Abstract
Far-red (FR) insensitive 219 (FIN219) was previously shown to be involved in phytochrome A-mediated FR light signaling. To further understand its function and regulatory relation with other light-signaling components, a yeast two-hybrid approach was used to isolate FIN219-interacting partners. Here, we demonstrate that FIN219-interacting protein 1 (FIP1) interacts with FIN219 in vitro and in vivo and is composed of 217 amino acids that belong to the tau class of the large glutathione S-transferase gene family. FIP1 was further shown to have glutathione S-transferase activity. The gain of function and partial loss of function of FIP1 resulted in a hyposensitive hypocotyl phenotype under continuous FR (cFR) light and a delayed flowering phenotype under long-day conditions, which suggests that FIP1 may exist in a complex to function in the regulation of Arabidopsis (Arabidopsis thaliana) development. In addition, FIP1 mRNA was down-regulated in the suppressor of phytochrome A-105 1 mutant and differentially expressed in constitutive photomorphogenic 1-4 (cop1-4) and cop1-5 mutants under cFR. Intriguingly, FIP1 expression was up-regulated in the fin219 mutant under all light conditions, except cFR. Furthermore, promoter activity assays revealed that FIP1 expression was light dependent, mainly associated with vascular tissues, and developmentally regulated. Subcellular localization studies revealed that the β-glucuronidase-FIP1 fusion protein was localized in the nucleus and cytoplasm. Taken together, these data indicate that FIP1 may interact with FIN219 to regulate cell elongation and flowering in response to light.
Light has a profound effect on plant growth and development. It not only provides an energy source for plant photosynthesis, but also acts as an important signal to regulate gene expression and various aspects of plant development (Kendrick and Kronenberg, 1994). Plants are equipped with different photoreceptors to sense changes in light. At least four different photoreceptor classes are found in Arabidopsis (Arabidopsis thaliana): phytochromes for red (R) and far-red (FR) light, cryptochromes and phototropins for blue (B) and UV-A light, and an unknown photoreceptor for UV-B light. Phytochromes are the most extensively studied among these photoreceptors and exist in phytochrome R-absorbing (Pr) and phytochrome FR-absorbing (Pfr) forms.
Research into light signal transduction by molecular genetics, cell biology, and DNA microarray approaches has made great progress (Tepperman et al., 2001, 2004; Jiao et al., 2003; Liscum et al., 2003; Matsushita et al., 2003; Parks, 2003; Bauer et al., 2004). In particular, the phytochrome A (phyA)-mediated signaling pathway in continuous FR (cFR) light has been intensively studied. Many intermediate transducers have been isolated (Soh et al., 1998, 2000; Hoecker et al., 1999; Hudson et al., 1999; Bolle et al., 2000; Fairchild et al., 2000; Fankhauser and Chory, 2000; Hsieh et al., 2000; Ballesteros et al., 2001; Desnos et al., 2001; Dieterle et al., 2001; Zeidler et al., 2001; Wang and Deng, 2002), most localized in the nuclei (Ni et al., 1998; Hoecker et al., 1999; Hudson et al., 1999; Fairchild et al., 2000; Fankhauser and Chory, 2000; Soh et al., 2000; Ballesteros et al., 2001; Wang and Deng, 2002), some in the cytosol (Bolle et al., 2000; Hsieh et al., 2000), and several in both subcellular locations (Desnos et al., 2001; Zeidler et al., 2001). So phyA can transduce the light signal through these components or directly interact with transcription factors, such as phytochrome-interacting factor 3 (PIF3) to turn on gene expression (Martinez-Garcia et al., 2000). In addition, recent evidence indicated that phyA-mediated signaling is desensitized through ubiquitination of the downstream positive regulators LONG AFTER FAR-RED LIGHT 1 (LAF1) and LONG HYPOCOTYL 5 (HY5) by CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) (Hardtke et al., 2000; Seo et al., 2003), a key repressor of photomorphogenesis in Arabidopsis. Furthermore, SUPPRESSOR OF PHYTOCHROME A-105 1 (SPA1), a nuclear-localized suppressor of phyA activity, can modulate expression of both LAF1 and HY5 by interacting with COP1, thus desensitizing the phyA-mediated signaling pathway (Saijo et al., 2003; Seo et al., 2003).
Protein-protein interactions play a critical role in signal transduction. For light signaling, a yeast two-hybrid method has been used to search for interaction partners of the phytochrome photoreceptors. A group of basic helix-loop-helix transcription factors, PIFs, isolated by this approach in Arabidopsis, have been found to interact with the C-terminal domain of phyA or phyB and are involved in different aspects of phytochrome-mediated phenotypes, such as chlorophyll biosynthesis (Huq et al., 2004), chloroplast development (Monte et al., 2004), light-responsive gene expression (Ni et al., 1998), and hypocotyl elongation (Kim et al., 2003). Moreover, a family of PIF-like (PIL) proteins, such as PIL5 and PIL6, which belong to basic helix-loop-helix factors, also play a crucial role in seed germination and circadian-controlled R light signaling, respectively (Fujimori et al., 2004; Oh et al., 2004). In addition, phytochrome kinase substrate 1 (PKS1), isolated from a yeast two-hybrid screen with the C-terminal 160 amino acids of PHYA used as bait, interacted with Pr and Pfr forms of both PHYA and PHYB and acted in the cytosol as a substrate of both photoreceptor kinases. Another PHYA-interacting protein, nucleoside diphosphate kinase 2 (NDPK2), has a preference for binding the Pfr form and functions as a positive regulator in both phyA and phyB signaling. Recent studies indicate that NDPK2 appears to be involved in auxin-regulated processes, such as cotyledon development, by modulating auxin transport (Choi et al., 2005). Therefore, phytochromes, through interacting with different factors either in the cytosol or in the nucleus, are able to regulate a number of aspects of plant development.
However, interacting partners of the intermediate components and their regulatory relations in light signaling remain to be elucidated. The far-red insensitive 219 (fin219) mutant was derived from the extragenic suppressor screening of the cop1-6 mutant in Arabidopsis and exhibited less sensitivity specifically to cFR. Its gene was cloned by a map-based method and the derived product shares 36% to 47% identity with a GH3 gene family of 19 members in the Arabidopsis genome (Hsieh et al., 2000). The GH3 gene was originally isolated from soybeans (Glycine max) in a rapid induction by auxin (Hagen et al., 1984). Recent studies found that JASMONATE INSENSITIVE 1 (JAR1), responsible for the jar1-1 mutation, has the same locus as FIN219 and belongs to the firefly (Photinus pyralis) luciferase family of adenylate-forming enzymes (Staswick et al., 2002). Further results indicated that JAR1 is actually a jasmonic acid (JA)-amino synthetase and mediates the formation of JA conjugation with various amino acids. Interestingly, the JA-Ile conjugate can complement the jasmonate insensitivity of the jar1-1 mutant (Staswick and Tiryaki, 2004). In addition, FIN219 has been shown to be induced rapidly by auxin and localized constitutively in the cytosol without changes in subcellular location by light. Moreover, FIN219 was demonstrated to be a suppressor of COP1 (Hsieh et al., 2000), which suggests that FIN219/JAR1 is an important regulator in modulating the integration of phytohormone signaling through auxin, jasmonate, and light signaling. However, its physiological function in light signaling and plant development remains to be elucidated.
To further understand the function of FIN219, we show that FIN219 is indeed involved in phyA-mediated FR light signaling and report on the isolation of a FIN219-interacting partner from a yeast two-hybrid library obtained from the Arabidopsis Biological Resource Center (ABRC). This gene, encoding a glutathione S-transferase (GST; At1g78370/AtGSTU20), was demonstrated to interact with FIN219 in vitro and in vivo and thus was named FIN219-interacting protein 1 (FIP1). Transgenic studies revealed that FIP1 may exist in a complex to regulate hypocotyl elongation and flowering. Promoter activity assays indicated that FIP1 expression was highly associated with the vascular tissues of hypocotyls, cotyledons, leaves, and floral organs. Taken together, these data reveal that FIP1, through interaction with FIN219 in response to light, may play a crucial role in cell elongation and plant development.
