et al., 2000; Zhao et al., 2001, 2002).
In contrast to CYP79B2/CYP79B3, whose counterparts in rice have not been identified, rice has at least seven genes encoding proteins similar to Arabidopsis YUCCA proteins. Expression analysis of these YUCCA-like genes indicated that one of them, OsYUCCA1, functionally predominates in various organs of rice, although other OsYUCCAs were also expressed in various organs and may confer redundant functions in these organs (Fig. 3B). Several results have led us to conclude that OsYUCCA1 probably plays an important role in IAA biosynthesis. First, free IAA levels increased in a DEX-dependent manner in transgenic rice plants carrying a DEX-inducible OsYUCCA1 construct (Fig. 5E). Second, when GUS was expressed under control of the DR5 promoter, DEX treatment led to enhanced GUS staining in the root tips of the transgenic rice plants carrying a DEX-inducible OsYUCCA1 construct (Fig. 5D). Third, OsYUCCA1 antisense plants exhibited abnormal phenotypes associated with auxin, such as shoot dwarfism and inhibited development and elongation of root (Fig. 6B). Fourth, OsYUCCA1-overexpressing rice plants exhibited phenotypes typical of IAA overproduction, including root hair production (Figs. 4, B and C, and 5B) and agravitropism (Figs. 4I and 5C). Finally, OsYUCCA1 was found to preferentially express in the top coleoptiles region (Fig. 3D), which is considered to be one of the most active sites of IAA production (Fig. 3C; Koshiba and Matsuyama, 1993; Ribnicky et al., 1998; Philippar et al., 1999).
As is often pointed out, phenotypic results from transgenic plants overexpressing a gene of interest must be interpreted with caution. Conclusions as to the biological function(s) of the overexpressed gene based on the transgene phenotype may be erroneous in the overexpression context. As an artifact of overexpression of the transgene, the gene may appear to have functions other than or in addition to its normal function(s). However, our results demonstrating preferential expression of OsYUCCA1 in the area almost overlapping with the DR5-GUS expression, and the abnormal phenotype associated with auxin deficiency in the OsYUCCA1 antisense plants, strongly support an important role for OsYUCCA1 in IAA synthesis in its natural context.
Recently, Cheng et al. (2006) demonstrated, through analysis of loss-of-function mutants of the YUCCA genes, that the YUCCA genes play a key role in IAA biosynthesis and development of various organs in Arabidopsis. According to their observation, more than two mutations of four YUCCA genes, YUCCA1, 2, 4, and 6, are essential for induction of abnormal phenotypes, whereas disruption of a single YUCCA gene causes no obvious developmental defects. Based on this observation and the fact the quadruple mutant is not seedling or embryo lethal, Cheng et al. (2006) discussed that not only four YUCCA genes they examined but also other genes are involved in auxin biosynthesis. Similarly, rice has at least seven YUCCA-like genes, including OsYUCCA1 (Fig. 2), and therefore it is very likely that these OsYUCCA genes are also redundantly involved in IAA biosynthesis in rice. The fact that antisense plants of OsYUCCA1 showed abnormal phenotypes associated with auxin deficiency indicates that genetic redundancy of the YUCCA genes may be less complex (Fig. 6B). OsYUCCA1 is a sole rice gene classified into the group of Arabidopsis YUCCA1 and 4; inactivation of both yuc1 and yuc4 affects flower development in Arabidopsis (Cheng et al., 2006).
Although the mechanism by which the product of YUCCA catalysis, NHT, is converted to IAA has not been conclusively demonstrated, a possible pathway leading from NHT to IAA in Arabidopsis involves conversion of NHT to IAOx, followed by conversion of IAOx to IAA via indole-3-acetonitrile (IAN) or indole-3-acetaldehyde (IAAld; Fig. 1; Zhao et al., 2002). If rice synthesizes IAA by the same pathway, then rice should contain the enzymes catalyzing these steps. Recently, Park et al. (2003) reported that the maize (Zea mays) nitrilase ZmNIT2 hydrolyzes IAN to IAA with efficiency at least 7- to 20-fold higher than that of Arabidopsis NIT enzymes. This result, and the fact that ZmNIT2 is expressed in kernels where high concentrations of IAA are synthesized by the Trp-dependent pathway, led them to suggest that ZmNIT2 is involved in IAA synthesis in maize. In our study, we found two genes similar to ZmNIT2 in the rice genome, and one of these genes encodes a protein 90% identical to ZmNIT2 at the amino acid sequence level (data not shown). Thus, this rice gene may be an ortholog of ZmNIT2 that functions in IAA synthesis in rice.
