Abstract
Angiosperm embryo sac development begins with a phase of free nuclear division followed by cellularization and differentiation of cell types. The indeterminate gametophyte1 (ig1) gene of maize (Zea mays) restricts the proliferative phase of female gametophyte development. ig1 mutant female gametophytes have a prolonged phase of free nuclear divisions leading to a variety of embryo sac abnormalities, including extra egg cells, extra polar nuclei, and extra synergids. Positional cloning of ig1 was performed based on the genome sequence of the orthologous region in rice. ig1 encodes a LATERAL ORGAN BOUNDARIES domain protein with high similarity to ASYMMETRIC LEAVES2 of Arabidopsis thaliana. A second mutant allele of ig1 was identified in a noncomplementation screen using active Mutator transposable element lines. Homozygous ig1 mutants have abnormal leaf morphology as well as abnormal embryo sac development. Affected leaves have disrupted abaxial–adaxial polarity and fail to repress the expression of meristem-specific knotted-like homeobox (knox) genes in leaf primordia, causing a proliferative, stem cell identity to persist in these cells. Despite the superficial similarity of ig1-O leaves and embryo sacs, ectopic knox gene expression cannot be detected in ig1-O embryo sacs.
INTRODUCTION
The plant life cycle has genetically active diploid and haploid phases, called the sporophyte and gametophyte, respectively. In green algae, such as Ulva lactuca, the two phases of the life cycle are morphologically similar. During land plant evolution, the extent of the gametophyte phase has become reduced. In primitive land plants, such as the bryophyte Physcomitrella patens, the gametophyte is the dominant phase of the life cycle and contains a leafy-shoot phase called the gametophore with at least superficial similarity to the vegetative shoots of seed plants, whereas the sporophyte is greatly reduced and dependent on the gametophyte (Cove and Knight, 1993). In seedless vascular plants such as ferns, the sporophyte is dominant and grows as a leafy shoot, whereas the gametophyte is free-living but greatly reduced and lacking a leafy-shoot phase (Banks, 1999). The gametophytes are smallest in angiosperms, consisting of only a few cells dependent on the sporophyte for growth and development.
The female gametophyte, or embryo sac, undergoes a stereotypical number of divisions to produce an eight-nuclei syncytium. The migration and position of these nuclei are highly regular. The embryo sac then cellularizes to produce four cell types: synergids, antipodals, an egg cell, and a homodiploid central cell (Drews and Yadegari, 2002). In wild-type maize (Zea mays), the antipodal cells are the only embryo sac cells that proliferate after cellularization. In Arabidopsis thaliana, there is no proliferation after cellularization of the embryo sac, as the antipodals degenerate during maturation (Murgia et al., 1993).
Recently, many mutations have been identified that act genetically during the haploid phase in angiosperms (Ebel et al., 2004; Johnson et al., 2004; Pagnussat et al., 2005). indeterminate gametophyte1 (ig1) in maize is unique among mutants that act in the haploid gametophytes; mutants are viable and have an increased number of nuclei before cellularization of the embryo sac (Huang and Sheridan, 1996; Ebel et al., 2004). retinoblastoma-related1 (rbr1) in Arabidopsis is the only other mutant reported with extra rounds of free nuclear divisions, but unlike ig1, it also affects the male gametophyte, and rbr1 embryo sacs do not produce viable progeny.
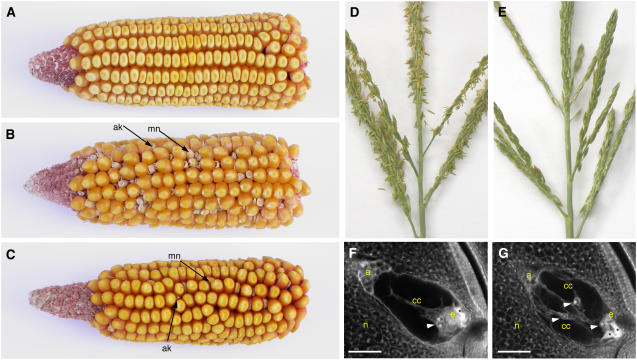
In ig1 mutant embryo sacs, the proliferative phase is prolonged, suggesting that wild-type ig1 function promotes the switch from proliferation to differentiation in the embryo sac (Lin, 1978, 1981; Huang and Sheridan, 1996). Consequently, mutant ig1 embryo sacs contain extra egg cells, extra central cells, and extra polar nuclei within central cells (Figure 1). The phenotypes of ig1 mutant embryo sacs suggest a position-based determination of cellular identity (Lin, 1978, 1981; Guo et al., 2004). The ability of the extra cells and nuclei to function as egg cells or polar nuclei, for example, appears to depend on their position in the embryo sac. Because of their abnormal structure, many of these defective embryo sacs give rise to abnormal seeds, which allowed the identification of the ig1-O reference allele (Figure 1B). These abnormalities include polyembryony, heterofertilization, miniature endosperms, and early abortion of seeds (Kermicle, 1971). Maize endosperm development is very sensitive to a deviation from the normal 2 maternal:1 paternal genome ratio in the endosperm. Consequently, when an embryo sac with three polar nuclei is fertilized by a standard haploid pollen grain, the resulting endosperm is miniature, and when an embryo sac with four or more polar nuclei is fertilized by a haploid pollen grain, the resulting endosperm aborts early in development and collapses (Lin, 1984). This leads to abnormal endosperm development when embryo sacs with extra polar nuclei are fertilized.