RESULTS
The jar1-1 Allele Has a Hyposensitive Phenotype under cFR
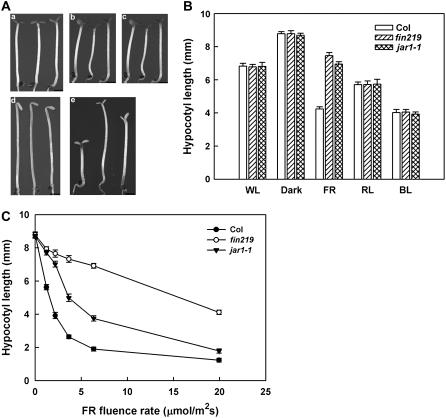
The fin219 mutant in Arabidopsis has been shown to have a long hypocotyl phenotype under cFR (Hsieh et al., 2000). Recently, the JAR1 gene was reported to have the same locus as FIN219; however, its corresponding mutant alleles did not show a hyposensitive phenotype under cFR (Staswick et al., 2002). To understand the discrepancy between the fin219 and jar1-1 mutants under cFR, we investigated the phenotype of the jar1-1 mutant and found a longer hypocotyl phenotype under cFR than under other light conditions as compared with the wild-type and fin219 mutant (Fig. 1, A and B). jar1-1 consistently displayed an intermediate hypocotyl phenotype under cFR of less than 20 μmol m−2 s−1. Especially under low fluence rates of FR at 1.47 μmol m−2 s−1, the hypocotyl length was close to that of fin219 (Fig. 1C). In addition, a null allele (SALK_059774) of FIN219/JAR1 (Supplemental Fig. S1), obtained from the ABRC, exhibited a similar pattern to that of the jar1-1 allele (data not shown). Thus, the jar1 allele, similar to the fin219 mutant, exhibits a long hypocotyl phenotype under cFR.
Figure 1.
jar1-1 mutation confers hyposensitivity to cFR light. A, Phenotype of wild-type, fin219 mutant, and jar1-1 mutant seedlings grown for 4 d in the dark or under different light conditions. a, White light (5.10 μmol m−2 s−1). b, R light (6.34 μmol m−2 s−1). c, B light (5.08 μmol m−2 s−1). d, Dark. e, FR light (1.47 μmol m−2 s−1). Sample order (left to right) in a to e, Wild type, fin219, jar1-1. All size bars = 1 mm. B, Measurement of hypocotyl length of wild-type Columbia, fin219 mutant, and jar1-1 mutant seedlings grown for 4 d in the dark or under different light conditions. Light intensity is the same as in A. Error bars indicate sd (n = 30). C, Fluence rate response curves for hypocotyls of wild-type Columbia, fin219 mutant, and jar1-1 mutant seedlings grown for 4 d in cFR light. Error bars indicate sd (n = 30).
Isolation of FIN219-Interacting Partners by the Yeast Two-Hybrid Method
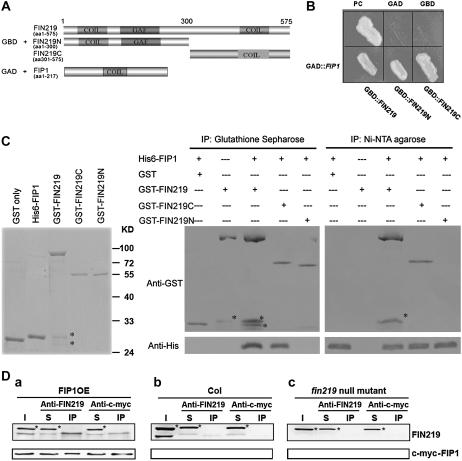
FIN219 is involved in the phyA-mediated light-signaling pathway and is induced rapidly by auxin. It shares 36% to 47% identity at the amino acid level with GH3 members in Arabidopsis (Hsieh et al., 2000) and contains two coiled-coil domains, one in the N terminus and the other in the C terminus (Fig. 2A), which implies that FIN219 may interact with other proteins via these domains. To further understand the FIN219 function in light signaling and plant development, we used a yeast two-hybrid method to isolate FIN219-interacting partners. A FIN219 full-length cDNA was cloned into yeast GAL4 DNA-binding domain (BD) vector pGBT9 (+2) and used as bait to screen a library CD4-10 obtained from the ABRC. Three clones recovered from medium plates lacking Trp, Leu, and His and were further checked by the use of a yeast mating approach. One of them, FIP1, showed potential interactions with the full-length, N-terminal, and C-terminal regions of FIN219 on the plates lacking Trp, Leu, and His (Fig. 2B). The interaction between FIP1 and FIN219 was further confirmed by in vitro pull-down assay (Fig. 2C) and in vivo coimmunoprecipitation (co-IP; Fig. 2D). The GST-FIN219 full-length, N-terminal 300 amino acids and C-terminal 274 amino acid fusions and the His-6-FIP1 fusion were expressed in Escherichia coli; purified recombinant proteins were used for pull-down assays (Fig. 2C, left). The results from pull-down assay with either glutathione (GSH) sepharose for the GST tag or nickel-nitrilotriacetic acid agarose (Ni-NTA) for the His tag revealed that FIP1 can interact with full-length and C-terminal regions of FIN219 rather than the N terminus (Fig. 2C, right). Interestingly, when His-6-FIP1 and GST tags were mixed, only the GST tag was pulled down by GSH sepharose resins, but not His-6-FIP1 (Fig. 2C, first lane of the middle image); similarly, when His-6-FIP1 and GST tag mixtures were pulled down with Ni-NTA resins, the pulled-down His-6-FIP1 was not recognized by monoclonal anti-GST antibodies (Fig. 2C, first lane of the right image), which is consistent with the notion that GST members in the large GST gene family are quite diverse among different classes (Dixon et al., 2002b). The interaction between FIP1 and FIN219 was further demonstrated in Arabidopsis. Co-IP was carried out with either FIN219 polyclonal antibodies raised against the N-terminal 300 amino acids of FIN219 or c-myc antibodies to precipitate total proteins isolated from 3-d-old cFR-grown transgenic seedlings overexpressing FIP1 cDNA with a c-myc tag. Although FIN219 antibodies recognized two bands, the lower one is the correct one because it was compared with that of wild-type Columbia and a fin219 null mutant, fin219T (SALK_059774; Supplemental Fig. S1). The cross-hybridized band was not immunoprecipitated by FIN219 antibodies in vivo, which indicates that the lower band recognized by antibodies was specific enough to show its interaction with FIP1 (Fig. 2D, a). Consistently, anti-c-myc antibodies were able to immunoprecipitate c-myc-FIP1 as well as FIN219 (Fig. 2D, a). In contrast, FIN219 antibodies can immunoprecipitate FIN219, but not c-myc-FIP1 from wild-type Columbia (Fig. 2D, b), whereas the fin219 null mutant as a control showed negative results (Fig. 2D, c). Thus, FIP1 is indeed capable of interacting with the C-terminal region of FIN219.
Figure 2.
FIP1 interacts with the C terminus of FIN219. A, Three different versions of FIN219 as bait and FIP1 as a prey used for interaction studies. FIN219 full length, FIN219N containing 300 amino acids in the N terminus, and FIN219C consisting of 274 amino acids in the C terminus of FIN219 are shown with the locations of the coiled-coil (Coil) and GAF domains. B, In vivo interaction analysis of FIP1 and FIN219 by the yeast GAL4 two-hybrid system. Three versions of FIN219 proteins expressed as bait fusion proteins and FIP1 protein as activator domain fusions were cotransformed into a yeast host AH109; transformants were selected on synthetic dextrose medium lacking Trp, Leu, and His (−trp −leu −his). PC, Positive control using COP1 as bait and HY5 as prey; GAD, negative control containing FIN219 as bait and an empty AD vector pACT; GBD, negative control containing an empty DNA BD vector pGBT9 (+2) and FIN219 as prey. Right, β-galactosidase activity assay using the filter-lift method. Filter paper was used to lift the yeast colonies from the plate shown in the left and carry out β-galactosidase activity assay by the filter method. C, Pull-down assay to confirm the interaction between FIP1 and FIN219, with HY5 used as prey. GAD, Negative control containing FIN219 as bait and an empty AD vector pACT; GBD, negative control containing an empty DNA BD vector pGBT9 (+2) and FIN219 used as prey. C, Pull-down assay to confirm the interaction between FIP1 and FIN219. Five micrograms of purified GST and recombinant proteins His-6-FIP1, GST-FIN219, GST-C274, and GST-N300 were used for pull-down assay shown with Coomassie Blue-stained gel (left). Purified respective recombinant proteins were mixed as depicted above the image and incubated at 4°C overnight, then immunoprecipitated with either GSH sepharose against GST tags (middle) or Ni-NTA resins against His tags (right), and resulting pellets were washed and underwent protein gel-blot analyses with anti-GST or anti-His antibodies. C, Asterisk (*) indicates breakdown products of the GST-FIN219 full length. D, Co-IP assay of FIP1 interaction with FIN219. One milligram of protein extracts isolated from FIP1 as a FIP1-c-myc fusion overexpressor (a), wild-type Columbia (b), and fin219 null mutant (c) of transgenic seedlings grown in cFR light for 4 d were mixed with either FIN219 polyclonal antibodies or c-myc antibodies and then coimmunoprecipitated. After SDS-PAGE and protein blotting, membranes were probed with FIN219 and c-myc antibodies. I, Input proteins; S, supernatants after precipitation; IP, pellets after precipitation. D, Asterisk (*) indicates a nonspecifically cross-hybridized band.
