Abstract
In Brassica self-incompatibility, the recognition of self/nonself pollen grains, is controlled by the S-locus, which encodes three highly polymorphic proteins: S-locus receptor kinase (SRK), S-locus protein 11 (SP11; also designated S-locus Cys-rich protein), and S-locus glycoprotein (SLG). SP11, located in the pollen coat, determines pollen S-haplotype specificity, whereas SRK, located on the plasma membrane of stigmatic papilla cells, determines stigmatic S-haplotype specificity. SLG shares significant sequence similarity with the extracellular domain of SRK and is abundant in the stigmatic cell wall, but its function is controversial. We previously showed that SP11 binds directly to its cognate SRK with high affinity (Kd = 0.7 nM) and induces its autophosphorylation. We also found that an SLG-like, 60-kD protein on the stigmatic membrane forms a high-affinity binding site for SP11. Here, we show that the 60-kD stigmatic membrane protein is a truncated form of SRK containing the extracellular domain, transmembrane domain, and part of the juxtamembrane domain. A transiently expressed, membrane-anchored form of SRK exhibits high-affinity binding to SP11, whereas the soluble SRK (eSRK) lacking the transmembrane domain exhibits no high-affinity binding, as is the case with SLG. The different binding affinities of the membrane-anchored SRK and soluble eSRK or SLG will be significant for the specific perception of SP11 by SRK.
INTRODUCTION
Many flowering plants have evolved systems of self-incompatibility (SI) to prevent self-fertilization and to promote outbreeding. In the case of the SI in Brassica, the recognition of self and nonself pollen at the stigmatic surface is genetically controlled by a multiallelic S-locus. The S-locus generally contains three highly polymorphic genes: S-locus protein 11 (SP11; or S-locus Cys-rich protein [SCR]), S-locus glycoprotein (SLG), and S-locus receptor kinase (SRK).
SP11 encodes a secreted form of a small basic protein that localizes to the pollen coat (Takayama et al., 2001; Iwano et al., 2003). The function of SP11 as a determinant of pollen S-haplotype specificity was definitively shown using a transgenic approach and a pollination bioassay (Schopfer et al., 1999; Takayama et al., 2000, 2001; Shiba et al., 2001). SRK encodes a membrane-spanning Ser/Thr receptor kinase that localizes to the plasma membrane of stigmatic papilla cells. Gain-of-function experiments clearly demonstrated that SRK is the sole determinant of stigma S-haplotype specificity (Takasaki et al., 2000; Silva et al., 2001). SLG encodes a secreted glycoprotein that accumulates abundantly (∼100-fold of SRK) in the stigmatic cell wall (Kandasamy et al., 1989; Kishi-Nishizawa et al., 1990). Transgenic experiments demonstrated that SLG is not directly involved in determining the S-haplotype specificity of the stigma (Takasaki et al., 2000; Silva et al., 2001). In one transgenic experiment, transgenic plants of Brassica rapa into which SLG28 (SLG from the S28-haplotype) was cointroduced with SRK28 (SRK from the S28-haplotype) exhibited an enhanced SI response against pollen from S28-haplotype plants, suggesting a supporting role for SLG in the SI response (Takasaki et al., 2000). However, this enhancing role of SLG was not observed when SRK910 and SLG910 genes from the self-incompatible Brassica napus W1 line were cointroduced into the self-compatible B. napus cv Westar line (Silva et al., 2001).
Recent biochemical studies have revealed that SP11 is a direct ligand for SRK. Our previous cross-linking experiments using 125I-labeled S8-SP11 suggested that S8-SP11 bound directly to SRK8 in the stigmatic microsomal membranes (Takayama et al., 2001). Furthermore, in an in vitro phosphorylation assay, S8-SP11 induced the autophosphorylation of SRK8 in an S-haplotype–specific manner. The induction of SRK autophosphorylation was also observed after self-pollination in vivo (Cabrillac et al., 2001). Together, these data suggest that during self-pollination, SP11 localized in the pollen coat spreads to papilla cell plasma membranes and binds the cognate SRK, inducing its autophosphorylation. Therefore, the S-haplotype–specific SP11–SRK interaction and SRK activation are responsible for self-pollen recognition and the subsequent triggering of the signaling cascade that results in self-pollen rejection.
Our previous reports have also suggested that 125I-labeled S8-SP11 binds directly not only to SRK8 but also to a 60-kD protein that is predicted to be a component of a high-affinity (Kd = 0.7 nM) SP11 binding site (Takayama et al., 2001; Takayama and Isogai, 2003). Immunoprecipitation experiments have suggested that the 60-kD protein is an SLG-like protein, most likely SLG8 or an extracellular domain of SRK8 (eSRK8), which was shown to be produced by alternative splicing of SRK (Stein et al., 1991; Giranton et al., 1995; Suzuki et al., 2003). Indeed, eSRK has been detected as a soluble protein in the S3-haplotype of Brassica oleracea (Giranton et al., 1995). Because of the high sequence homology of eSRK with SLG of the same haplotype (75 to 99%) and the fact that many other SLG-like proteins (termed the S-multigene family) are expressed in the stigma (Suzuki et al., 1995), the 60-kD protein has not yet been identified precisely.
Here, we report the purification and characterization of the S8-SP11 high-affinity binding site from stigmatic microsomal membranes. The purified S8-SP11 high-affinity binding site contained both SRK8 and the 60-kD protein, as suggested by our previous cross-linking experiments. Extensive immunological and biochemical characterization revealed that the 60-kD protein is neither SLG8 nor eSRK8 but rather a truncated form of SRK8 (here termed tSRK) containing the extracellular domain, the transmembrane domain, and part of the juxtamembrane domain of SRK8. Transient expression analyses using cultured plant cells suggested that tSRK8 is produced by posttranslational processing of SRK8. Interestingly, although both the artificially expressed membrane-anchored form of SRK8 (mSRK8) and the integral form of SRK8 bound to S8-SP11 with high affinity, the soluble form of SRK8 (eSRK8) exhibited no high-affinity binding to S8-SP11. The artificially dimerized form of eSRK8, however, has been shown to exhibit high-affinity binding, suggesting that the membrane anchorage is necessary for SRK to acquire the high-affinity dimeric configuration. Our findings resemble the interactions between mammalian receptor tyrosine kinases (RTKs) and their ligands, and we discuss the implications of our observations for SI signal transduction.
RESULTS
Affinity Purification of the SP11 High-Affinity Binding Site from Stigmatic Membranes
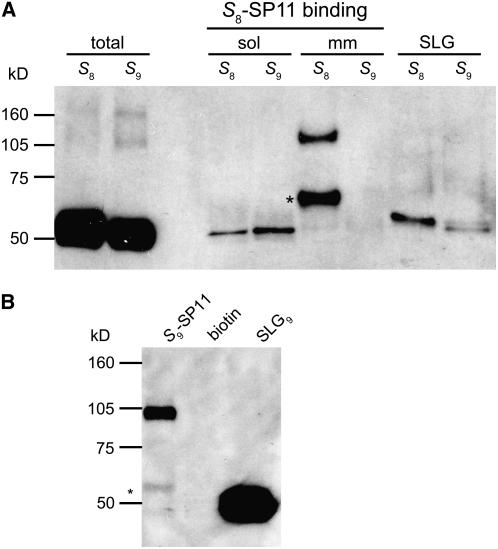
To identify the components of the SP11 high-affinity binding site on the stigmatic membrane, we performed affinity purification using a chemically synthesized, biotin-labeled S8-SP11 (biotin-S8-SP11). The stigma homogenate was incubated with biotin-S8-SP11 and then separated into soluble and microsomal membrane fractions by ultracentrifugation. After detergent solubilization of the microsomal membrane fraction with Triton X-100, biotin-S8-SP11 binding proteins were pulled down from each fraction with streptavidin–Sepharose. The S8-SP11 binding proteins were detected by immunoblotting with an SLG8 antibody (Ab-SLG8-C1) after electrophoretic separation (SDS-PAGE). Ab-SLG8-C1 is a polyclonal antibody raised against an SLG8 C-terminal peptide and detects not only SLG but also SRK from both the S8- and S9-haplotypes (Figure 1A).
Figure 1.
Purification of the SP11 High-Affinity Binding Site from Collected Stigmas.
