Abstract
Dicer is an essential component of RNA interference (RNAi) pathways, which have broad functions in gene regulation and genome organization. Probing the consequences of tissue-restricted Dicer loss in mice indicates a critical role for Dicer during meiosis in the female germline. Mouse oocytes lacking Dicer arrest in meiosis I with multiple disorganized spindles and severe chromosome congression defects. Oogenesis and early development are times of significant post-transcriptional regulation, with controlled mRNA storage, translation, and degradation. Our results suggest that Dicer is essential for turnover of a substantial subset of maternal transcripts that are normally lost during oocyte maturation. Furthermore, we find evidence that transposon-derived sequence elements may contribute to the metabolism of maternal transcripts through a Dicer-dependent pathway. Our studies identify Dicer as central to a regulatory network that controls oocyte gene expression programs and that promotes genomic integrity in a cell type notoriously susceptible to aneuploidy.
Keywords: Dicer, RNAi, oocyte, meiosis
It is estimated that up to 20% of human embryos have karyotypic aberrations due to chromosome segregation abnormalities that arise during oocyte maturation (Hassold and Hunt 2001). Although female meiosis begins prenatally, oocytes remain arrested in the prolonged dictyate stage of prophase I until post-pubertal hormonal signals initiate resumption of meiosis. Upon ovulation, oocytes complete the first meiotic division and enter metaphase of meiosis II, a transition performed in the absence of transcription and dependent on judicious translation of stockpiled maternal transcripts (Solter et al. 2004). The regulation of maternal messages during meiotic maturation requires exquisite control of mRNA stability and translation, and the breakdown of these processes, particularly with advancing maternal age, probably contributes to the high frequency of oocyte aneuploidy. However, many aspects of the mechanisms that control maternal mRNA storage, recruitment to the translational machinery and degradation are not well understood.
Dicer is a conserved ribonuclease whose function in the biogenesis of small RNAs is central to many RNA interference (RNAi)-related pathways. Studies in several organisms have implicated Dicer in transcriptional and post-transcriptional control of mRNA levels, as well as in regulation of translation. A member of an ancient and diversified biological system, Dicer’s many functions include virus and transposon defense, chromatin regulation, centromere integrity, and control of gene expression due to its role in biogenesis of microRNAs (miRNAs) (Murchison and Hannon 2004). In many organisms, Dicer plays an integral role in maintaining and organizing the germline (Knight and Bass 2001; Hatfield et al. 2005).
Here we uncover an essential function for Dicer in regulating mouse oogenesis. By disrupting Dicer specifically in growing oocytes, we find that oocytes lacking Dicer are unable to complete meiosis, and instead arrest with defects in meiotic spindle organization and chromosome congression. In addition, we show that small interfering RNAs (siRNAs) and miRNAs may be involved in turnover of many maternal transcripts whose degradation may be essential for successfully completing meiotic maturation.
Results
Dicer is expressed in the growing oocyte, but is dispensable for oocyte growth and development
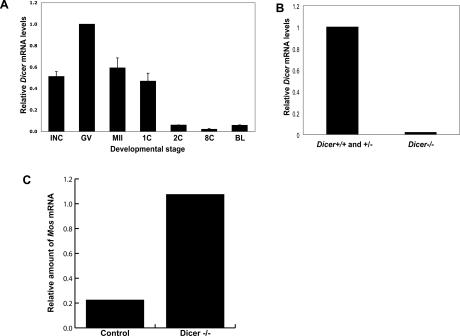
Although general roles for Dicer in maintaining and organizing the germline have been described in several organisms, similar functions in mammals have not been reported (Knight and Bass 2001; Hatfield et al. 2005). We therefore sought to probe potential roles for RNAi in the mammalian germline. Using RT–PCR, we detected robust Dicer expression in growing oocytes and early embryos (Fig. 1A). Interestingly Dicer expression appeared most intense in fully grown germinal vesicle (GV) oocytes. This suggested a potential role for Dicer during meiotic maturation, a time when development is driven largely by the regulated use of stored mRNAs. The decrease in Dicer mRNA that initiates with the onset of oocyte maturation is common to many maternal mRNAs (Schultz 2002).
Figure 1.
Dicer is expressed in growing oocytes, and is depleted in oocytes from Dicerflox/flox; Zp3-Cre mice. (A) Temporal pattern of Dicer mRNA expression. Quantitative RT–PCR was performed in mouse oocytes and preimplantation embryos. Metaphase II eggs; one-cell, two-cell, eight-cell embryos; and blastocysts were harvested 13 h, 20 h, 44 h, 68 h, and 96 h post-hCG, respectively. The values are normalized to the amount of Dicer mRNA in GV oocytes. The experiment was performed three times, and the data are presented as the mean ± SEM. (INC) Meiotically incompetent oocyte; (GV) fully grown GV-intact oocyte; (MII) metaphase II-arrested egg; (1C) one-cell stage embryo; (2C) two-cell stage embryo; (8C) eight-cell stage embryo; (BL) blastocyst. (B) Dicer mRNA levels in wild-type and mutant oocytes. Quantitative real-time PCR was performed. The experiment was performed five times, and the data are presented as the mean ± SEM. (C) Dicer is functionally depleted in Dicerflox/flox; Zp3-Cre oocytes. Twenty-four hours after Mos dsRNA injection into Dicerflox/flox; Zp3-Cre or control oocytes, Mos mRNA levels were measured by quantitative RT–PCR.
To probe its functional relevance in the female germline, we disrupted Dicer in oocytes by combining a conditional Dicer allele (Dicerflox) with a Cre recombinase transgene expressed from the Zp3 promoter (de Vries et al. 2000; Murchison et al. 2005; Andl et al. 2006). The Zp3-promoter begins to drive transcription shortly after the initiation of oocyte growth and provides its highest expression ∼10–12 d post-partum (Lira et al. 1990). Zp3-Cre activity results in an oocyte-specific null allele through deletion of the exons encoding the catalytic RNase III domains of Dicer, an outcome that we confirmed by quantitative real-time RT–PCR on GV oocytes (Fig. 1B). To test the perdurance of Dicer activity in Dicerflox/flox; Zp3-Cre cells, we injected these oocytes, as well as control oocytes, with double-stranded RNA (dsRNA) targeting Mos (Fig. 1C). Following injection, oocytes were cultured in milrinone-containing medium to inhibit oocyte maturation. After 24 h, the amount of Mos mRNA relative to similarly treated but uninjected oocytes was then determined by quantitative RT–PCR. We found no change in the amount of Mos mRNA in Dicerflox/flox; Zp3-Cre oocytes, whereas an 80% decrease was observed in control oocytes. Thus, Dicerflox/flox; Zp3-Cre GV oocytes are unable to initiate RNAi in response to long dsRNAs, indicating a substantial loss of Dicer activity.
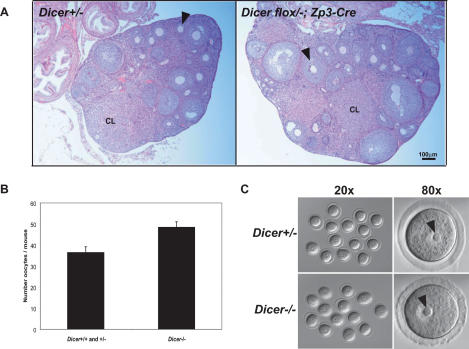
During an 11-wk breeding period with males of proven fertility, Dicerflox/flox; Zp3-Cre females failed to produce any offspring, whereas their wild-type and heterozygous siblings had several litters. This observation strongly indicated that Dicerflox/flox; Zp3-Cre females are infertile. Nevertheless, we found that Dicerflox/flox; Zp3-Cre ovaries were morphologically and histologically indistinguishable from those of controls, with normal numbers of follicles containing readily identifiable growing and fully grown oocytes (Fig. 2A). Dicerflox/flox;Zp3-Cre ovaries were capable of responding to gonadotropins as normal numbers of GV oocytes were recovered from PMSG-primed females regardless of genotype (Fig. 2B). Moreover, Dicer−/− GV oocytes appeared normal and had characteristic nucleolar structures (Fig. 2C). Considered together, these results indicate that although Dicer is enriched in developing oocytes, its function is dispensable for oocyte growth, development and response to hormonal signals.