FIP1 Encodes a Plant GST with Affinity on GSH and 1-Chloro-2,4-Dinitrobenzene Substrates
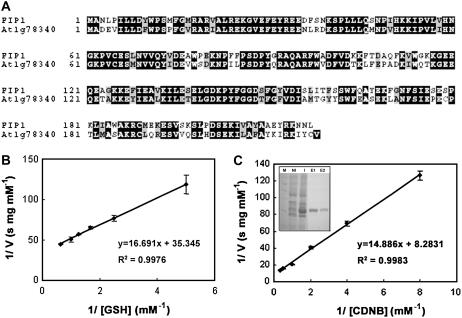
Sequencing and BLAST searching revealed that the FIP1 clone isolated from yeast two-hybrid screening encodes a plant GST with 217 amino acids (AtGSTU20/At1g78370) and belongs to the tau class of a large GST gene family composed of 53 members grouped into six classes in Arabidopsis (Dixon et al., 2002a; Wagner et al., 2002). The FIP1 gene consists of two exons and one intron and shares 67% identity at the amino acid level over the whole coding region with At1g78340 (AtGSTU22) within the same tau class (Fig. 3A). However, the similarity of FIP1 to other GSTs of different classes in the same gene family was below 25%. All plant GSTs display a conserved tertiary structure, although the primary amino acid sequences are quite diverse, which indicates that binding of the ligand such as GSH was important for their activities and functions during evolution (Dixon et al., 2002b). To further understand whether FIP1 has GST activity implied from its primary amino acid sequence, we expressed it in E. coli and obtained the recombinant at about 95% purity for activity assay (Fig. 3C, inset). The result showed that FIP1 can use GSH and 1-chloro-2,4-dinitrobenzene (CDNB) as substrates with Km = 0.467 mm and 1.794 mm, respectively (Fig. 3, B and C), which is similar to the value reported in the literature (Singhal et al., 1991; Prapanthadara et al., 1996). Thus, FIP1 interacting with FIN219 does have GST activity.
Figure 3.
An alignment of FIP1 primary sequence and a GST member At1g78340 at the amino acid level as well as activity assays of FIP1. A, Alignment of FIP1 with the closest GST member At1g78340 having 67% identity at the amino acid level. B and C, GST activity assays of FIP1 with different concentrations of GSH (B) and CDNB (C) used as substrates. C, Inset indicates 95% purity of the recombinant protein FIP1 used for enzymatic activity assays. M, Protein marker; NI, no IPTG induction; I, with IPTG induction; E1 and E2, eluted protein.
Transgenic Seedlings Overexpressing or Showing Reduced FIP1 Expression Exhibit a Hyposensitive Phenotype under cFR
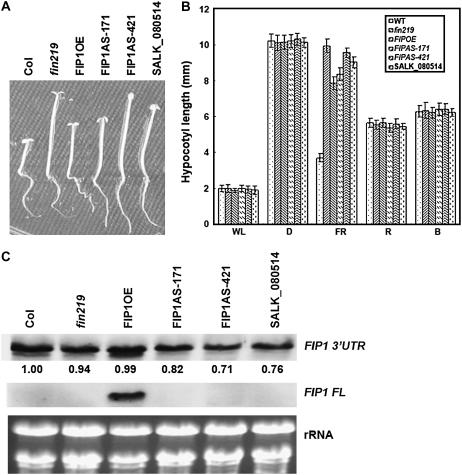
To determine whether FIP1 is involved in the light-signaling pathway, we overexpressed the FIP1 full-length cDNA as a 35S∷c-myc-FIP1 construct into wild-type Columbia to examine any effect on the phenotype and its response to light signals. Seven of 14 T2 transgenic lines showed a hyposensitive phenotype under cFR whereby the hypocotyl length is intermediate to that of wild-type Columbia and the fin219 mutant (Fig. 4, A and B). Under other light conditions, no obvious hypocotyl phenotype was observed (Fig. 4B). In addition, we introduced another translational fusion construct, 35S∷pRTL2-GUS-FIP1, into wild-type Columbia. Twelve of 30 T2 transgenic lines displayed the same phenotype as those containing the construct 35S∷c-myc-FIP1, showing longer hypocotyls than those of the wild type under cFR (data not shown). To further confirm whether the longer hypocotyl phenotype of transgenic seedlings was indeed caused by the overexpression of FIP1, RNA gel-blot analysis revealed that transgenic seedlings grown in the same condition as the phenotype described above showed highly expressed FIP1 mRNA (Fig. 4C, middle image), which confirms that FIP1 overexpression resulted in a less sensitive hypocotyl phenotype to cFR. Moreover, transgene expression did not affect endogenous FIP1 expression detected by the 3′-untranslated region (UTR) of FIP1 as a riboprobe (Fig. 4C, top). In addition, we generated transgenic plants harboring an antisense construct of FIP1 and obtained one T-DNA insertion line from the ABRC (SALK_080514). This T-DNA insertion line contains one T-DNA inserted in the promoter region of FIP1, which was confirmed by a FIP1 gene-specific primer and T-DNA border sequences. Homozygous FIP1 antisense transgenic seedlings and T-DNA-inserted seedlings underwent phenotypic investigation under various light conditions. Unexpectedly, all these transgenic seedlings displayed a long hypocotyl phenotype under cFR, with no obvious phenotype under other light conditions (Fig. 4, A and B). RNA gel-blot analysis further indicated that FIP1 transcripts were slightly reduced in amount, by 20% to 30%, in transgenic seedlings of the FIP1 antisense lines FIP1AS-171 and FIP1AS-421, as well as the T-DNA inserted line. When the same blot was reprobed by full-length FIP1 cDNA, the signal was barely detected (Fig. 4C) and became visible under longer exposure (data not shown), which indicates that FIP1 transcripts were low in amount under normal conditions in terms of sensitivity detectable by RNA gel-blot analysis. Given that transgenic seedlings overexpressing or showing decreased FIP1 expression exhibit a similar long hypocotyl phenotype under cFR, FIP1 might exist in a complex to perform its function, such as regulation of cell elongation. A similar case is also found in EARLY BOLTING IN SHORT DAYS (EBS). Overexpression of EBS results in phenotypic effects similar to those of recessive ebs mutations, namely, early flowering, dwarf phenotype, and reduced fertility (Pineiro et al., 2003). Therefore, pursuing further physiological functions of the potential complex containing FIP1 will be interesting.
Figure 4.
Investigation of phenotype and gene expression of FIP1 transgenic seedlings grown under various light conditions. A, Phenotypes of transgenic seedlings overexpressing or inhibiting FIP1 expression under cFR irradiation. Wild-type Columbia was used as a control. fin219 mutant is another control. FIPOE, FIP1 overexpressor; FIP1AS-171 and FIP1AS-421, FIP1 antisense lines. SALK_080514, T-DNA insertion line of FIP1. Homozygous transgenic seedlings were grown under cFR for 4 d. B, Measurement of hypocotyl length of wild-type Columbia, fin219 mutant (fin219), FIP1 overexpressor (FIP1OE), and FIP1 antisense line (FIP1AS-171 and FIP1AS-421) seedlings grown for 4 d in the dark or under different light conditions. Light intensity, White light (WL) 77 μmol m−2 s−1; FR light, 1.47 μmol m−2 s−1; R light, 93.72 μmol m−2 s−1; and B light, 3.75 μmol m−2 s−1. D, Darkness. Error bars indicate sd (n = 30). C, RNA gel-blot analysis of FIP1 gene expression in transgenic seedlings grown in cFR for 4 d; 20 μg total RNA were loaded onto a gel, then blotted to a nylon membrane (Roche) and probed with a gene-specific region at the 3′-UTR of the FIP1 gene (FIP1 3′-UTR) and the full length of FIP1 cDNA (FIP1 FL).