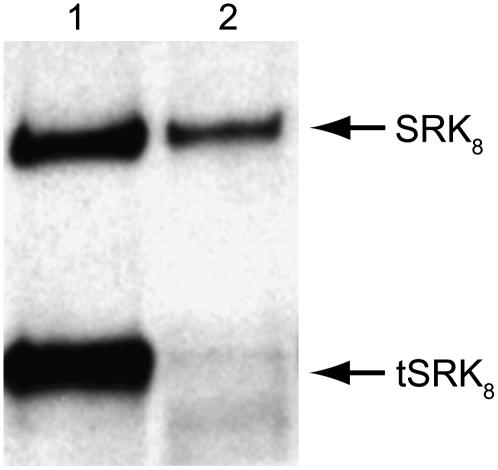
Stigma homogenates from B. rapa S8- and S9-homozygotes were incubated with biotin-S8-SP11 or biotin-S9-SP11 and separated into the soluble and microsomal membrane fractions. The microsomal fraction was detergent-solubilized with 1% Triton X-100. The biotin-SP11 binding proteins were precipitated from each fraction by streptavidin–Sepharose and separated by SDS-PAGE. Asterisks indicate the 60-kD protein bands.
(A) Immunoblot analysis of the biotin-S8-SP11 binding proteins from soluble (sol) and microsomal membrane (mm) fractions (equivalent to ∼1000 stigmas) using an anti-SLG8-C1 polyclonal antibody (Ab-SLG8-C1). Total stigma extracts (equivalent to one stigma; two left lanes) and the purified SLG (100 fmol; two right lanes) were also analyzed as controls.
(B) Immunoblot analysis of the biotin-S9-SP11 binding proteins (S9-SP11) and the biotin binding proteins (biotin; control) from the microsomal membrane fractions of S9-homozygotes. The proteins were detected using Ab-SLG8-C1. Purified SLG9 (1 pmol) was analyzed as a control.
Figure 1 shows the results of the affinity purification of SP11 binding proteins. The total stigma extract contains large amounts of Ab-SLG8-C1–reactive 50- to 60-kD proteins, which produced broad bands after applying the sample equivalent of only one stigma (“total” lanes in Figure 1A). A large percentage of these proteins is SLG, which is a soluble protein contained specifically in the stigma at a high concentration (∼0.2% [w/w] of total protein). The extracellular domain of SRK8 (eSRK8), which is produced by alternative splicing, also must be present in this fraction, although the precise amount is unknown (Giranton et al., 1995; Suzuki et al., 1996). There are many other SLG-like proteins (termed the S-multigene family) also expressed in the stigma (Suzuki et al., 1995, 1997; Kai et al., 2001). However, these soluble proteins exhibit little to no S8-SP11 binding activity, and the affinity-bound fraction showed narrow bands even after applying a sample equivalent to 1000 stigmas, suggesting a <1:10,000 reduction by this single affinity purification step (“sol” lanes in Figure 1A).
By contrast, two Ab-SLG8-C1 antibody-reactive proteins were specifically affinity-purified from the microsomal membrane fraction of S8-haplotype plants (“mm” lanes in Figure 1A). These two proteins, with apparent molecular masses of 110 and 60 kD, were expected to correspond to two high-affinity receptor components detected previously in cross-linking experiments with 125I-labeled S8-SP11 (Takayama et al., 2001). In previous work, we suggested that the 110-kD protein was SRK8 and that the 60-kD protein was SLG8 or eSRK8. In this work, we reconfirmed, by immunoblotting with several peptide antibodies raised against SRK8, that the 110-kD protein is integral SRK8 (see below).
What, then, is the 60-kD protein? Here, we found that the 60-kD protein recovered from the microsomal membrane fraction had a slightly larger molecular mass than SLG (∼55 kD) purified from the soluble stigma fraction (“SLG” lanes in Figure 1A). In our previous cross-linking experiments, the 125I-S8-SP11 cross-linked protein exhibited an ∼65-kD band after SDS-PAGE, but we could not precisely determine its original (non-cross-linked) size. In this experiment, the original S8-SP11 binding protein is clearly ∼5 kD larger than SLG8. Compared with SLG8, eSRK8 has a smaller polypeptide backbone (45.7 kD for eSRK8 and 46 kD for SLG8) and fewer potential glycosylation sites (eight for eSRK8 and nine for SLG8). Therefore, we predicted that the 60-kD protein has a different (and larger) molecular structure than the soluble forms of SLG8 and eSRK8.
To test whether the 60-kD protein is also present in other S-haplotypes, we repeated the pull-down experiment described above with stigmatic membranes from another S9-haplotype of B. rapa. In addition to the integral SRK9, biotin-labeled S9-SP11 precipitated a small amount of 60-kD protein that was larger than SLG9 (Figure 1B). Thus, we consider the presence of the 60-kD protein in the stigmatic membrane to be a general feature, although its relative abundance differs between S-haplotypes.
Identification of the 60-kD Protein
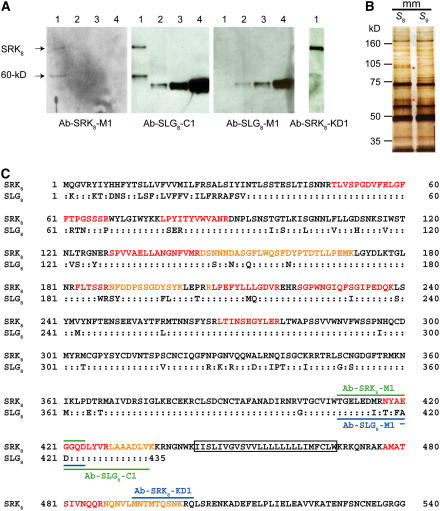
To determine whether the 60-kD protein is an SLG- or SRK-related protein, we immunoblotted with polyclonal antibodies that discriminate between SLG8 and SRK8. Because the sequence similarity between the extracellular domains of SRK and SLG is relatively low in the S8-haplotype (84% amino acid identity), we obtained these antibodies by immunizing rabbits with distinct peptide fragments from SLG8 and SRK8 (Figure 2C).
Figure 2.
Immunochemical Analysis of the 60-kD Protein.
(A) Comparative immunoblot analysis of the S8-SP11 binding proteins. The purified biotin-S8-SP11 binding proteins (lane 1; equivalent to ∼1000 stigmas) and the purified SLG8 (lane 2, 62.5 fmol; lane 3, 125 fmol; lane 4, 500 fmol) were immunoblotted using polyclonal antibodies against SLG8 or SRK8 polypeptides.
(B) Silver staining of the biotin-S8-SP11 binding proteins isolated from the stigmatic microsomal membranes of S8-homozygotes. The asterisks indicate the SRK and 60-kD protein bands, which were excised for LC-MS/MS analysis.
(C) Comparison of amino acid sequences between SLG8 and SRK8. Colons indicate identical amino acid residues. The solid lines above SRK8 and below SLG8 indicate the positions of the peptide sequences used for the antibody production. The tryptic fragments identified by LC-MS/MS analysis of the 60-kD protein are shown in red or orange characters. The putative transmembrane domain of SRK8 is boxed.
Immunoblot analysis was performed using affinity-purified S8-SP11 binding proteins and purified SLG8 as a control. The reactivities of the specific antibodies suggest that the 60-kD protein is not related to SLG8 but rather to SRK8 (Figure 2A). For example, the anti-SRK8-M1 antibody (Ab-SRK8-M1), which does not bind SLG8, clearly detects both SRK8 and the 60-kD protein. By contrast, the anti-SLG8-M1 antibody (Ab-SLG8-M1), which binds strongly to SLG8, detects neither SRK8 (110 kD) nor the 60-kD protein. Collectively, the immunoreactivities of SRK8 and the 60-kD protein always change in parallel according to the antibodies used, strongly suggesting that the 60-kD protein is related to SRK8.
To identify the 60-kD protein directly, we analyzed its proteolytic fragments using nanoscale liquid chromatography–coupled quadrupole time-of-flight tandem mass spectrometry. The affinity-purified 60-kD protein was separated by SDS-PAGE and excised after visualization by silver staining (Figure 2B). The protein bands collected from 40,000 stigmas were in-gel digested with trypsin, and the obtained tryptic peptides were subjected to liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis. Fourteen tryptic peptides were identified as fragments of the extracellular domain of SRK8 from acquired MS/MS spectra based on the MASCOT and ProteinLynx 2.0 searching algorithms (Table 1). Although 2 of these fragments were common to both SLG8 and SRK8, the other 12 fragments were specific to SRK8 (Figure 2C).