Figure 2.
Dicer is dispensable for oocyte growth and hormone response. (A) Ovarian histology in Dicerflox/flox; Zp3-Cre or wild-type (Dicer+/−) female mice. The ovaries were fixed overnight in Bouin’s fixative, embedded in paraffin, and sliced into 10-μm sections and stained with hematoxylin and eosin. The arrowheads indicate secondary follicles. (CL) Corpus luteum. (B) Number of oocytes recovered from Dicerflox/flox; Zp3-Cre (Dicer−/−) or wild-type (Dicer+/+ and Dicer+/−) female mice. Fully grown GV-intact oocytes were collected from the ovaries of PMSG-primed mice as described in Materials and Methods. The data are presented as the mean ± SEM; 19 wild-type and 12 mutant mice were analyzed. The difference between the two groups is not statistically significant (Student’s two-tailed t-test, p = 0.05). (C) Morphology of Dicer+/− and Dicer−/− oocytes. Oocytes were collected as described and bright-field microscopy was performed. Arrows indicate the prominent nucleoli characteristic of GV oocytes.
Dicer is required for meiotic spindle integrity and completion of meiosis I
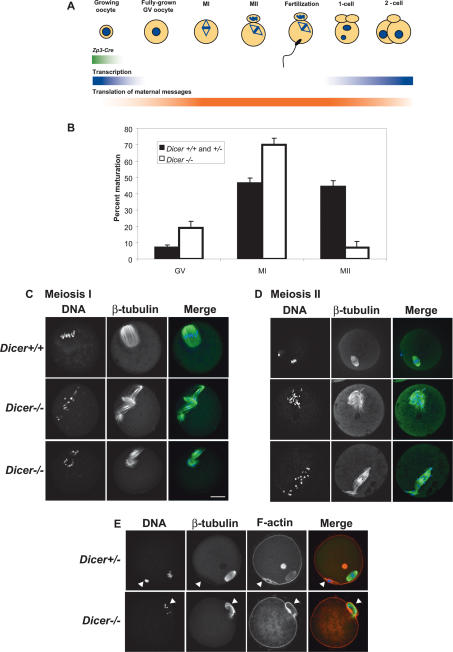
Meiotic maturation denotes the period during which the fully grown GV oocyte completes meiosis and is ovulated. In mice, this process takes ∼14 h as the oocyte undergoes GV breakdown (GVBD), completes the first meiotic division, and arrests at metaphase of meiosis II (MII) (Fig. 3A). Completion of meiosis I is morphologically marked by the extrusion of the first polar body (Brunet and Maro 2005). To determine whether Dicer is required for meiotic maturation, we cultured Dicer−/− GV oocytes from Dicerflox/flox; Zp3-Cre females and wild-type or heterozygous oocytes from their littermates for 16 h. Disappearance of the GV and appearance of the first polar body were used to score the maturation status of oocytes, revealing a metaphase I (MI) arrest in in vitro matured Dicer−/− oocytes. Although normal numbers of Dicer−/− oocytes underwent GVBD, notably fewer Dicer−/− oocytes produced a polar body (Fig. 3B). We obtained similar results from in vivo matured oocytes, with only 10% of Dicer−/− oocytes collected from the oviduct possessing a polar body in contrast to 54% for wild-type and heterozygous controls. We conclude that Dicer is required for efficient completion of meiosis I.
Figure 3.
Dicer is required for completion of meiotic maturation. (A) Schematic of oocyte maturation and early embryo development. The transition from fully grown GV oocyte to the late one-cell embryo occurs in the absence of significant transcription and relies on translation of maternal mRNA transcripts. The first part of this transition, when an oocyte completes meiosis I and arrests at metaphase of meiosis II is known as meiotic maturation. Zp3-Cre is expressed only in growing oocytes. (B) The maturation stage of Dicer−/− and control oocytes was scored by the presence of a GV (GV oocyte), a polar body (MII oocyte), or neither a GV nor a polar body (MI oocyte). Six-hundred-ninety-four Dicer+/+ and Dicer+/− and 582 Dicer−/− oocytes were analyzed in total. Error bars denote standard error of the mean. (C,D) Spindle morphology in Dicer−/− oocytes matured in vitro to metaphase I (C) or metaphase II (D). Oocytes were fixed in 3.7% paraformaldehyde and stained with a β-tubulin antibody; DNA was counterstained with DAPI or Sytox green. Representative images are shown. Bar, 25 μm. (E) Cortical actin and polar body morphology in mutant oocytes. Oocytes were fixed in 3.7% paraformaldehyde and stained with a β-tubulin antibody; F-actin was labeled with phalloidin-Alexa fluor 635 and DNA was counterstained with DAPI. The arrowheads indicate the position of the first polar body. β-Tubulin is often present inside the polar body in both wild-type and mutant oocytes.
The meiotic spindle is a highly dynamic microtubular structure that orchestrates chromosome movements during meiotic maturation (Brunet and Maro 2005). Nucleated in several cytoplasmic mictrotubule organizing centers (MTOCs) upon GVBD, spindle microtubules capture condensing chromosome arms and orient them onto the metaphase I plate. Following spindle migration to the cortex, anaphase I and polar body extrusion occur, whereupon the spindle reforms around sister chromatids and reorients for MII (Brunet and Maro 2005). To ascertain if a defect in spindle dynamics contributes to the failure of Dicer−/− oocytes to complete maturation, we used a β-tubulin antibody to visualize the spindle in Dicer−/− oocytes matured to MI. Although control oocytes formed single barrel-shaped MI spindles with chromosomes congregated at the metaphase plate, Dicer−/− oocytes frequently had multiple spindles with misaligned chromosomes strewn around the spindle axes (Fig. 3C). When staining was performed on oocytes matured to MII, we observed similar catastrophic spindle disorganization in Dicer−/− oocytes (Fig. 3D).
The first polar body contains one fully replicated haploid genome and forms under a cortical actin cap that is evaginated by the elongating anaphase I spindle (Brunet and Maro 2005). As 7% of Dicer−/− oocytes did form a polar body, we examined such oocytes for polar body morphology and content. Interestingly, while the cortical actin cap appeared normal in Dicer−/− MII oocytes, anaphase chromosomes were dispersed and lagging chromosomes were frequently observed (Fig. 3E). As chromosomes in Dicer−/− oocytes were often highly scattered it was sometimes possible to observe their fine structure. We found that while many Dicer−/− oocytes contained bivalents, some did apparently complete anaphase I because they contained univalents, even in the absence of an obvious polar body (data not shown).
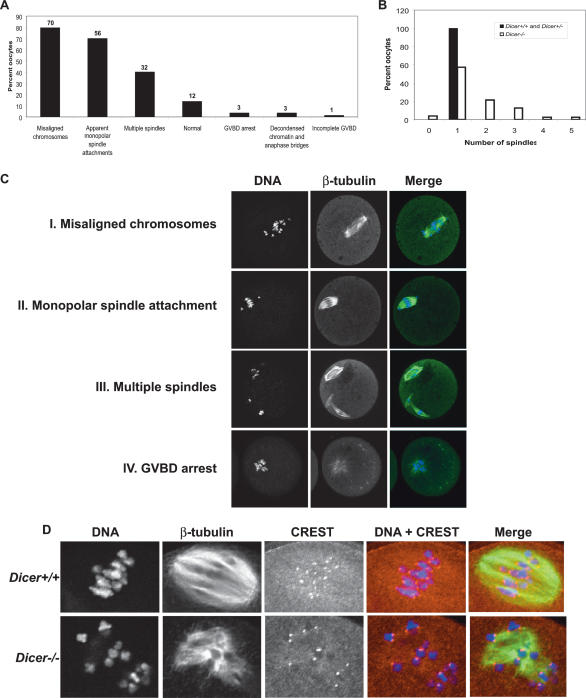
Almost 90% of Dicer−/− oocytes that had matured to either MI or MII had a clear defect in spindle organization and/or chromosome congression. These displayed a diverse array of spindle phenotypes including multiple spindles (40%), misaligned chromosomes (80%), apparent monopolar spindle attachments (affecting either one or many chromosomes) (70%), decondensed chromatin and anaphase bridges (3%), and premature arrest at GVBD (3%) (Fig. 4). Although most multispindled oocytes contained only one extra spindle, we observed oocytes with up to five spindles (Fig. 4B). We did not encounter any of these phenotypes in >50 control oocytes.