FIP1 Expression Is Regulated by Light-Signaling Transducers
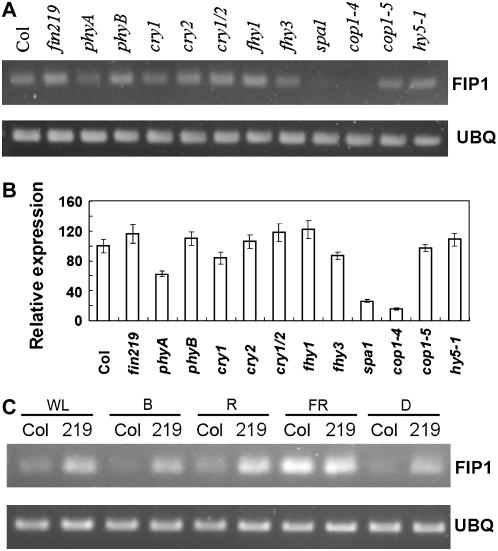
Because transgenic plants overexpressing or reducing FIP1 expression exhibit a hyposensitive long-hypocotyl phenotype in cFR (Fig. 4, A and B) and FIP1 has been shown to interact with FIN219 in vivo under cFR (Fig. 2), a positive regulator involved in the phyA-mediated FR light-signaling pathway, we tested whether various photoreceptors and light-signaling components regulate FIP1 expression. FIP1 expression was examined by reverse transcription (RT)-PCR with gene-specific primers in various mutants grown in cFR for 4 d. FIP1 mRNA expression was comparably expressed among the wild type, phyA mutant, and fin219 mutant. In contrast, its transcripts were significantly decreased in spa1, a suppressor of phyA, and in the weak allele cop1-4, which produced N-terminal 282 amino acids of COP1, but in a wild-type level in the null allele cop1-5 (Fig. 5, A and B), which implies that FIP1 was positively regulated by SPA1 and negatively controlled by COP1. Recent evidence shows that SPA1 and COP1 work together to repress photomorphogenesis (Yang and Wang, 2006). It will be interesting to see how SPA1 and COP1 regulate FIP1 expression at the protein level. To further understand the functional features of FIP1 expression regulated by light, we then tested FIP1 mRNA levels by RT-PCR with gene-specific primers in the wild type and the fin219 mutant under different light conditions. Intriguingly, FIP1 expression in the wild type was greatly increased in cFR as compared to other light conditions (Fig. 5C). Moreover, its expression was up-regulated in the fin219 mutant under all light conditions, except for cFR (Fig. 5C), under which FIP1 expression was comparable to that of the wild type. This result suggests that FIN219 may negatively control FIP1 expression under most light conditions.
Figure 5.
FIP1 gene expression in photoreceptor mutants and various light-signaling mutants under different light conditions. RT-PCR analysis was performed in the seedlings of different photoreceptor and light-signaling component mutants grown under cFR (A) and various light conditions (C) for 4 d. B, Quantitative expression data of A. Error bars indicate sd based on two independent experiments. One microgram of total RNA isolated from the seedlings underwent RT-PCR with gene-specific primers in the 3′-UTR of FIP1. UBQ, Ubiquitins used for internal control; WL, white light; D, dark; Col, wild-type Columbia. RT-PCR experiments were repeated twice with similar results. Images in A and C were taken under different sensitivity conditions of the fluorescent gel image system.
FIP1 Was Mainly Expressed in Vascular Tissues and Developmentally Regulated
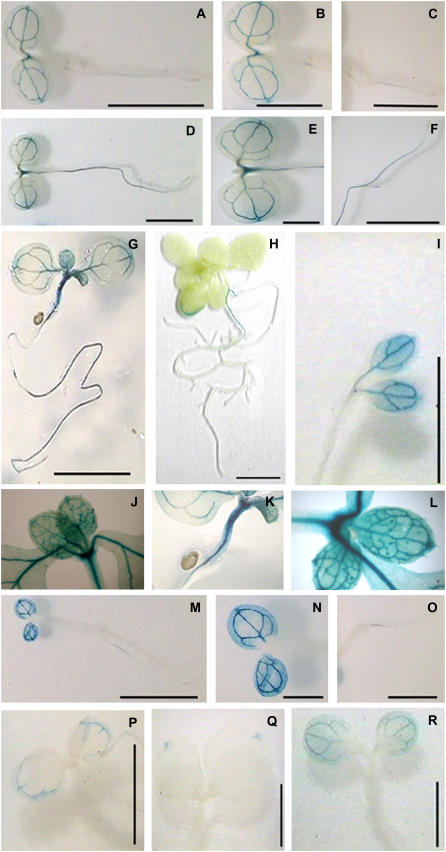
To further explore the possible involvement of FIP1 in plant development, we examined its expression patterns and regulation by promoter fusion with the β-glucuronidase (GUS) reporter gene. Approximately 1-kb FIP1 promoter sequence was fused with GUS transcriptionally and introduced into wild-type Arabidopsis (ecotype Columbia). FIP1 activity indicated by GUS staining was primarily located in the vascular tissues of cotyledons in very early seedling development, such as at 2 d-white light (Fig. 6, A and B) and cFR (Fig. 6, M and N), then appeared at the shoot apex and the upper part of hypocotyls and in roots at 4- and 7-d white light (Fig. 6, D–G). FIP1 was highly expressed in the basal portion of trichomes on the surface of true leaves, veins, shoot apex, and whole hypocotyls (Fig. 6, J–L) of 7-d-old seedlings grown in white light. Thereafter, its expression was found only in the margins of leaves and in roots at 14-d white light (Fig. 6H). In addition, FIP1 expression was also seen in the vascular tissues of cotyledons of 4-d-old seedlings grown in darkness (Fig. 6I); however, it appeared only in restricted regions of vascular tissues near the hydathode of cotyledons in 4-d-old seedlings grown under cFR (Fig. 6P). In continuous R light, its expression was restricted to the hydathode of cotyledons (Fig. 6Q); in B light, it was still expressed in the vascular tissues of the cotyledons (Fig. 6R). The site of free auxin production was recently reported to be in the hydathode of leaf tips, later in the lobes of leaves, then leaf margins and basal regions of trichomes, and finally in the central regions of the lamina (Aloni et al., 2003). Thus, the expression pattern of FIP1 seems to coincide with the sites of auxin production.
Figure 6.
Histochemical GUS staining of FIP1 promoter activity at the seedling stage under different light conditions. A, Two-day-old seedlings grown in white light. B, Close-up view of the cotyledons in A. C, Close-up view of the roots in A. D, Four-day-old seedlings grown in white light. E, Close-up view of cotyledons in D. F, Close-up view of roots in D. G, Seven-day-old seedlings grown in white light. H, Fourteen-day-old seedlings grown in white light. I, Four-day-old seedlings grown in darkness. J to L, Close-up view of different portions in G. M, Two-day-old seedlings grown in FR light. N, Close-up view of cotyledons in M. O, Close-up view of roots in M. P, Four-day-old seedlings grown in FR light. Q, Four-day-old seedlings grown in R light. R, Four-day-old seedlings grown in B light. Scale bar: A, C, D, F, M, and O, 2.5 mm; B, E, and N, 1.25 mm; G and H, 5 mm; I, P, Q, and R, 1 mm.