The alternative pathway leading from IAOx to IAA occurs via an IAAld intermediate. Seo et al. (1998) reported that aldehyde oxidase (AAO1), which catalyzes the conversion of IAAld to IAA, may be involved in IAA synthesis in Arabidopsis seedlings. We found three homologs of Arabidopsis AAO1 in the rice genome (data not shown). Although distinguishing between IAA and ABA synthesis-related AAO1s by their primary structures alone is difficult, owing to their highly similar sequences, one or more of these apparent AAO1 homologs may be involved in conversion of IAAld to IAA in rice.
The above results are consistent with the proposed biosynthetic pathway for IAA in rice shown in Figure 1. In the first two steps of this proposed pathway, which occur in the cytoplasm, Trp is converted to TAM by a decarboxylase reaction, and TAM is then oxygenized by OsYUCCA1 to produce NHT. The direct conversion of Trp to IAOx by CYP79B2/CYP79B3, an alternative pathway occurring in chloroplasts, appears not to contribute to IAA production in rice. In the third and fourth steps, the product of the YUCCA-catalyzed reaction, NHT, is converted to IAOx and then to IAN. In the final two steps, nitrilase converts IAN to IAAld, and AAO1 converts IAAld to IAA, as in Arabidopsis.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Wild-type rice (Oryza sativa L. cv Taichung 65) and transgenic rice plants were grown in Murashige and Skoog medium (Murashige and Skoog, 1962) containing 3% (w/v) Suc and 0.3% (w/v) Gelite, unless otherwise indicated. A stock solution of DEX (Wako Pure Chemical Industries) was prepared at 100 mm in dimethyl sulfoxide (Wako Pure Chemical Industries) and used in growth medium at 100 μm.
Construction of pAct-OsYUCCA1
To obtain the correct full-length cDNA for OsYUCCA1 (AK105488), which had two annotations in silico, two segments of OsYUCCA1 were PCR amplified. The 5′ segment was amplified with the forward primer 5′-acctcgagcgctacctacacacacaaccg-3′ (an XhoI linker site [underlined] was inserted) and the reverse primer 5′-gtgaacagtactccggcatccttgagccaagatggcacg-3′ (a ScaI site was introduced in frame). The 3′ segment was amplified with the forward primer 5′-gatgccggagtactgttcacgaggg-3′ (a ScaI site was introduced in frame) and the reverse primer 5′-cacacgtatattgtggtacaagatgatcaca-3′ (a SpeI site was added as a linker). The resulting PCR products were ligated into the cloning site of pCR-TOPO (Invitrogen). After the two cloned fragments were digested with XhoI and ScaI or ScaI and SpeI, then they were ligated into pBluescript SK+ (pBS-SK), and the insert was subjected to DNA sequencing to confirm their nucleotide sequences. The insert DNA was redigested with XhoI and SpeI. The resulting fragment ends were blunted and ligated into the binary vector pBI101-Hm2 (Ohta et al., 1990), which contains the Act1 promoter-NOS terminator fused into the SmaI site.
Construction of 6×UAS-OsYUCCA1
The cloned fragments were ligated into the vector pTA7001-6×GAL4 UAS, which harbors amplified DNA fragments encoding the two-component GAL4 (amino acids 1–77)-VP16 (amino acids 413–490)-rat glucocorticoid-binding domain (amino acids 519–795) construct between the 35S promoter from pBI221 (Aoyama and Chua, 1997) and the pea (Pisum sativum) rbcs-E9 terminator.
Construction of pAct1-antisense OsYUCCA1
The cloned fragments were ligated inversely into the binary vector pBI101-Hm2 (Ohta et al., 1990), which contains the Act1 promoter-NOS terminator fused into the SmaI site.
GUS Reporter Gene Constructs
To place the GUS reporter gene under control of the OsYUCCA1 promoter, we PCR amplified nucleotides −2097 to 1029 of the OsYUCCA1 gene sequence from the rice genome. The PCR product was then inserted in front of the GUS gene of pBI-Hm (kindly provided by Prof. Kenzo Nakamura, Nagoya University) to make the chimeric OsYUCCA1 promoter-GUS reporter gene construct (OsYUCCA1-GUS). The chimeric DR5 promoter-GUS reporter gene construct (DR5-GUS) was generated as previously reported (Scarpella et al., 2003).
Expression of GUS protein in whole plants was detected by staining for GUS activity. The plants were submerged in 2 to 3 mL of GUS buffer (0.05 m sodium phosphate, pH 8.0, 7% (v/v) methanol, 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-glucuronic acid) and incubated at 37°C in the dark. Stained plants were washed once in sterile water and then destained in several changes of 70% (v/v) ethanol until the chlorophyll was bleached. The plants were stored in 50% (v/v) glycerol.