Table 1.
Tryptic Peptides of the 60-kD Protein Identified by LC-MS/MS
| Observed Mass | Charge State | Experimental Massa | Calculated Massb | Peptide Sequences |
|---|---|---|---|---|
| 1100.53 | 2 | 2199.04 | 2199.08 | TLVSPGDVFELGFFTPGSSSR |
| 734.02 | 3 | 2199.05 | 2199.08 | TLVSPGDVFELGFFTPGSSSR |
| 747.91 | 2 | 1493.80 | 1493.80 | LPYITYVWVANR |
| 866.96 | 2 | 1731.90 | 1731.90 | SPVVAELLANGNFVMR + oxidation (M) |
| 710.39 | 1 | 709.38 | 709.38 | FLTSSR |
| 747.80 | 2 | 1493.58 | 1493.59 | NFDDPSSGDYSYK |
| 1035.79 | 3 | 3104.34 | 3104.37 | DSNNNDASGFLWQSFDYPTDTLLPEMK |
| 795.94 | 2 | 1589.87 | 1589.89 | RLPEFYLLLGDVR |
| 717.90 | 2 | 1433.78 | 1433.79 | LPEFYLLLGDVR |
| 930.43 | 2 | 1858.88 | 1858.85 | SGPWNGIQFSGIPEDQK |
| 647.85 | 2 | 1293.68 | 1293.66 | LTINSEGYLER |
| 692.84 | 2 | 1383.67 | 1383.64 | NYAEGGQDLYVR |
| 400.75 | 2 | 799.49 | 799.48 | LAAADLVK |
| 617.81 | 2 | 1233.61 | 1233.61 | AMATSIVNQQR + oxidation (M) |
| 811.88 | 2 | 1621.75 | 1621.76 | NQNVLMNTMTQSNK |
Calculated relative molecular mass of the matched peptide.
Experimental m/z converted to relative molecular mass.
Among the 14 detected tryptic peptide fragments, the ion intensity of NQNVLMNTMTQSNK (N488 to K501), a juxtamembrane domain peptide from the 60-kD protein, was lower (approximately one-tenth) than that derived from the 110-kD protein (SRK8). This observation suggests that the C terminus of the 60-kD protein is heterogeneous and that some extends beyond this juxtamembrane domain, but for the most part it terminates before the downstream end (K501) of this tryptic peptide. The fact that the adjacent upstream tryptic peptide fragment, AMATSIVNQQR (A477 to R487), was detected at similar intensities in both SRK8 and the 60-kD protein suggests that the majority of the 60-kD protein terminates within the region spanning N488 to K501. In support of this idea, the peptide antibody Ab-SRK8-KD1, raised against the peptide M493 to Q503 (Figure 2C), strongly detected SRK8 but only faintly stained the 60-kD protein (Figure 2A). The faint staining of the 60-kD protein band with the Ab-SRK8-KD1 antibody must be attributable to the fact that the proposed C termini of these forms lie within the N488 to K501 region, which largely overlaps the SRK8-KD1 epitope; therefore, some of the shorter species may lack the epitope required for recognition by the Ab-SRK8-KD1 antibody. The expected shorter C-terminal peptide fragment(s), however, was not detected by LC-MS/MS analyses. This is probably attributable to the low ionization efficiencies of peptides lacking the C-terminal Lys or Arg residue.
The presence of a membrane-anchored, truncated form of SRK was also suggested by protein gel blotting in the S3-haplotype of B. oleracea (Giranton et al., 1995). The amount of the truncated form of SRK3 was much lower than that of integral SRK3, similar to our observations with the S9-haplotype of B. rapa (Figure 1B). Thus, the occurrence of truncated forms seems to be a common feature of SRKs, although the relative abundance differs between S-haplotypes.
The Membrane-Anchored Form of Truncated SRK Is Sufficient for SP11 High-Affinity Binding
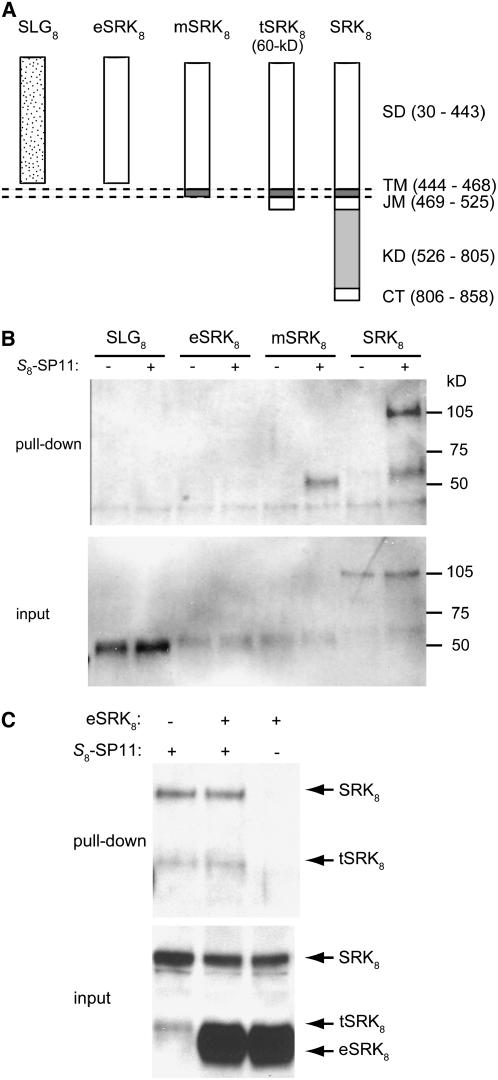
We investigated whether the membrane-anchored form of SRK8 is active solely in S8-SP11 high-affinity binding. To test this notion, we transiently expressed three forms of SRK8 in tobacco (Nicotiana tabacum) BY-2 cell protoplasts using a tomato mosaic virus expression system. These forms were eSRK8 (the extracellular domain alone), mSRK8 (the extracellular and transmembrane domains), and full-length SRK8. SLG8 was also expressed in the same system as a control (Figure 3A).
Figure 3.
SP11 Binding Activities of SLG and SRK Derivatives Expressed in Tobacco Cells.
(A) Schemes of the structures of mature SLG8 (SLG8), the extracellular domain of SRK8 (eSRK8), the membrane-anchored form of SRK8 (mSRK8), the truncated form of SRK8 (tSRK8 = 60-kD protein), and full-length SRK8 (SRK8). SD, extracellular domain (= S domain); TM, transmembrane domain; JM, juxtamembrane domain; KD, kinase domain; CT, C-terminal domain. The numbers represent amino acid positions relative to the initiating Met.
(B) SLG8, eSRK8, mSRK8, and full-length SRK8 were transiently expressed in tobacco BY-2 cell protoplasts. Biotin-S8-SP11 (+) or biotin (−) was incubated with the cell extracts, and the interacting proteins were recovered using streptavidin–Sepharose (see Methods). The recovered proteins (pull-down) and the BY-2 cell extracts before precipitation (input) were separated by SDS-PAGE and probed by immunoblotting with the antibody Ab-SLG8-C1.
(C) Pull-down assays in the presence of both full-length SRK8 and eSRK8. The microsomal membrane fraction of BY-2 cells expressing SRK8 was incubated with biotin-S8-SP11 (+) or biotin (−) in the presence of large quantities of soluble eSRK8. Interacting proteins (pull-down) and the proteins before precipitation (input) were immunoblotted with the antibody Ab-SLG8-C1.
In the BY-2 cell protoplasts, SLG8 and eSRK8 were expressed as soluble proteins and recovered from the culture medium, whereas SRK8 and most of mSRK8 were expressed as membrane-bound proteins and recovered from the microsomal membrane fractions. The binding affinities of these proteins for S8-SP11 were tested using in vitro pull-down assays similar to the purification of the high-affinity S8-SP11 binding site. The pull-down assays demonstrated that not only SRK8, but also mSRK8, has high-affinity S8-SP11 binding activity, whereas no binding activity was detected in the soluble forms of SLG8 and eSRK8 (Figure 3B). The binding ability of mSRK8 indicated that the cytoplasmic kinase domain of SRK8 is not essential for the binding of S8-SP11.