FIP1 Was Highly Expressed in Flower Organs and Associated with Vascular Bundles
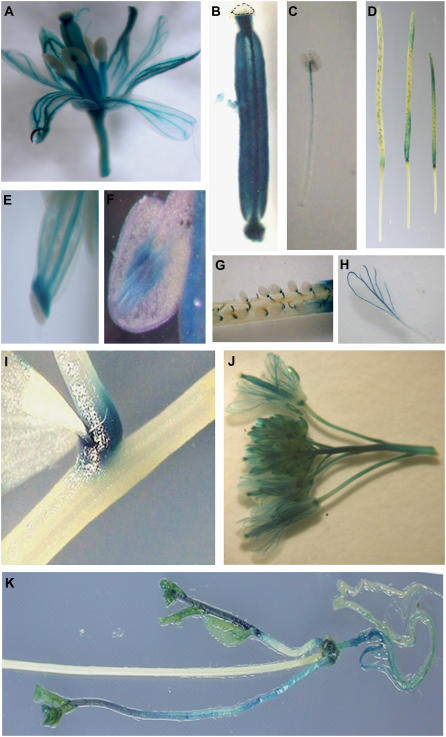
Because GUS-staining patterns of the FIP1 promoter activity were primarily in the vascular tissues at the seedling stage and regulated by light and development (Fig. 6), we investigated FIP1 expression at the adult stage. Histochemical GUS staining revealed that FIP1 was highly expressed in the vascular tissues of flower organs, including sepals, petals, stamens, and carpels (Fig. 7, A, J, H, C, F, B, and E). For stamens, GUS staining was seen in the vascular bundles of the anther (Fig. 7F) and the upper part of the filament of the stamen (Fig. 7C). GUS staining was prominent in the stylar xylem underneath the stigma of carpels and in the two medial and lateral vascular bundles of the ovary (Fig. 7, B and E), as well as the internode right beneath the ovary (Fig. 7B). In addition, GUS staining was seen in both ends of younger siliques, especially at the basal region, but disappeared when siliques matured gradually (Fig. 7D). GUS staining was also found in the funiculus, which connects seeds and siliques (Fig. 7G). Moreover, heavy GUS staining was seen at the branch point of stems and young pedicels, as well as the basal region of the stem and the root and at the newly formed inflorescence, rather than the old one (Fig. 7, I and K). Thus, FIP1 was highly expressed in flower organs, especially vascular bundles.
Figure 7.
Histochemical GUS staining of FIP1 promoter activity at adult stage under white light. A, Complete flower including sepals, petals, stamens, and carpel. B, Carpel. Dashed line indicates the stigma. C, Stamen. D, Siliques. E, Close-up view of the upper region of the carpel. F, Anther. G, Part of a silique. H, Petal. I, Branch point of the stem and the pedicel. J, Multiple flowers. K, Primary and secondary inflorescence. Leaves have been removed to see the stains at the basal region of the stem and root.
Transgenic Plants Overexpressing or Reducing FIP1 Expression Displayed a Delayed Flowering Phenotype under Long-Day Conditions
To further elucidate the physiological functions of FIP1 implied from promoter activity assays showing that FIP1 is highly expressed in flower organs, we investigated the possible effect of the FIP1 gain of function or loss of function on Arabidopsis flowering under long-day conditions. Two different indices, days to flowering and leaf number at bolting, showed that transgenic plants showing overexpressed or underexpressed FIP1 grown to the adult stage displayed a delayed flowering phenotype compared to the wild type under long-day conditions (Table I). Also, the fin219 mutant exhibited slightly delayed flowering. Thus, gain of function or loss of function of FIP1 results in similar phenotypic effects, including long hypocotyls and late flowering, which points to the possibility that FIP1 may be a component of a complex that functions in various aspects of Arabidopsis development.
Table I.
Flowering time of wild-type Arabidopsis Columbia (Col), fin219 mutant, and transgenic plants overexpressing FIP1 (FIP1OE) or showing reduced FIP1 expression (FIP1AS-171)
Plants were grown in 16-h light and 8-h dark conditions. The values shown here are means ± sd (n > 10).
| Genotype | Days at Bolting | No. of Leaves at Bolting |
|---|---|---|
| Col | 22.88 ± 1.36 | 8.57 ± 1.27 |
| fin219 | 25.25 ± 1.67 | 10.38 ± 2.33a |
| FIP1OE | 28.50 ± 2.56a | 10.88 ± 1.13a |
| FIP1AS-171 | 28.43 ± 2.50a | 11.86 ± 1.77a |
Differences between mutants and wild-type plants are significant at the P < 0.01 level.
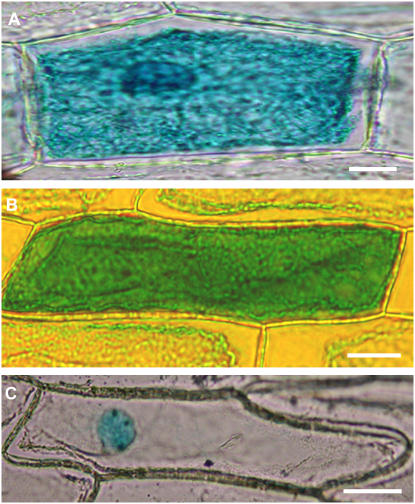
GUS-FIP1 Fusion Protein Was Localized in Both the Cytoplasm and the Nucleus and Its Subcellular Location Was Not Changed by Light
To further understand the FIP1 function implied from its location at the subcellular level, FIP1 full-length cDNA was fused with a pRTL2-GUS vector to become a translational fusion construct and subjected to a subcellular localization study in onion (Allium cepa) epidermal cells by particle bombardment. As compared with controls, bombarded cells showed the GUS-FIP1 fusion protein localized in the cytoplasm and the nucleus (Fig. 8B). Moreover, under light, the fusion protein did not change its subcellular location (data not shown). Accordingly, FIN219 is also localized in the cytoplasm and remains there regardless of light conditions (Hsieh et al., 2000); interaction between FIN219 and FIP1 as shown in Figure 2 may occur in the cytoplasm to complete their physiological functions.
Figure 8.
Subcellular localization of GUS-FIP1 fusion proteins in onion epidermal cells in a transient assay by particle bombardment. A, GUS-FIP1 fusion proteins are located in the cytosol and the nucleus. B, Construct containing GUS only is distributed evenly in both the cytosol and nucleus. C, Construct containing GUS-Nia (nuclear localization signal) is restricted only in the nucleus. Scale bar = 100 μm in A to C.
DISCUSSION
Here, we report that a FIN219-interacting protein, FIP1, isolated from a screening of a yeast two-hybrid library CD4-10 from the ABRC, interacted with the C terminus of FIN219 on pull-down assay and with the full length of FIN219 from FIP1-overexpressed plant extracts obtained from FR light-grown transgenic seedlings (Fig. 2). FIP1 may work together with FIN219 under cFR to regulate hypocotyl elongation of Arabidopsis seedlings. This observation was also implied by the result that the gain of function or partial loss of function of FIP1 resulted in similar phenotypic effects, such as hyposensitive hypocotyls under cFR and delayed flowering under long-day conditions (Fig. 4; Table I). A similar case was found in the EBS gene. Its gain-of-function and loss-of-function transgenic plants shared the same phenotypes, including early flowering, dwarf phenotype, and reduced fertility (Pineiro et al., 2003), probably resulting from a failure to form an accurate complex when the target protein is either overexpressed or abolished.
In addition, we found that c-myc-FIP1 fusion proteins isolated from FIP1-overexpressed transgenic seedlings grown under cFR existed in two bands in a native gel, one 70.2 kD and the other 96.2 kD (Supplemental Fig. S3). The 70.2-kD protein band is probably a homodimer of c-myc-FIP1 (a monomer is about 32 kD), whereas the 96.2-kD band is probably a heterodimer consisting of one c-myc-FIP1 and one FIN219 (64.5 kD), which is consistent with the co-IP result showing that FIP1 interacts with FIN219 under cFR (Fig. 2D). In addition, the 96.2-kD band is about 50% the amount of the 70.2-kD band, which implies that most of the FIP1 exists in homodimers and the rest is associated with FIN219. Thus, the heterodimer of FIP1 associated with FIN219 might be responsible for the control of hypocotyl elongation and flowering time.