Rice Transformation
The chimeric reporter gene constructs were introduced into Agrobacterium tumefaciens strain EHA101 and used to infect rice calli according to the method of Hiei et al. (1994). Transformed cells and plants were screened by selection for hygromycin (Wako Pure Chemical Industries) and were maintained in sterile culture. Regenerated plants were then grown to maturity in pots in a greenhouse. The primary transformants were self pollinated, and the resulting seeds (T1) were collected.
RNA Isolation and RT-PCR Analysis
To estimate mRNA expression levels for OsYUCCA genes 1 to 7 in rice organs, qRT-PCR analysis was performed. Rice plants in vegetative phase were grown for 2 weeks in Murashige and Skoog medium containing 3% (w/v) Suc and 0.3% (w/v) Gelite, unless otherwise indicated, and plants in reproductive phase were grown for 2 months in soil-filled pots in a greenhouse. Total RNA was extracted with TRIzol regent (Invitrogen), and the first-strand cDNA was synthesized from 2 μg of total RNA using an Omniscript RT kit (Qiagen). The PCR parameters for the detection of the OsYUCCA genes were 94°C for 5 min followed by 25, 30, 35, and 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min; the primers used for PCR are shown in Table I.
Table I.
Primers used for qRT-PCR analysis of OsYUCCA gene expression in rice organs
| Gene | Forward Primer Sequence (5′ → 3′) | Reverse Primer Sequence (5′ → 3′) |
|---|---|---|
| OsYUCCA1 | tcatcggacgccctcaacgtcgc | ggcagagcaagattatcagtc |
| OsYUCCA2 | gtccaaagggaggagtcgtccag | gcatgatgtttacacccggcctt |
| OsYUCCA3 | gtgagaacgggctctactcggtcg | gcttatgcatgaccgatgaacacg |
| OsYUCCA4 | gcagaatggcctgtacgctgttgg | cagaccagcacatgacgtgtctac |
| OsYUCCA5 | acctcctacgacgccgccatgatc | ctcccaacacagcgacgacagaac |
| OsYUCCA6 | ccattcccagatggttggaagg | catgttgcgcctcaagatatttg |
| OsYUCCA7 | cactgctgtgtcctacaatatcac | ggaggtgcatctccgtcatcttc |
To assess the induction kinetics of OsYUCCA1-7 and OsIAA3, plants were grown for 2 weeks and then treated with 20 μm IAA (Wako Pure Chemical Industries). The OsIAA3 gene was analyzed using the forward primer 5′-gggttctccaagacatgcaatc-3′ and the reverse primer 5′-ggatggaaatcaaagcattgaagc-3′. The PCR parameters for the detection of OsIAA3 were 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min.
Phenotypic Analysis
Plants were photographed using a digital camera (Nikon). To examine the response of plants to gravity, seedlings were grown vertically for 10 d under continuous light and dark conditions, rotated 90° to the horizontal plane, and then grown for two more days.
Histological Analysis
Node and shoot apex were embedded in Tissue-Tek OCT compound (Sakura Finetechnical) and sectioned at 30 μm intervals.
Quantification of Endogenous IAA
The preparation of samples and procedure of IAA quantification were according to Mori et al. (2005) with minor modifications. Rice leaves of shoots (100 mg) grown in light for 1 week were used for the analysis. The gas chromatography-mass spectrometry analyses were carried out by a mass spectrometer (JMS-K9, JEOL) with a gas chromatograph (6890 N, Agilent Technology) fitted with a capillary column (HP-5, 0.32 mm i.d. × 30 m, 0.25-μm film thickness; Agilent Technology).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers OsYUCCA1; BAD68007, OsYUCCA2; AAU43964, OsYUCCA3; BAD87432, OsYUCCA4; BAB32703, OsYUCCA5; ABA99096, OsYUCCA6; BAC80117, OsYUCCA7; CAE76085, YUCCA1; At4g32540, YUCCA2; At4g13260, YUCCA3; At1g04610, YUCCA4; At5g11320, YUCCA5; At5g43890, YUCCA6; At5g25620, YUCCA7; At2g33230, YUCCA8; At4g04610, YUCCA9; At1g04180, YUCCA10; At1g48910, YUCCA11; At1g21430.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of rice and Arabidopsis YUCCA and YUCCA-like proteins. Black and gray boxes indicate identical and similar amino acids, respectively. The putative FAD and NADPH binding motifs are boxed.
Supplementary Material
Acknowledgments
We thank Drs. Takashi Aoyama for providing the pTA7001 plasmid, Tomokazu Koshiba for suggestions concerning IAA measurement, and Hidemi Kitano for discussion.