Figure 4.
Loss of Dicer causes a range of spindle and chromosome congression defects. (A) Frequency distribution of Dicer−/− oocyte phenotypes. The absolute number of oocytes observed with a given phenotype is indicated above the bar. One oocyte may fall into more than one phenotype class. (B) Quantification of number of spindles per oocyte in Dicer−/− and wild-type oocytes. (C) β-Tubulin immunofluorescence staining of Dicer−/− oocytes exhibiting several common phenotypes. DNA was counterstained with DAPI or Sytox green. (D) Centromere staining in Dicer−/− oocytes. Oocytes were fixed in 3.7% paraformaldehyde and stained with a CREST antiserum together with a β-tubulin antibody; DNA was counterstained with DAPI.
Centromeres are sites of chromosome constriction during mitosis and meiosis and serve as a platform for formation of the kinetochore, a structure that interacts with microtubules during chromosome segregation. Defects in kinetochore assembly can create problems in chromosome congression and chromosome segregation (Carroll and Straight 2006). Because this was one potential underlying cause of defects we observed in Dicer−/− oocytes, we examined the integrity of centromeres by staining with CREST antisera (Fig. 4D). Although centromeres in Dicer−/− oocytes had normal CREST patterns, we observed that some bivalent chromosomes in Dicer−/− oocytes appeared to have decreased cohesion, with centromeres of some paired homologs lacking attachment (Fig. 4D). Of course, such an arrangement would be expected when chromosomes undergo anaphase I, but this arrangement was also observed on chromosomes that were clearly not in anaphase. Moreover, many “free” centromeres were observed that were not attached to microtubules, a phenotype indicative of kinetochore or kinetochore-attachment defects. These results suggest that the inner centromere is maintained normally in Dicer−/− oocytes, but that chromosome cohesion and kinetochore microtubule attachment may be disrupted. Altogether, these observations demonstrate that Dicer is required for spindle organization, chromosome attachment and meiotic maturation in mouse oocytes.
Small RNA biogenesis pathways are required for regulation of maternal transcripts
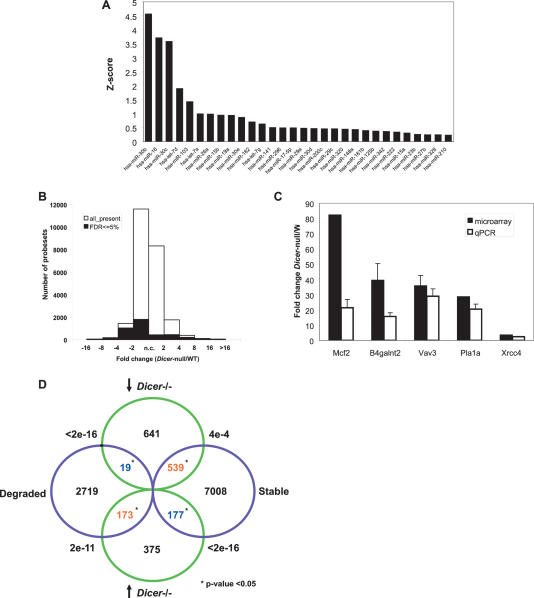
During the course of meiotic maturation the oocyte is transcriptionally quiescent. This state persists through fertilization and pronucleus formation until the zygotic genome becomes active. Although transcription initiates in the one-cell embryo during mid-S phase, it is not required for cleavage to the two-cell stage. Cleavage to the four-cell stage, however, does require transcription (Schultz 2002). Thus, the period of development from the oocyte to two-cell stage is supported exclusively by maternal mRNAs (Fig. 3A; Solter et al. 2004). Progression through these stages depends on timely translation and degradation of maternal transcripts accumulated during oocyte growth. MiRNAs are small Dicer-dependent noncoding RNAs that regulate stability and translation of their target mRNAs by recruiting RNA-induced silencing complexes (RISCs) (Murchison and Hannon 2004). The long coevolution of miRNAs and their targets has culminated in a dependence on animal miRNAs to confer robustness to gene expression profiles and streamline developmental transitions (Bartel 2004). Although miRNAs have been detected in mouse oocytes, their role in regulating maternal transcripts has not been explored (Amanai et al. 2006; Watanabe et al. 2006). We first sought to characterize the miRNAs present in mouse GV oocytes by performing quantitative RT–PCR on a subset of known miRNAs. We readily detected a complex population of miRNAs, with members of the miR-30, miR-16, and let-7 families being detected at the highest levels in our set (Fig. 5A). This raised the possibility of a role for miRNAs in controlling the pattern of mRNAs that are present and expressed in maturing oocytes.
Figure 5.
miRNAs and siRNAs may accelerate degradation of maternal transcripts. (A) Relative expression levels of the 30 most abundantly detected miRNAs in GV oocytes (out of a set of 103 tested miRNAs). miRNAs were quantified as described in Materials and Methods. Z-score expresses the divergence of the experimental result from the most probable result (mean) as a number of standard deviations. The larger the value of Z, the less probable the experimental result is due to chance. (B) Distribution of probe changes in the microarray comparing Dicer−/− to wild-type oocytes. FDR ≤5% (shaded region) denotes the probes whose changes hold greatest confidence. (C) Confirmation of selected microarray changes in Dicer−/− oocytes by quantitative RT–PCR. Fold change between Dicer−/− and wild-type oocytes is indicated on the Y-axis. Error bars denote standard error of the mean. (D) Superposition of probes that change in Dicer−/− with those that are degraded during meiotic maturation (Su et al. 2006). Only probe sets called present in both experiments (19,301 probe sets) were used in this analysis. Green circles show the number of probe sets significantly changed at least twofold in Dicer−/− oocytes, and violet circles show the probe sets degraded or kept stable from GV to MII (Su et al. 2006). The overlapping probe sets are shown in the overlapping region, together with the p-value by χ2 test. Statistical significance (p-value < 0.05) is indicated by an asterisk (*). Blue indicates a negative correlation between overlapping probe sets and red means two sets are positively correlated.
To determine whether maternal transcripts are misregulated in Dicer−/− ooyctes, we performed microarrays on Dicer−/− and control oocytes matured in vitro for 16 h. We found that many transcripts had different expression levels in these two genetic backgrounds. Of the 24,021 probe sets defined as present in either wild-type or Dicer−/− oocytes, 4420 (18.40%) were identified to be significantly changed using the criteria of FDR q-value ≤ 0.05, with 1295 up-regulated and 3125 down-regulated probe sets. Particularly, 1300 probe sets showed at least twofold down-regulated expression in Dicer−/−, while 861 probe sets showed more than twofold increases in the mutant (Fig. 5B). Of the probes that were more highly represented in Dicer−/− oocytes, many increased between two- and fourfold, while others went up drastically, with the greatest changes being several-hundred-fold (Supplementary Table 1). The increased abundance of several transcripts that was indicated by microarray analysis was confirmed by quantitative RT–PCR (Fig. 5C). Changes in transcript levels in Dicer−/− oocytes could be an indirect consequence of Dicer loss prior to the decrease in global transcription that initiates around the time of antrum formation. It is likely, however, that many transcripts also change as a direct consequence of Dicer ablation and reflect a role for Dicer in post-transcriptional gene regulation in oocytes, either in the deposition and maintenance of stored messages or in maternal transcript turnover.
Table 1.
Misregulation of transcripts involved in spindle regulation may contribute to the spindle organization defect in Dicer−/− oocytes
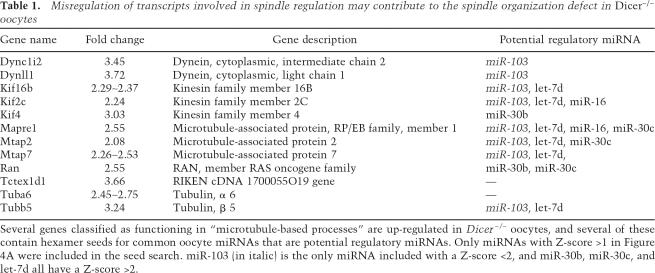
Several genes classified as functioning in “microtubule-based processes” are up-regulated in Dicer−/− oocytes, and several of these contain hexamer seeds for common oocyte miRNAs that are potential regulatory miRNAs. Only miRNAs with Z-score >1 in Figure 4A were included in the seed search. miR-103 (in italic) is the only miRNA included with a Z-score <2, and miR-30b, miR-30c, and let-7d all have a Z-score >2.