The fin219 mutant was derived from a suppressor screen of the cop1-6 allele and shown to have a long hypocotyl phenotype specifically under cFR; its gene product belongs to a GH3-like gene family (Hsieh et al., 2000). jar1 was isolated as a jasmonate-resistant mutant and could be involved in a jasmonate-signaling pathway (Staswick et al., 1992). Recently, JAR1 was isolated at the same locus as FIN219, but all the jar1 alleles had no hypocotyl phenotype under cFR (Staswick et al., 2002). Here, we demonstrated that the jar1-1 allele showed a long hypocotyl phenotype under FR below 20 μmol m−2 s−1 (Fig. 1, A and C); in particular, its hypocotyl length was close to that of the fin219 mutant at approximately 1.47 μmol m−2 s−1 of FR (Fig. 1C). Moreover, the jar1-1 mutant, like the fin219 mutant, displayed a long hypocotyl phenotype only under cFR (Fig. 1B). That previous results of the jar1 alleles did not show any phenotype under FR is probably due to a single higher FR fluence rate used, resulting in no obvious phenotype. Also, a null mutant of the fin219/jar1 allele with a T-DNA inserted in the second exon displayed a long hypocotyl phenotype with a similar FR fluence-dependent pattern (Supplemental Fig. S1; data not shown). Moreover, FIN219-overexpressed transgenic seedlings exhibited a hypersensitive phenotype at FR below 10 μmol m−2 s−1 (Hsieh et al., 2000). Thus, FIN219/JAR1 plays a role in FR light signaling to regulate cell elongation.
Although JAR1 has been shown to be a JA amino synthetase and may play a vital role in the homeostasis of jasmonate, leading to the regulation of plant metabolic, reproductive, and defensive processes (Staswick and Tiryaki, 2004), the function of FIN219/JAR1 in phyA-mediated FR signaling remains to be elucidated. Because FIN219/JAR1 is induced by auxin and methyl jasmonate (Hsieh et al., 2000; data not shown), FIN219/JAR1 plays a key role in integrating both hormone signaling and light signaling, especially phyA-mediated signal transduction. Here, we showed that FIP1 interacting with FIN219 was also involved in FR light signaling, which was further supported by transgenic studies showing that the gain of function or partial loss of function of FIP1 exhibited a long hypocotyl phenotype only under cFR condition. In addition, FIP1 transcripts were substantially affected in the spa1 mutant, a suppressor of phyA, and in the cop1-4 allele. Intriguingly, FIP1 transcripts in a null allele of COP1, cop1-5, remain in a similar level to wild type, which indicates that COP1 can repress FIP1 expression. Recent evidence has revealed that SPA1 and COP1 work together to repress photomorphogenesis (Saijo et al., 2003; Yang and Wang, 2006). Previous studies also indicated that FIN219 is a suppressor of COP1 on a genetic basis. Therefore, regulatory relationships among these players become complicated. Examining further how SPA1 and COP1 affect FIP1 at the protein level might be interesting. In addition, FIP1 transcripts were up-regulated in the fin219 mutant under white light, B light, R light, and darkness, whereas its transcripts were comparable in expression in the wild type and the fin219 mutant under FR light (Fig. 5), which implies that FIN219 may regulate the levels of FIP1 through different mechanisms dependent on light conditions. Moreover, we found that FIP1 expression was not induced by methyl jasmonate on histochemical staining of FIP1 promoter∷GUS transgenic seedlings and by GUS activity assay for FIP1 promoter activity under cFR (data not shown). Therefore, FIP1 interacts with FIN219 under cFR without the effect of jasmonate.
FIP1 is a member of the plant tau class of the large GST gene family composed of 53 members involved in diverse functions, including detoxification, transport of flavonoid compounds, reduction of oxidative stress, prevention of Bax-induced cell death, and induction of chalcone synthase expression to deter UV damage. So far, no GST members have been reported to be involved in light signaling at the molecular level. Tepperman et al. (2001) used a DNA microarray approach to examine the gene expression profiles induced rapidly by FR irradiation. One of the up-regulated genes is GST (AAD32887) and its expression increased promptly; however, the induction was inhibited by the phyA mutation. Here, we show that FIP1 interacts with FIN219 under cFR and is also involved in phyA-mediated FR signaling, which indicates that some members of GST participate in light signaling to regulate cell elongation. FIP1 was highly expressed in the vascular bundles of male and female flower tissues (Fig. 7, A, F, B, and E) and vascular tissues of seedling stages under different light conditions (Fig. 6). Furthermore, FIP1 was substantially expressed at the branching points of stems and young pedicels (Fig. 7I). It was also highly expressed at the basal region of young siliques, but reduced in expression in mature siliques (Fig. 7D). All these data indicate that FIP1 responds to light signals and is developmentally regulated. Also, FIP1 did not bind to GSH sepharose and was not recognized by anti-GST monoclonal antibodies (Fig. 2C, lane 1, middle and right sections), which is consistent with the notion that plant GSTs differ from animal GSTs in terms of tertiary structure as well as different classes.
The fin219 mutant displayed a slightly delayed flowering phenotype under long-day conditions (Table I), and its late-flowering phenotype was even more prominent under short-day treatment (data not shown). FIN219/JAR1 is a jasmonate-conjugating enzyme (Staswick and Tiryaki, 2004) and may play a crucial role in regulating homeostasis of jasmonate levels that mediate various signaling, including flowering. FIN219 promoter activity studies revealed that FIN219 was also expressed in floral organs, trichomes, and funiculus (Supplemental Fig. S2), which indicates that FIP1 and FIN219/JAR1 share partially overlapped expression patterns. Moreover, FIP1 transcripts were up-regulated by the fin219 mutation under white light (Fig. 5C), possibly leading to lower levels of GSH that may trigger a delayed flowering phenotype (Cobbett, 2000; Ogawa et al., 2001), which is consistent with fin219 null mutants showing a late-flowering phenotype (data not shown). Alternatively, FIP1 acts as a ligand to interact with FIN219 and then binds to jasmonate, thus mobilizing the resulting complex to regulate floral organ development, which leads to control of flowering time. To further explore the involvement of both FIP1 and FIN219/JAR1 in flowering, obtaining the double mutant fip1fin219 and measuring GSH levels in the flowering stage may be worthwhile.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Throughout this article, the wild type is Arabidopsis (Arabidopsis thaliana) ecotype Columbia. B light photoreceptor mutants cry1 (cry1-304), cry2 (cry2-1), and cry1cry2 (cry1-304cry2-1) double mutants are in the Columbia ecotype as described (Mockler et al., 1999). All other mutants used in this study were described previously (Hsieh et al., 2000; Wang and Deng, 2002). The T-DNA insertion line SALK_080514 was obtained from the ABRC and T-DNA was inserted into the promoter region of FIP1, 498 nucleotides upstream from the translation start site.
Surface sterilization and cold treatment of the seeds and the seedling growth conditions were described previously (Hsieh et al., 2000). The light source used in this study was described previously (Peters et al., 1998) and light intensities used for FR, R, B, and white light were described in the figure legends. All transgenic lines used for phenotypic analysis and RNA gel-blot assays were the T3 homozygous generation.
Yeast Two-Hybrid Assay
A GAL4 yeast two-hybrid system was used for screens of FIN219-interacting partners and protein-protein interaction studies. For the pGBT9 (+2)-FIN219 construct, a full-length FIN219 cDNA was derived from BamHI digestion of the recombinant plasmid pPZPY122-FIN219 and then ligated into the BamHI site of the BD vector pGBT9 (+2) to form a bait construct. An activation domain (AD) fusion of yeast two-hybrid library CD4-10 from the ABRC was used as prey. AD fusion recombinant plasmids were isolated by a standard method (Sambrook and Russell, 2001). The bait fusion plasmid pGBT9 (+2)-FIN219 and the prey recombinant fusion plasmids were then cotransformed into the yeast host AH109 (CLONTECH) and screened for interacting partners on plates lacking Trp, Leu, and His. The pGBT9 (+2)-FIN219N fusion plasmid was obtained by ligating a BamHI DNA fragment of the recombinant plasmid pPZPY122-FIN219N containing the N-terminal 300 amino acids of FIN219 into the BD vector pGBT9 (+2). For the pGBT9 (+2)-FIN219C recombinant plasmid, a BamHI DNA fragment containing the C-terminal 274 amino acids of FIN219 that was amplified by PCR using the primer 219CR, 5′-TACGGATCCTACGGGATCATGACTGGCT-3′ and the primer 219CL, 5′-TACGGATCCTGAGTCAAAACGCTGTGCT-3′ (underline indicates the built-in BamHI site) was cloned into the BamHI site of pGBT9 (+2). COP1 and HY5 cDNAs were obtained by PCR amplification with the following primers: COP1-L, 5′-TGAGGATCCATGGAAGAGATTTCGACGG-3′, COP1-R, 5′-GCAGGATCCTTCCAAATGATGAACCCTACTT-3′ (underline indicates the built-in BamHI site); HY5-L, 5′-CAGCTCGAGATGCAGGAATGGCAAGCGACTAGC-3′, HY5-R, 5′-ATACTCGAGATCAAAGGCTTGCATCAGCA-3′ (underline indicates the built-in XhoI site). COP1 and HY5 cDNA fragments were cloned into the BamHI site of the BD vector pGBT9 (+2) and the XhoI site of the AD vector pACT and used for positive control in protein-protein interaction studies. All the above recombinant plasmids underwent sequencing to confirm their correctness and in-frame reading.