During the transient expression analyses, we noticed that the cells expressing full-length SRK8 also produced the truncated form of SRK8 in the microsomal membrane fraction and that this truncated SRK8 was slightly larger than mSRK8 (Figure 3B). Because we used the virus vector containing the full-length SRK8 cDNA, this truncated SRK8 must be posttranslationally generated from the integral SRK8. Because of this posttranslational processing, the pull-down assay using the cells expressing only full-length SRK8 resulted in patterns similar (110- and 60-kD bands) to those from stigmas. These observations suggested the possibility that stigmatic tSRK8 (the 60-kD protein) is also posttranslationally and probably proteolytically generated from SRK8.
Contrary to our previous predictions, the soluble form of eSRK8 displayed no detectable binding to S8-SP11, as was the case with SLG8. To confirm this result, we repeated the pull-down assay in the presence of both full-length SRK8 and eSRK8. S8-SP11 selectively bound to full-length SRK8 and its processing product tSRK8, but no binding was detected with eSRK8 (Figure 3C). These results strongly suggested that membrane anchorage is critical for the high-affinity binding of SRK to SP11.
Membrane-Anchored SLG Exhibits No High-Affinity Binding to SP11
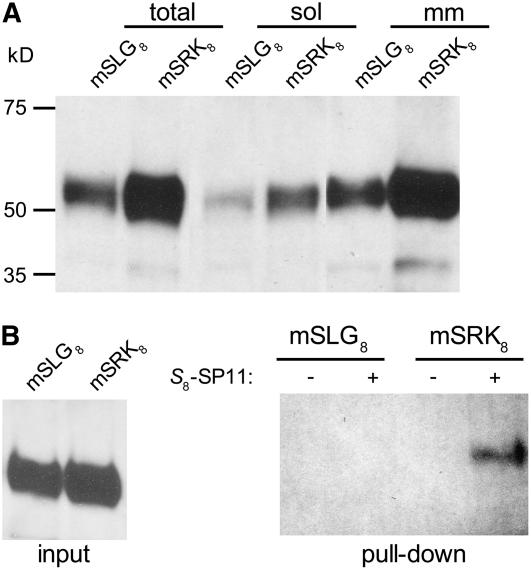
SLG8 in the stigma soluble fraction exhibited no high-affinity binding to S8-SP11. Previous interaction analyses using surface plasmon resonance also failed to detect an interaction between S8-SP11 and SLG8 (Takayama et al., 2001). However, because SLG shares significant homology with the extracellular domain of cognate SRK, SLG might potentially interact with SP11 under certain circumstances. To test this notion, we transiently expressed a membrane-anchored form of SLG8 (mSLG8), a chimeric protein consisting of SLG8 and the transmembrane domain of SRK8. Immunoblotting confirmed that the majority of mSLG8 and mSRK8 was expressed as similarly sized microsomal membrane proteins, although small portions shed from the membrane were also detected in the soluble fraction of the cell extract (Figure 4A). The microsomal membrane fraction was subjected to the biotin-S8-SP11 pull-down assay. Figure 4B illustrates that mSLG8, in contrast with mSRK8, does not acquire high-affinity binding activity with S8-SP11, despite its anchorage in the plasma membrane.
Figure 4.
SP11 Binding Activities of the Membrane-Anchored Forms of SLG and SRK.
(A) Expression of the membrane-anchored forms of SLG8 (mSLG8) and SRK8 (mSRK8). Whole protein extracts (total), soluble fractions (sol), and microsomal membrane fractions (mm) of mSLG8- or mSRK8-expressing BY-2 cells were analyzed by immunoblotting with the antibody Ab-SLG8-C1.
(B) The microsomal fractions of mSLG8- and mSRK8-expressing BY-2 cells were incubated with biotin-S8-SP11 (+) or biotin (−) and pulled down using streptavidin–Sepharose. Interacting proteins (pull-down) and the proteins before precipitation (input) were analyzed by immunoblotting with the antibody Ab-SLG8-C1.
Membrane Anchorage Is Necessary for Truncated SRK to Establish High-Affinity Binding to SP11
To better understand how the membrane anchorage of SRK facilitates high-affinity SP11 binding, we performed pull-down assays before and after solubilizing the stigmatic microsomal membranes. Before solubilization of the microsomal membranes with Triton X-100, both full-length SRK8 and the truncated tSRK8 exhibited high-affinity binding to S8-SP11 (Figure 5). However, most of the high-affinity binding activity of tSRK8 was lost after solubilization of the microsomal membrane. This observation suggested that the membrane anchorage is necessary for tSRK to establish high-affinity binding with S8-SP11. Nevertheless, the integral SRK8, at least in part, maintained its high-affinity binding activity even after solubilization of the microsomal membrane.
Figure 5.
SP11 Binding Activities of the Solubilized Stigmatic SRK.
Biotin-S8-SP11 was incubated with the stigmatic microsomal membranes of S8-homozygotes before (lane 1) or after (lane 2) solubilization of the membranes with Triton X-100. The biotin-S8-SP11–interacting proteins were analyzed by immunoblotting with the antibody Ab-SLG8-C1.
A Dimeric Form of eSRK Exhibits High-Affinity Binding to SP11
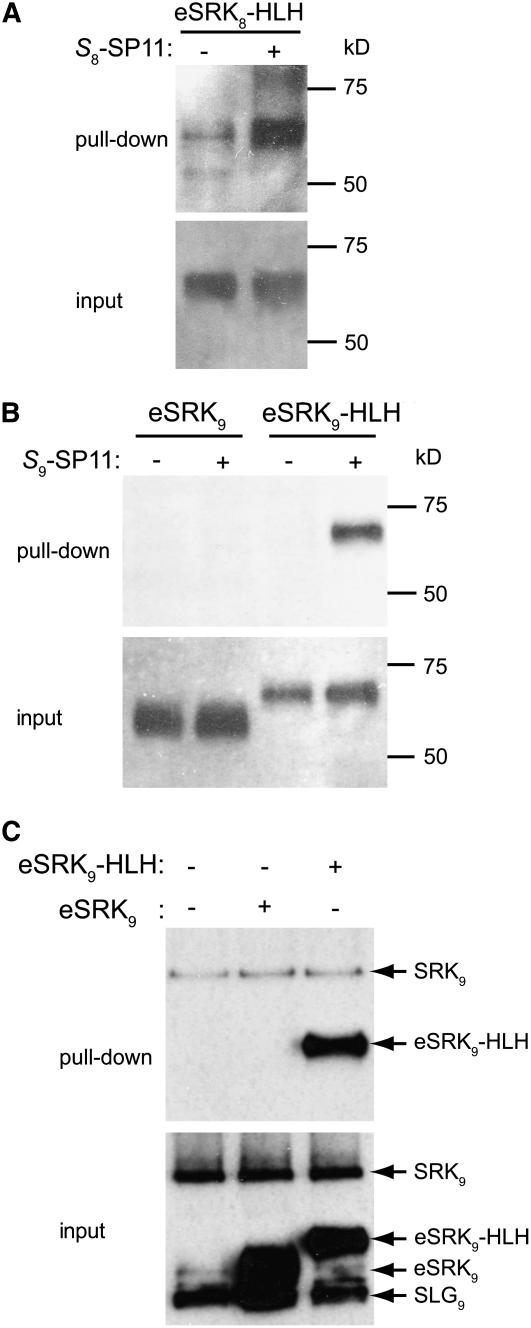
Previous cross-linking experiments have shown that SRK exists as a dimer on the stigmatic membrane of nonpollinated flowers (Giranton et al., 2000). Velocity sedimentation analytical ultracentrifugation also suggested that the integral form of stigmatic SRK exists as a dimer even after detergent solubilization of the plasma membrane, whereas SLG and eSRK exist primarily as monomers (Giranton et al., 2000). These data suggested that the high-affinity SP11 binding activity of SRK can be attributed to its dimeric condition. To test this hypothesis, we made a construct for the eSRK8-HLH-ZIP fusion protein that spontaneously dimerizes via an introduced C-terminal helix-loop-helix zipper (HLH-ZIP) domain from sterol-regulatory element binding protein 2 (Nagoshi and Yoneda, 2001). The eSRK8-HLH-ZIP fusion protein transiently expressed in BY-2 cells exhibited high-affinity binding to S8-SP11 (Figure 6A). To confirm this result in another S-haplotype, we expressed both eSRK9 and eSRK9-HLH-ZIP proteins in the stably transformed BY-2 cells. In contrast with eSRK9, which showed no high-affinity binding, the eSRK9-HLH-ZIP fusion protein exhibited high-affinity binding to S9-SP11 (Figure 6B). Furthermore, when these binding experiments were conducted in the presence of stigmatic membranes from S9-homozygotes, the eSRK9-HLH-ZIP fusion protein, but not eSRK9, exhibited high-affinity binding comparable to that of integral SRK9 on the stigmatic membrane (Figure 6C). These results suggest that dimerization can be a crucial event preceding the acquisition of high-affinity binding of SRK to SP11.