Oocyte maturation depends upon the timely, post-transcriptional control of mRNA content and expression. Previous expression analyses have identified a set of mRNAs that are specifically degraded during meiotic maturation (Su et al. 2006). Interestingly, we find a very strong correlation between these mRNAs and those whose levels increased upon Dicer loss, with 173 probes appearing in both datasets (χ2 test p-value < 10−10). In contrast, transcripts that are down-regulated in Dicer−/− oocytes tend to be stable during meiotic maturation (χ2 test p-value = 0.0004) (Fig. 5D).
To examine the hypothesis that miRNAs accelerate mRNA turnover during meiotic maturation we searched for miRNA target sites in transcripts that are up-regulated in Dicer−/− oocytes. We identified putative miRNA target sites in up-regulated transcripts, including seed sequences (6-base-pair [bp] sequences in positions two to seven of mature miRNAs) for the most abundant miRNA families. However, only three seed sequences, for mmu-miR-495, mmu-miR-126, and mmu-miR-302c* were enriched in the up-regulated Dicer−/− transcript set above the unchanged set (χ2 test p-value < 0.001). One hundred three hexamers not related to any known mouse miRNAs were also significantly enriched, although six of them were seed sequences for known human miRNAs (hsa-miR-570, hsa-miR-656, hsa-miR-548d, hsa-miR-496, hsa-miR-524*, and hsa-miR-576). We carried out a similar analysis for hepatmers and octamers (data not shown). With respect to octamers, we found those corresponding to seed matches for mmu-miR-706 and mmu-miR-495 were enriched in the up-regulated set (p < 0.005), as were 305 octamers unrelated to known miRNAs. Many of the most significantly enriched hexamers and octamers are AU rich and may reflect a general bias for AU richness in the 3′ untranslated regions (UTRs) of both up-regulated and down-regulated transcripts (p < 10−15) (Supplementary Table 2). Our observation that transcripts that are up-regulated in Dicer−/− oocytes are highly correlated with mRNAs that are degraded during oocyte maturation suggests that small RNAs may act, directly or indirectly, to accelerate transcript turnover during this developmental process. In contrast to the maternal-to-zygotic genome transition in zebrafish, whose success largely depends on the activity of a single miRNA (Giraldez et al. 2005, 2006), it appears that small RNA regulation of maternal mRNAs during meiotic maturation is more complex.
A role for Dicer in protecting the female germline from transposable elements
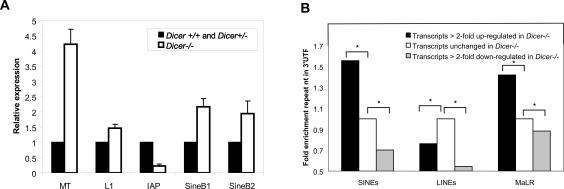
Dicer has been implicated in the control of transposable elements in a number of organisms. The recent discovery of repeat-derived small RNAs in mouse oocytes (Watanabe et al. 2006) raised the possibility that Dicer might be required to regulate transposons in these cells. To test this possibility, we measured changes in transposon levels in Dicer−/− oocytes by quantitative RT–PCR. Strikingly, we observed distinct changes in the abundance of several transposons, including a clear up-regulation of Mouse Transposon (MT) and short interspersed repetitive elements (SINEs) in the absence of Dicer (Fig. 6A). As MT has been reported to represent up to 13% of transcripts present in oocytes (Solter et al. 2004; Evsikov et al. 2006), such an increase is significant and suggests that Dicer−/− oocytes may contain abnormally high concentrations of transposon transcripts. Repeat sequences are often found embedded in the UTRs of genes, and transcripts in mouse ooctyes and early embryos often contain alternative 5′UTRs that are unusually rich in repeat-related sequences (Peaston et al. 2004).
Figure 6.
Transposons and mRNAs harboring transposon-derived sequences are misregulated in Dicer−/− oocytes. (A) Quantitation of retrotransposon levels in Dicer−/− oocytes by RT–PCR. Error bars indicate standard error of the mean. (B) Quantitation of repeat element representation in 3′UTRs of transcripts that are either up-regulated, down-regulated, or remain stable in the absence of Dicer. Values have been normalized to the percent 3′UTR repeat content of the unchanged set. Asterisk (*)denotes statistical significance (p < 1.0e−10) of repeat proportion difference (percent of 3′UTR contributed by repetitive elements) between two sets of transcripts calculated by prop.test in R package (http://www.r-project.org).
As we were unable to define a distinct miRNA signature that could explain the substantial expression changes observed in Dicer−/− oocytes, we examined the alternative hypothesis that repeat-derived small RNAs might subject a substantial fraction of oocyte mRNAs to Dicer-dependent regulation. We searched for repeat-related sequences in the 3′UTRs of transcripts that were increased in abundance in the absence of Dicer. Interestingly, we found that mRNAs that increased twofold or more in Dicer−/− oocytes have a highly significant enrichment for transposon-derived sequences in their 3′UTRs (Fig. 6B). SINE-derived motifs were particularly enriched in up-regulated transcripts, whereas sequences derived from long interspersed repetitive elements (LINEs) were not enriched. Interestingly, SINEs and MT elements are deregulated in Dicer−/− oocytes, while transcripts derived from LINEs are unaffected. Considered together, these data suggest that Dicer may regulate a considerable number of oocyte transcripts via transposon sequences embedded in their UTRs.
Discussion
Here we report the involvement of Dicer in ensuring spindle integrity in mammalian oocytes, thus uncovering a novel role for RNAi during mammalian meiosis. Although effects on spindle function and chromosome segregation are widely observed consequences of defects in RNAi, the mechanisms underlying the relationships between these pathways may not be identical in all contexts. In the fission yeast Schizosaccharomyces pombe RNAi is indispensable for centromere and kinetochore function through its direct role in heterochromatin maintenance (Hall et al. 2003; Volpe et al. 2003). In a number of other organisms including trypanosomes, Drosophila, Caenorhabditis elegans, chicken, and mammals, the nature of the role of RNAi in chromosome segregation is less clear (Durand-Dubief and Bastin 2003; Fukagawa et al. 2004; Deshpande et al. 2005; Kanellopoulou et al. 2005; Murchison et al. 2005; Yigit et al. 2006).
Dicer deficiency has been studied in some detail in several mouse cell types, tissues, and organs (Cobb et al. 2005, 2006; Harfe et al. 2005; Kanellopoulou et al. 2005; Muljo et al. 2005; Murchison et al. 2005; Andl et al. 2006; Harris et al. 2006; Yi et al. 2006). That such a severe spindle organization defect as is described here has not been previously observed suggests that Dicer may play a special role during meiosis. The primary source of the spectrum of spindle phenotypes we observe in Dicer−/− oocytes appears to be a failure of chromosomes to engage productively with microtubules either due to a defect in chromosome-derived signals for spindle organization, kinetochore assembly, or microtubule growth or responsiveness. The presence of unattached chromosomes in Dicer−/− oocytes may trigger the spindle checkpoint (Wassmann et al. 2003), leading to a prolonged arrest during which high MPF levels may favor inappropriate assembly of multiple spindles. With time, the spindle checkpoint-mediated arrest may be overcome, resulting in the failure to properly segregate chromosomes, the generation of defective polar bodies, and meiotic catastrophe.