The putative positive clones from yeast two-hybrid screening were further confirmed by yeast mating method (Fields and Song, 1989) and colony-lift filter assay (yeast protocols handbook; CLONTECH).
Pull-Down Assay
To construct the recombinant plasmid pGEX-4T-1-FIN219, a BamHI DNA fragment of the full-length FIN219 was released directly from pPZPY∷FIN219 and cloned into the expression vector pGEX-4T-1 (Amersham-Pharmacia). The first 300 residues in the N-terminal region of FIN219 and the remaining 274 residues in the C terminus were cloned as BglII DNA fragments into the same expression vector as the full-length FIN219. For the recombinant pRSET-B∷FIP1, a BglII DNA fragment of the full-length FIP1 was amplified by PCR with the primer for FIP1-L, 5′-TAGAGATCTATGGCGAACCTACCGAT-3′ and FIP1-R, 5′-CATAGATCTCAGAACACATTGGCTTAGCAACA-3′. The recombinant plasmids above were transformed into Escherichia coli DH5α and then induced by 1 mm isopropylthio-β-galactoside (IPTG) for expression in hosts BL21(DE3) codon plus (pGEX-4T-1-FIN219FL and pGEX-4T-1-FIN219N300) or BL21(DE3) pLysS (pGEX-4T-1-FIN219C274 and pRSET-B∷FIP1). Recombinant fusion proteins were purified by use of GSH sepharose (Amersham-Pharmacia) for GST-FIN219 full-length and GST-FIN219 C274 or electroelusion (Bio-Rad) for GST-FIN219 N300 or Ni-NTA (Qiagen) for His-6-FIP1 according to manufacturer's procedures. Five micrograms of purified recombinant proteins were mixed in 1 mL of interaction buffer (50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 0.1% NP-40) and then incubated at 4°C at 30 to 40 rpm for 1 h. Well-equilibrated GSH sepharose or Ni-NTA were added to the mixture and incubated at 4°C for another 30 min. After centrifugation (500g for 5 min), the pellet was washed with interaction buffer and underwent centrifugation again. Washing was repeated four times and then the pellet and supernatant were boiled with SDS loading dye and underwent SDS-PAGE. Western-blot analysis was performed by standard methods (Sambrook and Russell, 2001).
Protein Co-IP
Protein co-IP was performed as described (Staub et al., 1996). Protein A-sepharose 4B fast flow beads (Sigma) were coupled to FIN219 or c-myc antibodies and then incubated with 1 mg protein extract from FR light-treated FIP1-overexpressed, fin219 null mutant, and wild-type Arabidopsis seedlings. Pellets were analyzed on standard SDS-PAGE and subsequent western blotting.
Recombinant Plasmids for Plant Transformation
To overexpress FIP1 in Arabidopsis ecotype Columbia and the fin219 mutant, a NcoI-BglII DNA fragment containing full-length FIP1 cDNA was obtained by PCR amplification with the following primer pairs: FIP1-L, 5′-TTGCCATGGTTATGGCGAACCTACCGAT-3′ (underline indicates the built-in NcoI site); and FIP1-R, 5′-CATAGATCTCAGAACACATTGGCTTAGCAACA-3′ (underline indicates the built-in BglII site). The fragment was then ligated into the binary transformation vector pCAMBIA1390-c-myc or pPZP221 with a pRTL2-GUS expression cassette. The resulting constructs pCAMBIA1390-c-myc-FIP1 and pPZP221-GUS-FIP1 were introduced into the Agrobacterium GV3101 by the standard method (Sambrook and Russell, 2001) and then transformed into Arabidopsis ecotype Columbia and the fin219 mutant by floral dipping (Clough and Bent, 1998). Transgenic plants containing transgenes were selected with use of 25 μg/mL hygromycin for the c-myc-tagged construct and 100 μg/mL gentamycin for the GUS-tagged construct.
FIP1 antisense transgenic plants were generated from a binary vector pPZP221 harboring a PstI-digested DNA fragment that is an expression cassette of the pRTL2-GUS/NIA-FIP1 plasmid containing the FIP1 antisense cDNA. Two homozygous lines, AS171 and AS421, were selected for this study.
A total of approximately 40 independent T1 transgenic plants were selected and grown to the T2 generation for each transformation construct. Phenotypic analysis was conducted with single T-DNA insertion lines, as determined by a drug resistance test. All transgenic phenotypes reported here were observed in at least 10 independent lines.
GST Activity Assay
GST activity of FIP1 as purified recombinant proteins His-6-FIP1 was determined according to Habig et al. (1974) with use of the Microplate Spectrophotometer SPECTRAmax PLUS (Molecular Devices). The reaction solution contained 100 mm phosphate buffer, pH 6.5, 1 mm GSH, 1 mm CDNB, and an appropriate amount of samples. We read the absorbance change for 3 min. The extinction coefficient for the CDNB-GSH product is 9.6 mm−1 cm−1. We adjusted the concentration of CDNB or GSH to evaluate Km or Vmax. Lineweaver-Burk analysis was performed by plotting 1/V versus 1/[substrate]. Km and Vmax values were determined from the slope (Km/Vmax) and y intercept (1/Vmax) of the plot.
RNA Gel-Blot and RT-PCR Analyses
Total RNA was isolated from 4-d-old seedlings of various light photoreceptor and signaling mutants grown under different light conditions as described previously (Hsieh et al., 1996). Twenty micrograms of total RNA were loaded onto the gel and blotted to the positive-charged nylon membrane (Roche). Full-length FIP1 cDNA was prepared by PCR for digoxygenin labeling as a probe according to the manufacturer's procedure (Roche). Furthermore, a pair of gene-specific primers at the 3′-UTR of FIP1 was also used to investigate the specificity of FIP1 expression. The primer sequences were as follows: FIP1S, 5′-GCTGAGTATAGGAAGAACAA-3′ and FIP1F, 5′-AATTCCATACTTAGGATGCA-3′. Hybridization and washing conditions were performed as usual (Sambrook and Russell, 2001). For RT-PCR analyses, 1 μg of DNaseI-treated total RNA underwent RT at 42°C with ImProm-II reverse transcriptase (Promega). FIP1 was amplified from 1 μL of the 20 μL of cDNA by PCR for 35 cycles (94°C, 30 s; 50°C, 30 s; 72°C, 30 s) using a Peltier thermal cycler (MJ Research) with FIP1S and FIP1F primers described above and ubiquitin 10 for 26 cycles (94°C, 30 s; 56°C, 30 s; 72°C, 30 s) with the following primers for UBQ1, 5′-GATCTTTGCCGGAAAAGAATTGGAGGATGGT and UBQ2, 5′-CGACTTGTCATTAGAAAGAAAGAGATAACAGG.
Histochemical GUS Staining of FIP1 Promoter Activity
A 1,038-bp promoter region of FIP1 was cloned into the BamHI site of the binary vector pCambia1381Z to establish a transcriptional fusion construct and then introduced into wild-type Columbia. The resulting transgenic plants were grown to homozygous lines and underwent histochemical GUS staining as described previously (Jefferson et al., 1987).
Transient Assay of Subcellular Localization by Particle Bombardment
To construct the GUS-FIP1 fusion protein, a BglII DNA fragment was released from the recombinant plasmid pRSET-B-FIP1 and then cloned into pRTL2-GUS/Nia. The vectors pRTL2-GUS and pRTL2-GUS/Nia alone were also used as a control for particle bombardment conducted as described (von Arnim and Deng, 1994). After bombardment, onion (Allium cepa) epidermal cells were kept in darkness at 25°C overnight and then some were transferred to white light for 1 d, with the rest kept in darkness for one more day. GUS staining was performed as described (von Arnim and Deng, 1994).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression of the FIN219/JAR1 null mutant in Arabidopsis.