Figure 6.
SP11 Binding Activities of the Artificially Dimerized eSRK.
(A) The HLH-ZIP–fused extracellular domain of eSRK8 (eSRK8-HLH-ZIP) was incubated with biotin-S8-SP11 (+) or biotin (−). Interacting proteins (pull-down) and the proteins before precipitation (input) were analyzed by immunoblotting with the antibody Ab-SLG8-C1.
(B) The binding of eSRK9 and eSRK9-HLH-ZIP to biotin-S9-SP11 was analyzed as in (A).
(C) The stigmatic microsomal membrane fraction of S9-homozygotes was incubated with biotin-S9-SP11 in the presence of soluble eSRK9 or eSRK9-HLH-ZIP. Binding was analyzed as in (A).
DISCUSSION
In this study, we purified the high-affinity SP11 binding site on the stigmatic membrane using biotin-tagged SP11. The high-affinity binding site was suggested to exist at only 180 fmol/mg stigmatic microsomal membrane proteins, corresponding to ∼1 fmol/stigma (Takayama et al., 2001; Takayama and Isogai, 2003). The purified high-affinity binding site consisted of two proteins of 110 and 60 kD, as suggested previously by cross-linking experiment using radiolabeled SP11. Thorough immunochemical and LC-MS/MS analyses revealed that the 110-kD protein is integral SRK and that the 60-kD protein is neither SLG nor eSRK but rather the membrane-anchored form of tSRK. The tSRK is ∼5 kD larger than SLG or eSRK, which we had not noticed in our previous study (Takayama et al., 2001) because we only detected the protein band after cross-linking with 125I-labeled-SP11 and thus could not determine its original molecular size.
Another unexpected finding was that in contrast to the integral and membrane-anchored forms of SRK, which exhibited high-affinity binding to SP11, the soluble form of SRK (eSRK) exhibited no high-affinity binding. This finding suggests that the membrane anchorage is necessary for SRK to create a high-affinity binding site for SP11. This conclusion was further supported by the observation that the membrane-anchored form of tSRK lost its high-affinity binding activity after solubilizing the microsomal membrane with detergent, whereas the integral form of SRK retained its binding activity to a certain extent (see below for further discussion).
The pull-down method used in this study was designed to detect only high-affinity, stabilized interactions between SP11 and its binding site. We used nanomolar levels of biotin-SP11 to detect the interaction, and no chemical cross-linkers were used before membrane solubilization, during ultracentrifugation, or in the subsequent high-salt washes. Most mammalian RTKs, such as epidermal growth factor receptor and fibroblast growth factor receptor, are thought to exist in an autoinhibited state before stimulation by their ligands (Schlessinger, 2003). These autoinhibited RTKs exist in monomeric or inactive dimeric states and are in equilibrium with the small population of unoccupied receptors that are in an active dimeric configuration compatible with trans-autophosphorylation. The latter bind to ligands with high affinity, which shifts the equilibrium toward the stabilized dimeric active configuration, resulting in autophosphorylation (in trans) of Tyr residues in the catalytic core and kinase activation (Jiang and Hunter, 1999; Schlessinger, 2000). We have shown previously (Takayama et al., 2001) that SP11 interacts with its cognate SRK on the stigmatic membrane in an S-haplotype–specific manner. The SP11-interacting SRK is recovered from the plasma membrane in its activated form, which exhibits increased autophosphorylating activity. These results strongly suggest that SP11 also induces SRK dimerization or some rearrangement of a preformed SRK dimer and that the conformational change stabilizes the SP11–SRK interaction and the subsequent SRK activation.
In the context of this model, the membrane anchorage is necessary for SRK to maintain its configuration in the equilibrium state between the inactive monomeric or dimeric low-affinity form(s) and the dimeric active high-affinity form. Formation of the active dimer must be supported by the cell membrane, which restricts receptor diffusion in two dimensions (Tzahar et al., 1997). These arguments are strongly supported by the observation that the inactive soluble form of eSRK also acquired high-affinity binding activity by artificial dimerization via a C-terminal HLH-ZIP domain. Similar results were reported in mammalian transmembrane receptors, in which the soluble forms of extracellular domains often exhibited ligand binding affinities one order of magnitude lower than their integral forms (Nagao et al., 1992; Yet and Jones, 1993; Zhou et al., 1993). Structural studies have suggested that the soluble forms exist as inactive monomers whose dimerization is autoinhibited by intramolecular interactions (Schlessinger, 2002). It is also suggested that these inactive soluble receptors acquire high-affinity configurations by increasing their local densities, such as by increasing receptor concentrations, by chemically cross-linking monomeric receptors, or by providing membrane anchorage to the soluble receptors (Böni-Schnetzler and Pilch, 1987; Hurwitz et al., 1991; Zhou et al., 1993).
The oligomeric status of SRK has actually been observed in planta by cross-linking and velocity sedimentation experiments (Giranton et al., 2000). These data strongly suggested that SRK molecules do indeed exist as dimers or oligomers on nonpollinated stigmatic membranes, whereas SLG and eSRK exist primarily as monomers. It was also suggested that the integral form of stigmatic SRK retained its oligomeric status even after membrane solubilization by Triton X-100 but was resolved to a monomer after SDS treatment in the absence of a reducing agent (Giranton et al., 2000). These results are consistent with our observation that the integral form of SRK, at least in part, retained high-affinity binding activity for SP11 even after membrane solubilization, whereas most solubilized tSRK lost its high-affinity binding activity. These data suggest that most SRK on the nonpollinated stigmatic membrane spontaneously and noncovalently associates to form inactive dimers, which must still be an autoinhibited configuration. The kinase domain of SRK contributes to stabilizing the dimeric form, although the precise molecular mechanisms are unknown. The SP11 binding on these preformed dimers is supposed to induce specific, intersubunit conformational changes within a dimer, leading to a stabilized dimeric configuration and full activation, as was reported for RTKs (Jiang and Hunter, 1999). Despite the central importance of the receptor kinases in plant signal transduction, most of which are Ser/Thr kinases, unlike mammalian RTKs, their ligand perception and their activation mechanisms are poorly understood at the molecular level. In this study, we present an example of a plant receptor-like kinase being activated through a homodimerization process.
The actual SP11 binding affinities of eSRK and SLG remain unclear. We assumed that eSRK had a low binding affinity for SP11, although it could not be measured using our current assay system. Nasrallah's group demonstrated S-haplotype–specific interactions between eSRK6 (or eSRK13) and the cognate SCR/SP11s using recombinant proteins (Kachroo et al., 2001; Chookajorn et al., 2004). They measured interactions by ELISA or pull-down assay using eSRKs anchored to plates or resin. Although exact values are unknown because most of the binding data were not described by Kd but rather by binding maximum values, the binding affinities were assumed to be low based on the experimental SP11 concentrations. In addition, because they immobilized huge amounts of eSRK (0.5 to 0.75 μg) on wells or in the resin, the local densities of eSRKs were expected to be rather high—possibly high enough for the spontaneous oligomerization of the eSRKs. In our experiments, in which SP11 and eSRK were mixed in solution at levels <50 nM, no such interactions were detected. We show here that the artificially dimerized form of eSRK exhibited high-affinity binding to SP11 in solution. The use of this form of eSRK would help in investigations of the structural basis of SP11–SRK interactions (e.g., to assess the residues that are important for the interaction and to map sequences that determine recognition specificity between different S-haplotypes). Our findings also have potential implications for future studies on other plant receptor kinases, including the design of experimental strategies for ligand searches for the orphan receptor kinases. Affinity purification of the ligand using the extracellular domains of receptor kinases is expected to be unsuccessful in experiments with SRK-like receptor kinases.