The nature of the small RNAs acting downstream from Dicer to promote proper spindle function in oocytes remains uncertain. The two most likely possibilities, based upon known properties of the RNAi pathway, are that siRNAs mediating the construction of critical centromeric chromatin domains are not properly generated, or that a lack of proper miRNA or siRNA profiles leads to defects in gene expression that affect spindle function. With respect to the former model, centromeric siRNAs have not yet been identified in mammalian systems. The latter model is consistent with the dramatic changes in mRNA profiles in Dicer−/− oocytes. To investigate functional roles for miRNAs or other Dicer-dependent small RNAs during ooctye maturation, we analyzed Gene Ontology (GO) terms (Gene Ontology Consortium 2001) for transcripts that accumulate in Dicer−/− oocytes. We found that mRNAs related to several biological processes were significantly enriched, among these being “microtubule related processes.” This is of particular interest considering the profound effect of Dicer loss on the meiotic spindle. Strikingly, several of the 12 up-regulated genes with this GO term (including several tubulin subunits, chromokinesins, Eb1, and Ran) had putative binding sites for miRNAs present in oocytes (Table 1). Changes in any of a number of candidate targets could lead to the observed defects in chromosome segregation. One interesting candidate is the small GTPase Ran whose transcript accumulates 2.55-fold above normal in Dicer−/− oocytes (Table 1) and whose graded activity, directed by condensed chromosomes themselves, is required to organize the meiotic spindle (Brunet and Maro 2005).
The recent discovery of piwi-interacting RNAs (piRNAs) in the germlines of mammals and Drosophila has highlighted the important role of RNAi pathways in protecting germ cells from transposable elements (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Vagin et al. 2006; Watanabe et al. 2006). These studies have focused attention on the Piwi subfamily of Argonaute proteins, which may bind small RNAs generated in a Dicer-independent fashion (Vagin et al. 2006). However, there is also substantial evidence for the involvement of Dicer-dependent small RNAs in transposon regulation in animals (Robert et al. 2004; Svoboda et al. 2004; Kanellopoulou et al. 2005; Rehwinkel et al. 2006; Watanabe et al. 2006). Our genetic anaysis has clearly demonstrated that the MT and SINE families of retrotransposons are significantly up-regulated in Dicer−/− oocytes. Considered together with the marked lack of an effect of individual Piwi family mutations on the mammalian female germline, our studies indicate that both clades of Argonaute protein may participate in transposon control both through Dicer-dependent and Dicer-independent mechanisms.
A full understanding of the role of repeat elements and their corresponding small RNAs in controlling gene expression in oocytes awaits a complete catalog of Dicer-dependent and Dicer-independent species through oocyte development and early embryogenesis. Interestingly, our studies thus far suggest that repeat-derived small RNAs may control the abundance of some maternal mRNAs during oocyte maturation. This raises the possibility that the presence of repeat-derived sequences in the 3′UTRs of mRNAs might be far from neutral, instead allowing the subjugation of certain mRNAs to developmental control, by coincident post-transcriptional suppression of transposon transcripts and mRNAs sharing sequences with repetitive elements. This evolutionary strategy would be analogous to the coevolution of miRNAs and miRNA-binding sites in certain classes of mRNAs and would be a classical embodiment of the transposon as a “controlling element” as originally posited by McClintock (1950, 1953).
Materials and methods
Animals and oocytes
3A8 Dicer conditional animals (Andl et al. 2006) were crossed to Zp3-Cre mice (Jackson Laboratories) (de Vries et al. 2000) and their progeny were intercrossed to produce Dicerflox/flox; Zp3-Cre animals. To determine fertility, two female Dicerflox/flox; Zp3-Cre female mice were bred with several males of proven fertility for a period of 11 wk. Most mice used in this study were 4–6 wk old, and were primed by intraperitoneal injection of 5 IU of PMSG 48 h before oocyte collection. Fully grown, GV-intact cumulus-enclosed oocytes were collected as described (Schultz et al. 1983). Oocytes were collected in MEM/PVP (bicarbonate-free minimal essential medium [Earle’s salt] supplemented with pyruvate [100 μg/mL], gentamicin [10 μg/mL], polyvinylpyrrolidone [PVP; 3 mg/mL], 25 mM HEPES at pH 7.3) containing 2.5 μM milrinone and allowed to mature in milrinone-free CZB (Chatot et al. 1989) in an atmosphere of 5% CO2 in air at 37°C for 8 h (MI) or 16 h (MII). Metaphase II egg and embryo collection for the Dicer developmental profile and immunofluorescence were performed as previously described (Romanova et al. 2006). Meiotically incompetent oocytes were obtained from 13-d-old female mice as described (Svoboda et al. 2001). All animal experiments were approved by the Institutional Animal Use and Care Committee and were consistent with National Institutes of Health guidelines.
dsRNA preparation and microinjection
Mos dsRNA was prepared and injected (5 pL of 0.2 μg/μL solution) as previously described (Svoboda et al. 2000).
Immunohistochemistry and immunofluoresence
Ovaries prepared for immunohistochemistry were fixed overnight in Bouin’s fixative, embedded in paraffin, sliced to 10-μm sections, and stained with hematoxylin and eosin. Immunofluorescence was performed as previously described (Anger et al. 2005). Primary antibodies were used at the following dilutions: β-tubulin (Sigma, T4026) 1:500, CREST antiserum 1:50, and Alexa-fluor 635 phalloidin (Molecular Probes, A34054) 1:500. Secondary antibodies were used as follows: Cy5-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch, 715-175-150) 1:100, Cy5-conjugated donkey anti-human IgG (Jackson ImmunoResearch, 709-175-149) 1:100, and goat anti-mouse IgG1 fluorescein (FITC)-conjugated (SouthernBiotech, 1070-02) 1:100. Sytox green (Invitrogen, 1:5000) was included in the final wash. DAPI (Sigma, 1.5 μg/mL) was added to the mounting medium (Vectashield, Vector Laboratories).
Quantitative RT–PCR
Fifteen to 20 oocytes were pooled for RNA extraction using the Absolutely RNA Microprep kit (Stratagene). cDNA synthesis was performed using random hexamers, and quantitative PCR was performed using Sybr Green mix (Applied Biosystems) in a PTC-200 thermal cycler (MJ Research). Signals were normalized to β-actin. Each reaction was perfomed in duplicate and repeated on different samples at least twice. Primer sequences were Vav3F, TACCTAAACCAGTAGATTATTCTT; Vav3R, CTAATTGCATATTCTCCAGATTCT; Pla1aF, CTACTTTG TCAATGGAGGCCAAGA; Pla1aR, cCAGGGCACTGATGTA GAGGTGTA; Mcf2F, AGGCACTAGACCAGGACCAACTTT; Mcf2R, CACAGCTGCCTCCCTGACCTTTAT; B4galntF, ACA GACTGTACCCAAGTTGTATGA; B4galntR, CTGGGTAAT ATTTTCGAATACTCT; Xrcc4F, ACCTAAAATGGCTCCACAG GAGTT; Xrcc4R, AGGGTTTCTAAAGACATGTTTTCA; MT-F, TGTTAAGAGCTCTGTCGGATGTTG; MT-R, ACTGATTCTT CAGTCCCAGCTAAC; DicerF, TACACACGCCTCCTACCAC TACAA; DicerR, CCAAAATCGCATCTCCCAGGAATT; ActinF, CGGTTCCGATGCCCTGAGGCTCTT; ActinR, CGTCACACT TCATGATGGAATTGA; MosF, GGGAACAGGTATGTCTGAT GCA; MosR, CACCGTGGTAAGTGGCTTTATACA. Other transposon primers can be found in Martens et al. (2005). For the Dicer developmental profile, total RNA isolation, reverse transcription, and real-time PCR were performed as previously described (Romanova et al. 2006). TaqMan Gene Expression Assay Mm00521715_m1 (Applied Biosystems) was used to quantify Dicer expression that was normalized to GFP and expressed relative to the amount of Dicer mRNA in GV oocytes (Romanova et al. 2006).
miRNA expression profiling
Total RNA was isolated from fully grown oocytes using the mirVana miRNA isolation kit (Ambion), according to the manufacturer’s protocol. miRNAs were quantified using the TaqMan MicroRNA Assays Human Panel (Applied Biosystems) following the manufacturer’s protocol. Of the 157 miRNAs in the panel, 103 were assayed (the ones that are identical in human and mouse), as well as five negative controls. Five oocyte equivalents were used for reverse transcription and 0.44 oocyte equivalents for the real-time PCR reaction. The experiment was repeated two times and all the reactions were run in duplicate.