Supplemental Figure S2. Histochemical GUS staining of FIN219 promoter activity under white light conditions.
Supplemental Figure S3. Size determination of FIP1 in FIP1-overexpressed transgenic seedlings grown in FR light.
Supplementary Material
Acknowledgments
We are grateful to the ABRC (Ohio State University, Columbus) for the yeast two-hybrid library CD4-10. We also appreciate Ashok Kumar H.G. for reading and critical comments on the manuscript.
This work was supported by the National Science Council (grant no. NSC 92–2311–B-002–027), Taiwan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hsu-Liang Hsieh (hlhsieh@ntu.edu.tw).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aloni R, Schwalm K, Langhans M, Ullrich CI (2003) Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta 216 841–853 [DOI] [PubMed] [Google Scholar]
- Ballesteros ML, Bolle C, Lois LM, Moore JM, Vielle-Calzada J-P, Grossniklaus U, Chua N-H (2001) LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev 15 2613–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D, Viczian A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adam E, Fejes E, Schafer E, et al (2004) Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C, Koncz C, Chua N-H (2000) PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev 14 1269–1278 [PMC free article] [PubMed] [Google Scholar]
- Choi G, Kim J-I, Hong S-W, Shin B, Choi G, Blakeslee JJ, Murphy AS, Seo YW, Kim K, Koh E-J, et al (2005) A possible role for NDPK2 in the regulation of auxin-mediated responses for plant growth and development. Plant Cell Physiol 46 1246–1254 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip, a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cobbett CS (2000) Phytochelatins and their roles in heavy metal detoxification. Plant Physiol 123 825–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnos T, Puente P, Whitelam GC, Harberd NP (2001) FHY1: a phytochrome A-specific signal transducer. Genes Dev 15 2980–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle M, Zhou Y-C, Schäfer E, Funk M, Kretsch T (2001) EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev 15 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Davis BG, Edwards R (2002. a) Functional divergence in the glutathione transferase superfamily in plants—identification of two classes with putative functions in redox homeostasis in Arabidopsis thaliana. J Biol Chem 277 30859–30869 [DOI] [PubMed] [Google Scholar]
- Dixon DP, Lapthorn A, Edwards R (2002. b) Plant glutathione transferases. Genome Biol 3 reviews3004.1–3004.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild C, Schumaker MA, Quail PH (2000) HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev 14 2377–2391 [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chory J (2000) RSF1, an Arabidopsis locus implicated in phytochrome A signaling. Plant Physiol 124 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Song O (1989) A novel genetic system to detect protein-protein interactions. Nature 340 245–246 [DOI] [PubMed] [Google Scholar]
- Fujimori T, Yamashino T, Kato T, Mizuno T (2004) Circadian-controlled basic/helix-loop-helix factor, PIL 6, implicated in light-signal transduction in Arabidopsis thaliana. Plant Cell Physiol 45 1078–1086 [DOI] [PubMed] [Google Scholar]
- Habig WH, Pabst MJ, Jakoby WB (1974) Glutathione S-transferase: the first enzymatic step in mercapturic acid formation. J Biol Chem 249 7130–7139 [PubMed] [Google Scholar]
- Hagen G, Kleinschmidt A, Guilfoyle T (1984) Auxin-regulated gene expression in intact soybean hypocotyls and excised hypocotyl sections. Planta 162 147–153 [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng XW (2000) HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1-binding domain. EMBO J 19 4997–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH (1999) SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284 496–499 [DOI] [PubMed] [Google Scholar]
- Hsieh HL, Okamoto H, Wang M, Ang LH, Matsui M, Goodman H, Deng XW (2000) FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev 14 1958–1970 [PMC free article] [PubMed] [Google Scholar]
- Hsieh HL, Tong CG, Thomas C, Roux SJ (1996) Light-regulated mRNA abundance of a gene encoding a calmodulin-regulated, chromatin-associated NTPase in pea. Plant Mol Biol 30 135–148 [DOI] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH (1999) The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev 13 2017–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Hudson M, Kim C, Apel K, Quail PH (2004) Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305 1937–1941 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Yang H, Ma L, Sun N, Yu H, Liu T, Gao Y, Gu H, Chen Z, Wada M, et al (2003) A genome-wide analysis of blue-light regulation of Arabidopsis transcription factor gene expression during seedling development. Plant Physiol 133 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick RE, Kronenberg GHM (1994) Photomorphogenesis in Plants, Ed 2. Kluwer Academic Publishers, Dordrecht, The Netherlands
- Kim J, Yi H, Choi G, Shin B, Song P-S, Choi G (2003) Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Hodgson DW, Campbell TJ (2003) Blue light signaling through the cryptochromes and phototropins: so that's what the blues is all about. Plant Physiol 133 1429–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia J, Huq E, Quali PH (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288 859–863 [DOI] [PubMed] [Google Scholar]
- Matsushita T, Mochizuki N, Nagatani A (2003) Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424 571–574 [DOI] [PubMed] [Google Scholar]
- Mockler TC, Guo H, Yang H, Duong H, Lin C (1999) Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126 2073–2082 [DOI] [PubMed] [Google Scholar]
- Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, Ecker JR, Li X, Zhang Y, Quail PH (2004) The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc Natl Acad Sci USA 101 16091–16098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95 657–667 [DOI] [PubMed] [Google Scholar]
- Ogawa K, Tasaka Y, Mino M, Tanaka Y, Iwabuchi M (2001) Association of glutathione with flowering in Arabidopsis thaliana. Plant Cell Physiol 42 524–530 [DOI] [PubMed] [Google Scholar]
- Oh E, Kim J, Park E, Kim J-I, Kang C, Choi G (2004) PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16 3045–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM (2003) The red side of photomorphogenesis. Plant Physiol 133 1437–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JL, Széll M, Kendrick RE (1998) The expression of light-regulated genes in the high-pigment-1 mutant of tomato. Plant Physiol 117 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineiro M, Gomez-Mena C, Schaffer R, Martinez-Zapater JM, Coupland G (2003) EARLY BOLTING IN SHORT DAYS is related to chromatin remodeling factors and regulates flowering in Arabidopsis by repressing FT. Plant Cell 15 1552–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prapanthadara L-A, Koottathep S, Promtet N, Hemingway J, Ketterman AJ (1996) Purification and characterization of a major glutathione S-transferase from the mosquito Anopheles dirus (species B). Insect Biochem Mol Biol 26 277–285 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Seo HS, Yang J-Y, Ishikawa M, Bolle C, Ballesteros M, Chua N-H (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423 995–999 [DOI] [PubMed] [Google Scholar]
- Singhal SS, Tiwari NK, Ahmad H, Srivastava SK, Awasthi YC (1991) Purification and characterization of glutathione S-transferase from sugarcane leaves. Phytochemistry 30 1409–1414 [Google Scholar]
- Soh MS, Hong SH, Hanzawa H, Furuya M, Nam HG (1998) Genetic identification of FIN2, a far red light-specific signaling component of Arabidopsis thaliana. Plant J 16 411–419 [DOI] [PubMed] [Google Scholar]
- Soh MS, Kim Y-M, Han S-J, Song P-S (2000) REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell 12 2061–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH (1992) Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA 89 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I, Rowe ML (2002) Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub JM, Wei N, Deng X-W (1996) Evidence for FUS6 as a component of the nuclear-localized COP9 complex in Arabidopsis. Plant Cell 8 2047–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Hudson ME, Khanna R, Zhu T, Chang SH, Wang X, Quail PH (2004) Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J 38 725–739 [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang H-S, Wang X, Quail PH (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW (1994) Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79 1035–1045 [DOI] [PubMed] [Google Scholar]
- Wagner U, Edwards R, Dixon DP, Mauch F (2002) Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol Biol 49 515–532 [DOI] [PubMed] [Google Scholar]
- Wang H, Deng XW (2002) Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J 21 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wang H (2006) The central coiled-coil domain and carboxyl-terminal WD-repeat domain of Arabidopsis SPA1 are responsible for mediating repression of light signaling. Plant J 47 564–576 [DOI] [PubMed] [Google Scholar]
- Zeidler M, Bolle C, Chua NH (2001) The phytochrome A specific signaling component PAT3 is a positive regulator of Arabidopsis photomorphogenesis. Plant Cell Physiol 42 1193–1200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.