In stigmatic papilla cells, SLG accumulates abundantly in the cell wall, where eSRK is also present (Giranton et al., 1995). The quantity of SRK on the plasma membrane is approximately two orders of magnitude lower than that of these soluble proteins (Giranton et al., 1995; Kemp and Doughty, 2003). When the pollen grain lands on a papilla cell, SP11 from the pollen coat must pass through the SLG/eSRK-rich cell wall space to reach SRK on the plasma membrane to initiate SI signaling. Thus, it seems reasonable that SLG/eSRK would bind SP11 with lower affinity. What, then, are the biological functions of SLG and eSRK? Why are the SLG alleles distributed across so many Brassica S-haplotypes? Why are the splice site and the in-frame stop codon at the 5′ end of the first intron used to produce eSRK so well conserved in alleles of SRK (Giranton et al., 1995; Suzuki et al., 1996)? Plants are known to produce many soluble receptor-like proteins that are highly similar in sequence to the extracellular domains of their corresponding receptor kinases (Torii, 2000). These truncated forms are produced either from the same gene by alternative splicing, as in the case of eSRK, or from different genes, as in the case of SLG. In either case, the physiological significance of the soluble forms of receptor kinases is virtually unknown. One emerging hypothesis suggests that these soluble receptor-like proteins form heterodimers with corresponding receptor kinases in the presence of ligands, triggering signaling cascades within the cell. However, our previous prediction that SLG and/or eSRK would be involved in SP11 perception together with SRK was not borne out by this study. The biological functions of SLG and eSRK should be reexamined in future studies.
We revealed that the 60-kD stigma protein is a membrane-anchored form of tSRK that contains the extracellular domain, the transmembrane domain, and the short intracellular (juxtamembrane) region but that lacks the kinase domain. In this study, we demonstrated the presence of tSRKs in two S-haplotypes (S8- and S9-haplotypes) of B. rapa. The presence of tSRK was also suggested in the S3-haplotype of B. oleracea (Giranton et al., 1995), suggesting that the occurrence of the tSRK might be a general feature of SRKs. Because the corresponding protein was also produced in tobacco cells transformed with SRK cDNA, tSRK must be posttranslationally produced from integral SRK, probably by a protease that is present in both Brassica and tobacco cells. A similar proteolytic cleavage has been reported in XA21, a rice (Oryza sativa) receptor kinase that confers resistance against the phytopathogenic bacterium Xanthomonas oryzae pv oryzae (Xu et al., 2006). Although the precise cleavage site and the protease involved have not been determined, XA21 was shown to be proteolytically cleaved within the intracellular juxtamembrane domain by developmentally controlled proteolytic activity.
It remains unknown whether tSRK has a special function in SI signaling. In mammalian RTKs, certain truncated forms lacking the kinase domain play critical roles in signal transduction. For example, certain truncated neurotrophin receptor kinases of TrkB and TrkC have been shown to form dimers with, and thereby inhibit the activation of, the integral forms of these receptors (Biffo et al., 1995). On the other hand, one of these truncated Trks, TrkB-T1, has a specialized positive function that activates calcium release from intracellular stores by ligand binding in astroglial cells (Rose et al., 2003). To determine the physiological function of tSRK, we must design experiments in which the production of tSRK in the papilla cell can be artificially regulated. Determining the precise C-terminal tSRK cleavage site and identifying the protease involved in the cleavage will shed light on the molecular function of tSRK.
METHODS
Synthesis of Biotin-Labeled SP11
S8-SP11 and S9-SP11 were chemically synthesized by the Peptide Institute as described previously (Takayama et al., 2001). Biotin-S8-SP11 was chemically synthesized by assembling the biotinylated pentapeptide (biotinyl-QELEA) at the N terminus of S8-SP11. Biotin-S9-SP11 was prepared by chemically labeling S9-SP11 (17 nmol) with biotin-(AC5)2sulfo-OSu (20 nmol; Dojin) at room temperature for 90 min. Biotin-labeled SP11s were purified to a single peak by reverse-phase HPLC.
Purification of the High-Affinity SP11 Binding Site from Stigma
Stigmas were collected from S8- and S9-homozygotes of Brassica rapa on the day of anthesis and stored at −80°C until use. The stigmas were homogenized in binding buffer (50 mM HEPES-KOH, pH 7.4, 100 mM NaCl, 5 mM MgCl2, 2 mM MnCl2, 10 mM NaF, 10 mM β-glycerophosphate, 0.005% Triton X-100, 1 mM DTT, and EDTA-free protease inhibitor cocktail [Roche]). The homogenates were treated with 10 nM biotin-S8-SP11, biotin-S9-SP11, or biotin (control) for 60 min on ice and then separated into supernatants and microsomal membranes by ultracentrifugation (120,000g for 60 min at 4°C). The microsomal membranes were solubilized with 1% Triton X-100, and the extracts were collected by ultracentrifugation at 120,000g for 30 min at 4°C. The biotin-S8-SP11 binding proteins were recovered by streptavidin–Sepharose (Amersham Biosciences). After washing the resin three times with binding buffer and three times with wash buffer (50 mM HEPES-KOH, pH 7.4, 300 mM NaCl, 5 mM MgCl2, 2 mM MnCl2, 10 mM NaF, 10 mM β-glycerophosphate, 0.005% Triton X-100, and 1 mM DTT), the biotin-SP11–bound proteins were eluted by the addition of SDS-PAGE sample buffer (50 mM Tris-HCl, pH 6.8, 10% [v/v] glycerol, 2% [w/v] lithium dodecyl sulfate, and 10 mM DTT) and boiling. The eluted proteins were separated by SDS-PAGE (4 to 12% acrylamide gel) and visualized using a Silver Staining II kit (Wako).
Antibody Production and Immunoblotting
Polyclonal antibodies against SRK8 and SLG8 were generated in rabbits by immunization with the keyhole limpet hemocyanin–conjugated peptides (SRK8-M1, CTGELEDMRNYAEGGQD; SRK8-KD1, CMNTMTQSNKRQ; SLG8-M1, CTGELEDIRTYFADGQD; SLG8-C1, CADGQDLYVRLAAADLV). The IgG fractions of the obtained antisera were affinity-purified using the peptide-conjugated resin. For immunoblotting, the proteins were separated by SDS-PAGE (4 to 12% acrylamide) and transferred to polyvinylidene difluoride membranes. Blots were incubated with the affinity-purified antibodies and then developed with horseradish peroxidase–labeled anti-rabbit IgG and the ECL Plus chemiluminescence detection system (Amersham).
Identification of the SP11 Binding Protein by LC-MS/MS Analysis
The affinity-purified stigmatic proteins were separated by SDS-PAGE (4 to 12% acrylamide) and stained by MS-compatible silver staining (Shevchenko et al., 1996). The protein band (60-kD range) was manually excised and subjected to in-gel trypsin digestion (Shevchenko et al., 1996; Wilm and Mann, 1996). The extracted tryptic peptides were concentrated by vacuum centrifugation and analyzed by LC-MS/MS.
LC-MS/MS analyses were performed on a nanoESI-Qq-TOF Ultima mass spectrometer (Waters-Micromass) interfaced on-line with a capillary HPLC system (Waters-Micromass Modular CapLC) using a reverse-phase trapping column (PepMap microcolumn containing 5 μm of C18 100-Å PepMap, 0.3 mm inner diameter × 1 mm; LC Packings) and a reverse-phase capillary column (NanoEase capillary column containing Atlantis dC18 3 μm 100-Å, 75 μm inner diameter × 150 mm; Waters). The top two ions in each survey scan were subjected to automatic MS/MS fragmentation analysis in low-energy collision-induced dissociation. The acquired MS/MS spectra were searched against the National Center for Biotechnology Information database using the Mascot 2.0 searching algorithm (Matrix Science) or ProteinLynx Global Server 2.0 software (Waters).