Microarray analysis
RNA was isolated from 25 oocytes after 16 h in culture and amplified as previously described (Zeng et al. 2004; Pan et al. 2005). Three samples were generated for each group. The biotinylated cRNA was fragmented and hybridized to the Affymetrix MOE430 v2 chip, which contains ∼45,000 transcripts. We used gcRMA (GC robust multiarray average) (Wu et al. 2004) to perform the background adjustment and normalization, and used SAM (significance analysis of microarray) (Tusher et al. 2001) to pick up significantly changed probe sets and calculate the false discovery rate (FDR) for potential multiple-testing problems. Probe sets that were considered as absent in all six arrays based on Affymetrix MAS present/absent call (http://www.affymetrix.com/support/technical/whitepapers.affx) were removed and not used in later analysis. We finally used FDR <5% and at least twofold change to select significantly changed probe sets. We used EASE (Hosack et al. 2003) to identify the molecular function and biological process GO terms.
miRNA seed sequence analysis
For each set of up- or down-regulated genes in Dicer−/− oocytes, we searched all A, C, G, and T hexamers and octamers in the 3′UTRs, and we also selected genes with no change in expression between wild-type and Dicer−/− oocytes as controls to do the same hexamer or octamer search. Pearson’s χ2 test in R package (http://www.r-project.org) was performed to calculate the significance of the difference of the motif frequencies between up- or down-regulated gene sets and their control set. Each hexamer or octamer was then compared with the corresponding seed region of mature miRNAs to find the related miRNA.
Repeat analysis
We used RepeatMasker (http://www.repeatmasker.org) to identify repeat elements, including rodent-specific repeats, in 3′UTRs of transcripts. We compared the proportion of repeat regions in twofold up-reglated transcripts and a control set of transcripts whose expression levels do not change in the microarray analysis. Significance of proportion difference is calculated by using prop.test in R package for different repeat families, including SINEs, LINEs, and long terminal repeats (LTRs).
Acknowledgments
We thank Oliver Tam, Janet Partridge, Michelle Carmell, Supriya Prasanth, and Bruce Stillman for help, discussions, and critical reading of the manuscript. Thanks to all other members of the Hannon Laboratory for discussion and help with manuscript preparation. The CREST antisera was a gift from Rabindranath de la Fuente. Thanks to Joel Parker and Don Baldwin for assistance with microarrays and to Lisa Bianco, Jodi Coblentz, Lea Manzella, and Maria Mosquera for help with genotyping and histology. We are grateful to You-Qiang Su and John Eppig for sharing microarray data. E.P.M. is an Engelhorn Scholar of the Watson School of Biological Sciences and is supported by a predoctoral fellowship from the Department of Defense Breast Cancer Research Program (X81XWH-05-1-0256). R.M.S. is supported by NIH grant HD22681. Z.X. and M.Q.Z. were supported by grant number HG001696. G.J.H. is an Investigator of the Howard Hughes Medical Institute, and this work is supported in part by grants from the NIH to G.J.H.
Footnotes
Supplemental material is available at http//:www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1521307
References
- Amanai M., Brahmajosyula M., Perry A.C., Brahmajosyula M., Perry A.C., Perry A.C. A restricted role for sperm-borne microRNAs in mammalian fertilization. Biol. Reprod. 2006;75:877–884. doi: 10.1095/biolreprod.106.056499. [DOI] [PubMed] [Google Scholar]
- Andl T., Murchison E.P., Liu F., Zhang Y., Yunta-Gonzalez M., Tobias J.W., Andl C.D., Seykora J.T., Hannon G.J., Millar S.E., Murchison E.P., Liu F., Zhang Y., Yunta-Gonzalez M., Tobias J.W., Andl C.D., Seykora J.T., Hannon G.J., Millar S.E., Liu F., Zhang Y., Yunta-Gonzalez M., Tobias J.W., Andl C.D., Seykora J.T., Hannon G.J., Millar S.E., Zhang Y., Yunta-Gonzalez M., Tobias J.W., Andl C.D., Seykora J.T., Hannon G.J., Millar S.E., Yunta-Gonzalez M., Tobias J.W., Andl C.D., Seykora J.T., Hannon G.J., Millar S.E., Tobias J.W., Andl C.D., Seykora J.T., Hannon G.J., Millar S.E., Andl C.D., Seykora J.T., Hannon G.J., Millar S.E., Seykora J.T., Hannon G.J., Millar S.E., Hannon G.J., Millar S.E., Millar S.E. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr. Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anger M., Stein P., Schultz R.M., Stein P., Schultz R.M., Schultz R.M. CDC6 requirement for spindle formation during maturation of mouse oocytes. Biol. Reprod. 2005;72:188–194. doi: 10.1095/biolreprod.104.035451. [DOI] [PubMed] [Google Scholar]
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovina N., Morris P., Brownstein M.J., Kuramouchi-Miyaqawa S., Nakano T., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovina N., Morris P., Brownstein M.J., Kuramouchi-Miyaqawa S., Nakano T., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovina N., Morris P., Brownstein M.J., Kuramouchi-Miyaqawa S., Nakano T., Lagos-Quintana M., Landgraf P., Iovina N., Morris P., Brownstein M.J., Kuramouchi-Miyaqawa S., Nakano T., Landgraf P., Iovina N., Morris P., Brownstein M.J., Kuramouchi-Miyaqawa S., Nakano T., Iovina N., Morris P., Brownstein M.J., Kuramouchi-Miyaqawa S., Nakano T., Morris P., Brownstein M.J., Kuramouchi-Miyaqawa S., Nakano T., Brownstein M.J., Kuramouchi-Miyaqawa S., Nakano T., Kuramouchi-Miyaqawa S., Nakano T., Nakano T., et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Brunet S., Maro B., Maro B. Cytoskeleton and cell cycle control during meiotic maturation of the mouse oocyte: Integrating time and space. Reproduction. 2005;130:801–811. doi: 10.1530/rep.1.00364. [DOI] [PubMed] [Google Scholar]
- Carroll C.W., Straight A.F., Straight A.F. Centromere formation: From epigenetics to self-assembly. Trends Cell Biol. 2006;16:70–78. doi: 10.1016/j.tcb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Chatot C.L., Ziomek C.A., Bavister B.D., Lewis J.L., Torres I., Ziomek C.A., Bavister B.D., Lewis J.L., Torres I., Bavister B.D., Lewis J.L., Torres I., Lewis J.L., Torres I., Torres I. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J. Reprod. Fertil. 1989;86:679–688. doi: 10.1530/jrf.0.0860679. [DOI] [PubMed] [Google Scholar]
- Cobb B.S., Nesterova T.B., Thompson E., Hertweck A., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fishe A.G., Smale S.T., Nesterova T.B., Thompson E., Hertweck A., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fishe A.G., Smale S.T., Thompson E., Hertweck A., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fishe A.G., Smale S.T., Hertweck A., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fishe A.G., Smale S.T., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fishe A.G., Smale S.T., Godwin J., Wilson C.B., Brockdorff N., Fishe A.G., Smale S.T., Wilson C.B., Brockdorff N., Fishe A.G., Smale S.T., Brockdorff N., Fishe A.G., Smale S.T., Fishe A.G., Smale S.T., Smale S.T., et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J. Exp. Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb B.S., Hertweck A., Smith J., O’Connor E., Graf D., Smale S.T., Sakaquicki S., Livesey F.J., Fisher A.G., Merkenschlager M., Hertweck A., Smith J., O’Connor E., Graf D., Smale S.T., Sakaquicki S., Livesey F.J., Fisher A.G., Merkenschlager M., Smith J., O’Connor E., Graf D., Smale S.T., Sakaquicki S., Livesey F.J., Fisher A.G., Merkenschlager M., O’Connor E., Graf D., Smale S.T., Sakaquicki S., Livesey F.J., Fisher A.G., Merkenschlager M., Graf D., Smale S.T., Sakaquicki S., Livesey F.J., Fisher A.G., Merkenschlager M., Smale S.T., Sakaquicki S., Livesey F.J., Fisher A.G., Merkenschlager M., Sakaquicki S., Livesey F.J., Fisher A.G., Merkenschlager M., Livesey F.J., Fisher A.G., Merkenschlager M., Fisher A.G., Merkenschlager M., Merkenschlager M. A role for Dicer in immune regulation. J. Exp. Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries W.N., Binns L.T., Fancher K.S., Dean J., Moore R., Kimler R., Knowles B.B., Binns L.T., Fancher K.S., Dean J., Moore R., Kimler R., Knowles B.B., Fancher K.S., Dean J., Moore R., Kimler R., Knowles B.B., Dean J., Moore R., Kimler R., Knowles B.B., Moore R., Kimler R., Knowles B.B., Kimler R., Knowles B.B., Knowles B.B. Expression of Cre recombinase in mouse oocytes: A means to study maternal effect genes. Genesis. 2000;26:110–112. [PubMed] [Google Scholar]