This work was supported by the Center of Excellence and the Japan Rice Genome Project of the Ministry of Agriculture, Forestry and Fisheries (grant-in-aid).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Makoto Matsuoka (makoto@agr.nagoya-u.ac.jp).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aoyama T, Chua N-H (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11 605–612 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JD, Slovin JP, Hendrickson AM (2003) Two genetically discrete pathways convert tryptophan to auxin: more redundancy in auxin biosynthesis. Trends Plant Sci 8 197–199 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Estelle M (2004) Auxin signaling and regulated protein degradation. Trends Plant Sci 9 302–308 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6 271–282 [DOI] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Fujioka S, Takatsuto S, Yoshida S, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M (2005) The rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell 17 2243–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull AK, Vij R, Celenza JL (2000) Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci USA 97 2379–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Im KH, Savka MA, Wu MJ, DeWitt NG, Shillito R, Binns AN (1998) Auxin-dependent cell expansion mediated by overexpressed auxin-binding protein 1. Science 282 1114–1117 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 446–451 [DOI] [PubMed] [Google Scholar]
- Koshiba T, Matsuyama H (1993) An in vitro system of indole-3-acetic acid formation from tryptophan in maize (Zea mays) coleoptile extracts. Plant Physiol 102 1319–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O (2002) Molecular genetics of auxin signaling. Annu Rev Plant Biol 53 377–398 [DOI] [PubMed] [Google Scholar]
- McElroy D, Zhang W, Cao J, Wu R (1990) Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Hansen CH, Wittstock U, Halkier BA (2000) Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J Biol Chem 275 33712–33717 [DOI] [PubMed] [Google Scholar]
- Mori Y, Nishimura T, Koshiba T (2005) Vigorous synthesis of indole-3-acetic acid in the apical very tip leads to a constant basipetal flow of the hormone in maize coleoptiles. Plant Sci 168 467–473 [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant 15 473–498 [Google Scholar]
- Nakamura A, Umemura I, Gomi K, Hasegawa Y, Kitano H, Sazuka T, Matsuoka M (2006) Production and characterization of auxin-insensitive rice by overexpression of a mutagenized rice IAA protein. Plant J 46 297–306 [DOI] [PubMed] [Google Scholar]
- Ohta N, Masurekar M, Newton A (1990) Cloning and cell cycle-dependent expression of DNA replication gene dnaC from Caulobacter crescentus. J Bacteriol 172 7027–7034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov IA, Teale WD, Trebar M, Blilou I, Palme K (2005) The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends Plant Sci 10 170–177 [DOI] [PubMed] [Google Scholar]
- Park WJ, Kriechbaumer V, Moller A, Piotrowski M, Meeley RB, Gierl A, Glawischnig E (2003) The Nitrilase ZmNIT2 converts indole-3-acetonitrile to indole-3-acetic acid. Plant Physiol 133 794–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippar K, Fuchs I, Luthen H, Hoth S, Bauer CS, Haga K, Thiel G, Ljung K, Sandberg G, Bottger M, et al (1999) Auxin-induced K+ channel expression represents an essential step in coleoptile growth and gravitropism. Proc Natl Acad Sci USA 96 12186–12191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribnicky DM, Cooke TJ, Cohen JD (1998) A microtechnique for the analysis of free and conjugated indole-3-acetic acid in milligram amounts of plant tissue using a benchtop gas chromatograph-mass spectrometer. Planta 204 1–7 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99 463–472 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E, Rueb S, Meijer AH (2003) The RADICLELESS1 gene is required for vascular pattern formation in rice. Development 130 645–658 [DOI] [PubMed] [Google Scholar]
- Seo M, Koiwai H, Akaba S, Komano T, Oritani T, Kamiya Y, Koshiba T (1998) Abscisic aldehyde oxidase in leaves of Arabidopsis thaliana. Plant J 23 481–488 [DOI] [PubMed] [Google Scholar]
- Teale WD, Paponov IA, Palme K (2006) Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7 847–859 [DOI] [PubMed] [Google Scholar]
- Tobena-Santamaria R, Bliek M, Ljung K, Sandberg G, Mol JN, Souer E, Koes R (2002) FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev 16 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward C, Bemis SM, Hill EJ, Sawa S, Koshiba T, Torii KU (2005) Interaction of auxin and ERECTA in elaborating Arabidopsis inflorescence architecture revealed by the activation tagging of a new member of the YUCCA family putative flavin monooxygenases. Plant Physiol 139 192–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zazimalova E, Napier RM (2003) Points of regulation for auxin action. Plant Cell Rep 21 625–634 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291 306–309 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR, Normanly J, Chory J, Celenza JL (2002) (Trp)-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev 16 3100–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.