Transient Expression of SRK/SLG-Related Proteins in Tobacco BY-2 Cell Protoplasts
The integral forms of SRK8, SLG8, and their derivatives were produced in the protoplasts of tobacco (Nicotiana tabacum) BY-2 suspension cultured cells using a tomato mosaic tobamovirus system (Hori and Watanabe, 2003). The protoplasts of BY-2 cells were prepared as described (Watanabe et al., 1987). The cDNA fragments corresponding to eSRK8 (amino acids 1 to 443), mSRK8 (amino acids 1 to 468), and full-length SRK8 (amino acids 1 to 858) were amplified from an SRK8 cDNA clone by PCR and cloned into the tomato mosaic tobamovirus–derived vector TogJ, which is a modified TocJ vector (Hori and Watanabe, 2003). The chimeric mSLG8 construct was created by combining the full-length SLG8 coding region and the SRK8 transmembrane domain (amino acids 436 to 468) coding region. The HLH-ZIP fusion eSRK8 (eSRK8-HLH-ZIP) was prepared by fusing the HLH-ZIP (amino acids 343 to 403) coding region of SREBP-2 (Nagoshi and Yoneda, 2001) in-frame to an eSRK8 (amino acids 1 to 443) cDNA fragment. The combined fragments of mSLG8 and eSRK8-HLH-ZIP were also cloned into TogJ. In vitro transcripts were produced using a T7 kit (Promega) and introduced into the protoplasts by electroporation. The protoplasts were harvested after culturing for 15 h at 28°C and homogenized with a pestle in binding buffer. The homogenates were centrifuged for 1 h at 120,000g, and the supernatants were used as the soluble fraction. The pellets were resuspended in binding buffer and used as the microsomal fraction. Biotin-S8-SP11 (25 nM) was added to these fractions and incubated for 60 min on ice. The interacting proteins were recovered by streptavidin–Sepharose from the soluble fraction, and the microsomal fraction was solubilized with 1% Triton X-100.
Stable Expression of eSRK9 and eSRK9-HLH-ZIP Proteins in BY-2 Cultured Cells
The extracellular domain of SRK9 (eSRK9) and the eSRK9-HLH-ZIP fusion protein were stably expressed in BY-2 suspension cultured cells. The cDNA fragment corresponding to eSRK9 (amino acids 1 to 436) was amplified from the SRK9 cDNA clone by PCR and fused with cDNA encoding the FLAG epitope tag. The HLH-ZIP fusion eSRK9 (eSRK9-HLH-ZIP) was generated by sequentially annealing the eSRK9 (amino acids 1 to 436) open reading frame, the HLH-ZIP domain (amino acids 343 to 403) coding sequence, and the FLAG epitope tag. For greater protein accumulation, the coding sequence of the C-terminal propeptide (amino acids 335 to 353) of horseradish peroxidase C1a was fused to the eSRK9-FLAG and the eSRK9-HLH-ZIP-FLAG constructs. The C-terminal propeptide is responsible for the protein sorting to vacuoles and is removed before or concomitant with production of the mature protein (Lerouge et al., 1998; Matsui et al., 2003). The obtained constructs were subcloned into the binary vector pMSH1 under the control of the cauliflower mosaic virus 35S promoter and the 5′ untranslated region of the tobacco alcohol dehydrogenase gene. These binary vectors were introduced into BY-2 tobacco cells by Agrobacterium tumefaciens–mediated transformation (Nakayama et al., 2000). The transformed BY-2 cells were homogenized in extraction buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM CaCl2, 0.05% Tween 20, 1 mM DTT, and EDTA-free protease inhibitor cocktail [Roche]), and the recombinant proteins were purified from the supernatants using an anti-FLAG M2 affinity gel (Sigma-Aldrich). The recombinant proteins were eluted with 100 μg/mL FLAG peptide, dialyzed with binding buffer, and concentrated using Centricon YM30 concentrators (Pierce). These purified fusion proteins were incubated with biotin-S9-SP11 (25 nM) for 60 min on ice, and the interacting proteins were recovered by streptavidin–Sepharose. For binding analysis in the presence of stigmatic membranes from S9-homozygotes, the concentration of biotin-S9-SP11 was changed to 10 nM and incubated for 90 min on ice.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AB035504 (S8-SP11), AB022078 (S9-SP11), D84468 (SLG8), D88192 (SLG9), D38563 (SRK8), and D88193 (SRK9).
Acknowledgments
We thank Yuichiro Watanabe and Koichi Hori (University of Tokyo) for providing the viral TogJ vector and valuable technical advice. We thank Yoshihiro Yoneda (Osaka University) and Ryuichiro Sato (University of Tokyo) for providing the SREBP-2 clone. We thank Kazuya Yoshida and Takeshi Matsui (Nara Institute of Science and Technology) for providing the C-terminal propeptide–encoding pMSH1 vector. We thank Aya Ota, Hanae Sugita, Hitomi Ichikawa, Yoshie Ohnishi, and Humiyo Yamamoto for technical assistance. This work was supported in part by Grants-in-Aid for Creative Scientific Research (Grants 16GS0316 to A.I. and 13GS0023 to S.T.) and for the 21st Century Centers of Excellence Program to the Nara Institute of Science and Technology and Iwate University from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by Grants-in-Aid for Scientific Research (B) (Grants 16380072 and 18380069 to S.T. and 17380001 to M.W.) and for JSPS Fellows (to H.S.) from the Japan Society for the Promotion of Science.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Seiji Takayama (takayama@bs.naist.jp).
References
- Biffo, S., Offenhauser, N., Carter, B.D., and Barde, Y.A. (1995). Selective binding and internalisation by truncated receptors restrict the availability of BDNF during development. Development 121 2461–2470. [DOI] [PubMed] [Google Scholar]
- Böni-Schnetzler, M., and Pilch, P.F. (1987). Mechanism of epidermal growth factor receptor autophosphorylation and high-affinity binding. Proc. Natl. Acad. Sci. USA 84 7832–7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrillac, D., Cock, J.M., Dumas, C., and Gaude, T. (2001). The S-locus receptor kinase is inhibited by thioredoxins and activated by pollen coat proteins. Nature 410 220–223. [DOI] [PubMed] [Google Scholar]
- Chookajorn, T., Kachroo, A., Ripoll, D.R., Clark, A.G., and Nasrallah, J.B. (2004). Specificity determinants and diversification of the Brassica self-incompatibility pollen ligand. Proc. Natl. Acad. Sci. USA 101 911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giranton, J.L., Ariza, M.J., Dumas, C., Cock, J.M., and Gaude, T. (1995). The S locus receptor kinase gene encodes a soluble glycoprotein corresponding to the SKR extracellular domain in Brassica oleracea. Plant J. 8 827–834. [DOI] [PubMed] [Google Scholar]
- Giranton, J.L., Dumas, C., Cock, J.M., and Gaude, T. (2000). The integral membrane S-locus receptor kinase of Brassica has serine/threonine kinase activity in a membranous environment and spontaneously forms oligomers in planta. Proc. Natl. Acad. Sci. USA 97 3759–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori, K., and Watanabe, Y. (2003). Construction of a tobamovirus vector that can systemically spread and express foreign gene products in solanaceous plants. Plant Biotechnol. 20 129–136. [Google Scholar]
- Hurwitz, D.R., Emanuel, S.L., Nathan, M.H., Sarver, N., Ullrich, A., Felder, S., Lax, I., and Schlessinger, J. (1991). EGF induces increased ligand binding affinity and dimerization of soluble epidermal growth factor (EGF) receptor extracellular domain. J. Biol. Chem. 266 22035–22043. [PubMed] [Google Scholar]
- Iwano, M., Shiba, H., Funato, M., Shimosato, H., Takayama, S., and Isogai, A. (2003). Immunohistochemical studies on translocation of pollen S-haplotype determinant in self-incompatibility of Brassica rapa. Plant Cell Physiol. 44 428–436. [DOI] [PubMed] [Google Scholar]