- Deshpande G., Calhoun G., Schedl P., Calhoun G., Schedl P., Schedl P. Drosophila argonaute-2 is required early in embryogenesis for the assembly of centric/centromeric heterochromatin, nuclear division, nuclear migration, and germ-cell formation. Genes & Dev. 2005;19:1680–1685. doi: 10.1101/gad.1316805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Dubief M., Bastin P., Bastin P. TbAGO1, an argonaute protein required for RNA interference, is involved in mitosis and chromosome segregation in Trypanosoma brucei. BMC Biol. 2003;1:2. doi: 10.1186/1741-7007-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evsikov A.V., Graber J.H., Brockman J.M., Hampl A., Holbrook A.E., Singh P., Eppig J.J., Solter D., Knowles B.B., Graber J.H., Brockman J.M., Hampl A., Holbrook A.E., Singh P., Eppig J.J., Solter D., Knowles B.B., Brockman J.M., Hampl A., Holbrook A.E., Singh P., Eppig J.J., Solter D., Knowles B.B., Hampl A., Holbrook A.E., Singh P., Eppig J.J., Solter D., Knowles B.B., Holbrook A.E., Singh P., Eppig J.J., Solter D., Knowles B.B., Singh P., Eppig J.J., Solter D., Knowles B.B., Eppig J.J., Solter D., Knowles B.B., Solter D., Knowles B.B., Knowles B.B. Cracking the egg: Molecular dynamics and evolutionary aspects of the transition from the fully grown oocyte to embryo. Genes & Dev. 2006;20:2713–2727. doi: 10.1101/gad.1471006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakagama T., Oshimura M., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakagama T., Oshimura M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakagama T., Oshimura M., Ikeno M., Okazaki T., Takami Y., Nakagama T., Oshimura M., Okazaki T., Takami Y., Nakagama T., Oshimura M., Takami Y., Nakagama T., Oshimura M., Nakagama T., Oshimura M., Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium Creating the gene ontology resource: Design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A.J., Cinalli R.M., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Cinalli R.M., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Hammond S.M., Bartel D.P., Schier A.F., Bartel D.P., Schier A.F., Schier A.F. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Inoue K., Enright A.J., Schier A.F., Enright A.J., Schier A.F., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Girard A., Sachidanandam R., Hannon G.J., Carmell M.A., Sachidanandam R., Hannon G.J., Carmell M.A., Hannon G.J., Carmell M.A., Carmell M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grivna S.T., Beyret E., Wang Z., Lin H., Beyret E., Wang Z., Lin H., Wang Z., Lin H., Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes & Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall I.M., Noma K., Grewal S.I., Noma K., Grewal S.I., Grewal S.I. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl. Acad. Sci. 2003;100:193–198. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe B.D., McManus M.T., Mansfield J.H., Hornstein E., Tabin C.J., McManus M.T., Mansfield J.H., Hornstein E., Tabin C.J., Mansfield J.H., Hornstein E., Tabin C.J., Hornstein E., Tabin C.J., Tabin C.J. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc. Natl. Acad. Sci. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K.S., Zhang Z., McManus M.T., Harfe B.D., Sun X., Zhang Z., McManus M.T., Harfe B.D., Sun X., McManus M.T., Harfe B.D., Sun X., Harfe B.D., Sun X., Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc. Natl. Acad. Sci. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T., Hunt P., Hunt P. To err (meiotically) is human: The genesis of human aneuploidy. Nat. Rev. Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Hatfield S.D., Shcherbata H.R., Fischer K.A., Nakahara K., Carthew R.W., Ruohola-Baker H., Shcherbata H.R., Fischer K.A., Nakahara K., Carthew R.W., Ruohola-Baker H., Fischer K.A., Nakahara K., Carthew R.W., Ruohola-Baker H., Nakahara K., Carthew R.W., Ruohola-Baker H., Carthew R.W., Ruohola-Baker H., Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- Hosack D.A., Dennis G., Jr., Sherman B.T., Lane H.C., Lempicki R.A., Dennis G., Jr., Sherman B.T., Lane H.C., Lempicki R.A., Sherman B.T., Lane H.C., Lempicki R.A., Lane H.C., Lempicki R.A., Lempicki R.A. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C., Muljo S.A., Kung A.L., Ganesan S., Drapkin R., Jenuwein T., Livingston D.M., Rajewsky K., Muljo S.A., Kung A.L., Ganesan S., Drapkin R., Jenuwein T., Livingston D.M., Rajewsky K., Kung A.L., Ganesan S., Drapkin R., Jenuwein T., Livingston D.M., Rajewsky K., Ganesan S., Drapkin R., Jenuwein T., Livingston D.M., Rajewsky K., Drapkin R., Jenuwein T., Livingston D.M., Rajewsky K., Jenuwein T., Livingston D.M., Rajewsky K., Livingston D.M., Rajewsky K., Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes & Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S.W., Bass B.L., Bass B.L. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N.C., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Nakano T., Bartel D.P., Kingston R.E., Bartel D.P., Kingston R.E., Kingston R.E. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Lira S.A., Kinloch R.A., Mortillo S., Wassarman P.M., Kinloch R.A., Mortillo S., Wassarman P.M., Mortillo S., Wassarman P.M., Wassarman P.M. An upstream region of the mouse ZP3 gene directs expression of firefly luciferase specifically to growing oocytes in transgenic mice. Proc. Natl. Acad. Sci. 1990;87:7215–7219. doi: 10.1073/pnas.87.18.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J.H., O’Sullivan R.J., Braunschweig U., Opravil S., Radolf M., Steinlein P., Jenuwein T., O’Sullivan R.J., Braunschweig U., Opravil S., Radolf M., Steinlein P., Jenuwein T., Braunschweig U., Opravil S., Radolf M., Steinlein P., Jenuwein T., Opravil S., Radolf M., Steinlein P., Jenuwein T., Radolf M., Steinlein P., Jenuwein T., Steinlein P., Jenuwein T., Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. 1950;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. Induction of instability at selected loci in maize. Genetics. 1953;38:579–599. doi: 10.1093/genetics/38.6.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo S.A., Ansel K.M., Kanellopoulou C., Livingston D.M., Rao A., Rajewsky K., Ansel K.M., Kanellopoulou C., Livingston D.M., Rao A., Rajewsky K., Kanellopoulou C., Livingston D.M., Rao A., Rajewsky K., Livingston D.M., Rao A., Rajewsky K., Rao A., Rajewsky K., Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J. Exp. Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison E.P., Hannon G.J., Hannon G.J. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol. 2004;16:223–229. doi: 10.1016/j.ceb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Murchison E.P., Partridge J.F., Tam O.H., Cheloufi S., Hannon G.J., Partridge J.F., Tam O.H., Cheloufi S., Hannon G.J., Tam O.H., Cheloufi S., Hannon G.J., Cheloufi S., Hannon G.J., Hannon G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., O’Brien M.J., Wigglesworth K., Eppig J.J., Schultz R.M., O’Brien M.J., Wigglesworth K., Eppig J.J., Schultz R.M., Wigglesworth K., Eppig J.J., Schultz R.M., Eppig J.J., Schultz R.M., Schultz R.M. Transcript profiling during mouse oocyte development and the effect of gonadotropin priming and development in vitro. Dev. Biol. 2005;286:493–506. doi: 10.1016/j.ydbio.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Peaston A.E., Evsikov A.V., Graber J.H., de Vries W.N., Holbrook A.E., Solter D., Knowles B.B., Evsikov A.V., Graber J.H., de Vries W.N., Holbrook A.E., Solter D., Knowles B.B., Graber J.H., de Vries W.N., Holbrook A.E., Solter D., Knowles B.B., de Vries W.N., Holbrook A.E., Solter D., Knowles B.B., Holbrook A.E., Solter D., Knowles B.B., Solter D., Knowles B.B., Knowles B.B. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J., Natalin P., Stark A., Brennecke J., Cohen S.M., Izaurralde E., Natalin P., Stark A., Brennecke J., Cohen S.M., Izaurralde E., Stark A., Brennecke J., Cohen S.M., Izaurralde E., Brennecke J., Cohen S.M., Izaurralde E., Cohen S.M., Izaurralde E., Izaurralde E. Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster. Mol. Cell. Biol. 2006;26:2965–2975. doi: 10.1128/MCB.26.8.2965-2975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert V.J., Vastenhouw N.L., Plasterk R.H., Vastenhouw N.L., Plasterk R.H., Plasterk R.H. RNA interference, transposon silencing, and cosuppression in the Caenorhabditis elegans germ line: Similarities and differences. Cold Spring Harb. Symp. Quant. Biol. 2004;69:397–402. doi: 10.1101/sqb.2004.69.397. [DOI] [PubMed] [Google Scholar]
- Romanova L.G., Anger M., Zatsepina O.V., Schultz R.M., Anger M., Zatsepina O.V., Schultz R.M., Zatsepina O.V., Schultz R.M., Schultz R.M. Implication of nucleolar protein SURF6 in ribosome biogenesis and preimplantation mouse development. Biol. Reprod. 2006;75:690–696. doi: 10.1095/biolreprod.106.054072. [DOI] [PubMed] [Google Scholar]
- Schultz R.M. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum. Reprod. Update. 2002;8:323–331. doi: 10.1093/humupd/8.4.323. [DOI] [PubMed] [Google Scholar]
- Schultz R.M., Montgomery R.R., Belanoff J.R., Montgomery R.R., Belanoff J.R., Belanoff J.R. Regulation of mouse oocyte meiotic maturation: Implication of a decrease in oocyte cAMP and protein dephosphorylation in commitment to resume meiosis. Dev. Biol. 1983;97:264–273. doi: 10.1016/0012-1606(83)90085-4. [DOI] [PubMed] [Google Scholar]
- Solter D., Hiiragi T., Evsikov A.V., Moyer J., De Vries W.N., Peaston A.E., Knowles B.B., Hiiragi T., Evsikov A.V., Moyer J., De Vries W.N., Peaston A.E., Knowles B.B., Evsikov A.V., Moyer J., De Vries W.N., Peaston A.E., Knowles B.B., Moyer J., De Vries W.N., Peaston A.E., Knowles B.B., De Vries W.N., Peaston A.E., Knowles B.B., Peaston A.E., Knowles B.B., Knowles B.B. Epigenetic mechanisms in early mammalian development. Cold Spring Harb. Symp. Quant. Biol. 2004;69:11–17. doi: 10.1101/sqb.2004.69.11. [DOI] [PubMed] [Google Scholar]
- Su Y.Q., Sugiura K., Woo Y., Wigglesworth K., Kamdar S., Affourtit J., Eppig J.J., Sugiura K., Woo Y., Wigglesworth K., Kamdar S., Affourtit J., Eppig J.J., Woo Y., Wigglesworth K., Kamdar S., Affourtit J., Eppig J.J., Wigglesworth K., Kamdar S., Affourtit J., Eppig J.J., Kamdar S., Affourtit J., Eppig J.J., Affourtit J., Eppig J.J., Eppig J.J. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev. Biol. 2006;302:104–117. doi: 10.1016/j.ydbio.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P., Stein P., Hayashi H., Schultz R.M., Stein P., Hayashi H., Schultz R.M., Hayashi H., Schultz R.M., Schultz R.M. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development. 2000;127:4147–4156. doi: 10.1242/dev.127.19.4147. [DOI] [PubMed] [Google Scholar]
- Svoboda P., Stein P., Schultz R.M., Stein P., Schultz R.M., Schultz R.M. RNAi in mouse oocytes and preimplantation embryos: Effectiveness of hairpin dsRNA. Biochem. Biophys. Res. Commun. 2001;287:1099–1104. doi: 10.1006/bbrc.2001.5707. [DOI] [PubMed] [Google Scholar]
- Svoboda P., Stein P., Anger M., Bernstein E., Hannon G.J., Schultz R.M., Stein P., Anger M., Bernstein E., Hannon G.J., Schultz R.M., Anger M., Bernstein E., Hannon G.J., Schultz R.M., Bernstein E., Hannon G.J., Schultz R.M., Hannon G.J., Schultz R.M., Schultz R.M. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev. Biol. 2004;269:276–285. doi: 10.1016/j.ydbio.2004.01.028. [DOI] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani R., Chu G., Tibshirani R., Chu G., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin V.V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D., Li C., Seitz H., Gvozdev V., Zamore P.D., Seitz H., Gvozdev V., Zamore P.D., Gvozdev V., Zamore P.D., Zamore P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Volpe T., Schramke V., Hamilton G.L., White S.A., Teng G., Martienssen R.A., Allshire R.C., Schramke V., Hamilton G.L., White S.A., Teng G., Martienssen R.A., Allshire R.C., Hamilton G.L., White S.A., Teng G., Martienssen R.A., Allshire R.C., White S.A., Teng G., Martienssen R.A., Allshire R.C., Teng G., Martienssen R.A., Allshire R.C., Martienssen R.A., Allshire R.C., Allshire R.C. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 2003;11:137–146. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- Wassmann K., Niault T., Maro B., Niault T., Maro B., Maro B. Metaphase I arrest upon activation of the Mad2-dependent spindle checkpoint in mouse oocytes. Curr. Biol. 2003;13:1596–1608. doi: 10.1016/j.cub.2003.08.052. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Takeda A., Tsukiyama T., Mise K., Okuno T., Sasaki H., Minami N., Imai H., Takeda A., Tsukiyama T., Mise K., Okuno T., Sasaki H., Minami N., Imai H., Tsukiyama T., Mise K., Okuno T., Sasaki H., Minami N., Imai H., Mise K., Okuno T., Sasaki H., Minami N., Imai H., Okuno T., Sasaki H., Minami N., Imai H., Sasaki H., Minami N., Imai H., Minami N., Imai H., Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes & Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Irizarry R.A., Gentleman R., Martinez-Murillo F., Spencer F., Irizarry R.A., Gentleman R., Martinez-Murillo F., Spencer F., Gentleman R., Martinez-Murillo F., Spencer F., Martinez-Murillo F., Spencer F., Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 2004;99:909–917. [Google Scholar]
- Yi R., O’Carroll D., Pasolli H.A., Zhang Z., Dietrich F.S., Tarakhovsky A., Fuchs E., O’Carroll D., Pasolli H.A., Zhang Z., Dietrich F.S., Tarakhovsky A., Fuchs E., Pasolli H.A., Zhang Z., Dietrich F.S., Tarakhovsky A., Fuchs E., Zhang Z., Dietrich F.S., Tarakhovsky A., Fuchs E., Dietrich F.S., Tarakhovsky A., Fuchs E., Tarakhovsky A., Fuchs E., Fuchs E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat. Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- Yigit E., Batista P.J., Bei Y., Pang K.M., Chen C.C., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C., Batista P.J., Bei Y., Pang K.M., Chen C.C., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C., Bei Y., Pang K.M., Chen C.C., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C., Pang K.M., Chen C.C., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C., Chen C.C., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C., Mitani S., Simard M.J., Mello C.C., Simard M.J., Mello C.C., Mello C.C. Analysis of the C. elegansargonaute family reveals that distinct argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- Zeng F., Baldwin D.A., Schultz R.M., Baldwin D.A., Schultz R.M., Schultz R.M. Transcript profiling during preimplantation mouse development. Dev. Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]