- Jiang, G., and Hunter, T. (1999). Receptor signaling: When dimerization is not enough. Curr. Biol. 9 R568–R571. [DOI] [PubMed] [Google Scholar]
- Kachroo, A., Schopfer, C.R., Nasrallah, M.E., and Nasrallah, J.B. (2001). Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science 293 1824–1826. [DOI] [PubMed] [Google Scholar]
- Kai, N., Suzuki, G., Watanabe, M., Isogai, A., and Hinata, K. (2001). Sequence comparisons among dispersed members of the Brassica S multigene family in an S9 genome. Mol. Genet. Genomics 265 526–534. [DOI] [PubMed] [Google Scholar]
- Kandasamy, M.K., Paolillo, D.J., Faraday, C.D., Nasrallah, J.B., and Nasrallah, M.E. (1989). The S-locus specific glycoproteins of Brassica accumulate in the cell wall of developing stigma papillae. Dev. Biol. 134 462–472. [DOI] [PubMed] [Google Scholar]
- Kemp, B.P., and Doughty, J. (2003). Just how complex is the Brassica S-receptor complex? J. Exp. Bot. 54 157–168. [DOI] [PubMed] [Google Scholar]
- Kishi-Nishizawa, N., Isogai, A., Watanabe, M., Hinata, K., Yamakawa, S., Shojima, S., and Suzuki, A. (1990). Ultrastructure of papillar cells in Brassica campestris revealed by liquid helium rapid-freezing and substitution-fixation method. Plant Cell Physiol. 31 1207–1219. [Google Scholar]
- Lerouge, P., Cabanes-Mecheteau, M., Rayon, C., Fischette-Laine, A.C., Gomord, V., and Faye, L. (1998). N-Glycoprotein biosynthesis in plants: Recent developments and future trends. Plant Mol. Biol. 38 31–48. [PubMed] [Google Scholar]
- Matsui, T., Nakayama, H., Yoshida, K., and Shinmyo, A. (2003). Vesicular transport route of horseradish C1a peroxidase is regulated by N- and C-terminal propeptides in tobacco cells. Appl. Microbiol. Biotechnol. 62 517–522. [DOI] [PubMed] [Google Scholar]
- Nagao, M., Masuda, S., Abe, S., Ueda, M., and Sasaki, R. (1992). Production and ligand-binding characteristics of the soluble form of murine erythropoietin receptor. Biochem. Biophys. Res. Commun. 188 888–897. [DOI] [PubMed] [Google Scholar]
- Nagoshi, E., and Yoneda, Y. (2001). Dimerization of sterol regulatory element-binding protein 2 via the helix-loop-helix–leucine zipper domain is a prerequisite for its nuclear localization mediated by importin β. Mol. Cell. Biol. 21 2779–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, H., Yoshida, K., Ono, H., Murooka, Y., and Shinmyo, A. (2000). Ectoine, the compatible solute of Halomonas elongata, confers hyperosmotic tolerance in cultured tobacco cells. Plant Physiol. 122 1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, C.R., Blum, R., Pichler, B., Lepier, A., Kafitz, K.W., and Konnerth, A. (2003). Truncated TrkB-T1 mediates neurotrophin-evoked calcium signalling in glia cells. Nature 426 74–78. [DOI] [PubMed] [Google Scholar]
- Schlessinger, J. (2000). Cell signaling by receptor tyrosine kinases. Cell 103 211–225. [DOI] [PubMed] [Google Scholar]
- Schlessinger, J. (2002). Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell 110 669–672. [DOI] [PubMed] [Google Scholar]
- Schlessinger, J. (2003). Autoinhibition control. Science 300 750–752. [DOI] [PubMed] [Google Scholar]
- Schopfer, C.R., Nasrallah, M.E., and Nasrallah, J.B. (1999). The male determinant of self-incompatibility in Brassica. Science 286 1697–1700. [DOI] [PubMed] [Google Scholar]
- Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68 850–858. [DOI] [PubMed] [Google Scholar]
- Shiba, H., Takayama, S., Iwano, M., Shimosato, H., Funato, M., Nakagawa, T., Che, F.-S., Suzuki, G., Watanabe, M., Hinata, K., and Isogai, A. (2001). A pollen coat protein, SP11/SCR, determines the pollen S-specificity in the self-incompatibility of Brassica species. Plant Physiol. 125 2095–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, N.F., Stone, S.L., Christie, L.N., Sulaman, W., Nazarian, K.A., Burnett, L.A., Arnoldo, M.A., Rothstein, S.J., and Goring, D.R. (2001). Expression of the S receptor kinase in self-compatible Brassica napus cv. Westar leads to the allele-specific rejection of self-incompatible Brassica napus pollen. Mol. Genet. Genomics 265 552–559. [DOI] [PubMed] [Google Scholar]
- Stein, J.C., Howlett, B., Boyes, D.C., Nasrallah, M.E., and Nasrallah, J.B. (1991). Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc. Natl. Acad. Sci. USA 88 8816–8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, G., Kakizaki, T., Takada, Y., Shiba, H., Takayama, S., Isogai, A., and Watanabe, M. (2003). The S haplotypes lacking SLG in the genome of Brassica rapa. Plant Cell Rep. 21 911–915. [DOI] [PubMed] [Google Scholar]
- Suzuki, G., Watanabe, M., Kai, N., Matsuda, N., Toriyama, K., Takayama, S., Isogai, A., and Hinata, K. (1997). Three members of the S multigene family are linked to the S locus of Brassica. Mol. Gen. Genet. 256 257–264. [DOI] [PubMed] [Google Scholar]
- Suzuki, G., Watanabe, M., Toriyama, K., Isogai, A., and Hinata, K. (1995). Molecular cloning of members of the S-multigene family in self-incompatible Brassica campestris L. Plant Cell Physiol. 36 1273–1280. [PubMed] [Google Scholar]
- Suzuki, G., Watanabe, M., Toriyama, K., Isogai, A., and Hinata, K. (1996). Expression of SLG9 and SRK9 genes in transgenic tobacco. Plant Cell Physiol. 37 866–869. [Google Scholar]
- Takasaki, T., Hatakeyama, K., Suzuki, G., Watanabe, M., Isogai, A., and Hinata, K. (2000). The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403 913–916. [DOI] [PubMed] [Google Scholar]
- Takayama, S., and Isogai, A. (2003). Molecular mechanism of self-recognition in Brassica self-incompatibility. J. Exp. Bot. 54 149–156. [DOI] [PubMed] [Google Scholar]
- Takayama, S., Shiba, H., Iwano, M., Shimosato, H., Che, F.-S., Kai, N., Watanabe, M., Suzuki, G., Hinata, K., and Isogai, A. (2000). The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl. Acad. Sci. USA 97 1920–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama, S., Shimosato, H., Shiba, H., Funato, M., Che, F.-S., Watanabe, M., Iwano, M., and Isogai, A. (2001). Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 413 534–538. [DOI] [PubMed] [Google Scholar]
- Torii, K.U. (2000). Receptor kinase activation and signal transduction in plants: An emerging picture. Curr. Opin. Plant Biol. 3 361–367. [DOI] [PubMed] [Google Scholar]
- Tzahar, E., et al. (1997). Bivalence of EGF-like ligands drives the ErbB signaling network. EMBO J. 16 4938–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y., Meshi, T., and Okada, Y. (1987). Infection of tobacco protoplasts with in vitro transcribed tobacco mosaic virus RNA using an improved electroporation method. FEBS Lett. 219 65–69. [Google Scholar]
- Wilm, M., and Mann, M. (1996). Analytical properties of the nanoelectrospray ion source. Anal. Chem. 68 1–8. [DOI] [PubMed] [Google Scholar]
- Xu, W., Wang, Y.-S., Liu, G.-Z., Chen, X., Tinjuangjun, P., Pi, L.-Y., and Song, W.-Y. (2006). The autophosphorylated Ser686, Thr688, and Ser689 residues in the intracellular juxtamembrane domain of XA21 are implicated in stability control of rice receptor-like kinase. Plant J. 45 740–751. [DOI] [PubMed] [Google Scholar]
- Yet, M.G., and Jones, S.S. (1993). The extracytoplasmic domain of the erythropoietin receptor forms a monomeric complex with erythropoietin. Blood 82 1713–1719. [PubMed] [Google Scholar]
- Zhou, M., Felder, S., Rubinstein, M., Hurwitz, D.R., Ullrich, A., Lax, I., and Schlessinger, J. (1993). Real-time measurements of kinetics of EGF binding to soluble EGF receptor monomers and dimers support the dimerization model for receptor activation. Biochemistry 32 8193–8198. [DOI] [PubMed] [Google Scholar]