Figure 1.
Phenotypes of ig1 Mutants.
(A) Ear from a W64A wild-type plant.
(B) Ear from an ig1-O/ig1-O W64A mutant plant.
(C) Ear from an ig1-mum/ig1-mum W64A plant.
(D) Fertile wild-type tassel with anthers extruded.
(E) Sterile ig1/ig1 mutant tassel without extruded anthers.
(F) Wild-type embryo sac.
(G) ig1 embryo sac.
Asterisks indicate degenerated synergids. Arrowheads point to polar nuclei. a, antipodal cells; ak, aborted kernel; cc, central cell; e, egg cell; mn, miniature kernel; n, nucellus. Bars = 25 μm.
Because ig1 embryo sacs are frequently viable, homozygous lines can be established to examine the role of ig1 in sporophyte development. Homozygous mutants for ig1-O reportedly have normal vegetative morphology but in some genetic backgrounds have sporophytic male sterility, even though there is no gametophytic affect on pollen, either failing to shed pollen or extrude anthers (Kermicle, 1994).
Additionally, ig1 restricts the embryogenic potential of cells that lack one of the two parental genomes, so that mutant embryo sacs produce haploid progeny, of both maternal and paternal origin, at a higher rate than wild-type embryos (Kermicle, 1969). Paternal haploid production has been used in maize breeding to transfer germplasm from one variety of maize to the cytoplasm of another variety (e.g., a male sterility conditioning cytoplasm) (Kindiger and Hamann, 1993). The ability to produce embryos without a paternal genetic contribution bears some resemblance to apomixis, the production of seeds that are clones of the mother plant (Grimanelli et al., 2001); understanding ig1 may provide further insight into the development of apomictic plants.
Comparisons of genetic maps and DNA sequences demonstrate a significant conservation of gene order between maize and rice (Moore et al., 1995). ig1 was cloned by taking advantage of the conserved gene order between maize and rice in the ig1 region. This report describes the cloning of the indeterminate gametophyte1 gene and the phenotype of ig1-O and a second mutant allele, ig1-mum, which affect leaf development as well as embryo sac development. The phenotype of ig1 plants suggests that common mechanisms control the switch from proliferation to differentiation in both the gametophyte and sporophyte of angiosperms.
RESULTS
The Phenotype of ig1-O Is Affected by Genetic Background
The ig1-O mutation was backcrossed three to five times to different inbred lines and then pollinated to test the effect of genetic background on the frequency of mutant phenotypes (Table 1). The A158 inbred line was the most severe, with the highest degree of prefertilization failure of ig1 mutant embryo sacs and the lowest frequency of normal seeds. The W23 inbred line in which ig1-O was first identified had the highest frequency of seeds arising from embryo sacs with extra polar nuclei (miniature and aborted seeds), and W64A gave the next highest frequency for these classes along with A158. Interestingly, although Mo17 was a moderate background with regard to most phenotypic classes, it produced the highest percentage of kernels with more than one embryo (i.e., embryo sacs with more than one egg cell). B73 and W22 suppressed the ig1-O seed phenotypes the most. Interestingly, the presence of the homozygous male-sterile phenotype did not correlate with the severity of ig1 seed phenotypes, suggesting that different loci modify the sporophytic and gametophytic phenotypes of ig1.
Table 1.
Phenotypic Severity of ig1-O and ig1-mum
| Normal Endosperm
|
Absent or Abnormal Embryo | Miniature Endosperm
|
Aborted/Collapsed Endosperm | Ovules without Seeds | ||||
|---|---|---|---|---|---|---|---|---|
| One Embryo | Two or More Embryos | One Embryo | Two or More Embryos | Homozygous Phenotype | ||||
| ig1-O | ||||||||
| A158 (656) | 24 | 3 | 1 | 13 | 1 | 12 | 46 | Male-fertile |
| W23 (845) | 34 | 2 | 3 | 9 | 0 | 25 | 27 | Male-sterile |
| W64A (793) | 46 | 6 | 2 | 7 | 1 | 18 | 20 | Male-sterile |
| Mo17 (948) | 53 | 11 | 2 | 6 | 2 | 11 | 15 | Male-sterile |
| B73 (792) | 62 | 1 | 2 | 1 | 0 | 7 | 27 | ND |
| W22 (989) | 72 | 0 | 0 | 2 | 1 | 2 | 24 | Male-fertile |
| ig1-mum | ||||||||
| Mo17 (775) | 70 | 4 | 3 | 3 | 2 | 3 | 15 | Variable male sterility |
Values shown are percentages of mutant ovules. Values shown in boldface represent normal seeds. Numbers in parentheses indicate total ovules (with and without seeds) examined. Inbred lines are ordered from the most to the least severe based on lowest to highest frequency of normal seed production (i.e., highest to lowest penetrance of combined ovule failure and seed defects). ND, not determined.
Identifying a Second Allele of ig1
A noncomplementation screen for the male-sterile phenotype of ig1-O (Figure 1) was used to identify a second mutant allele, ig1-mum, from an active Mutator (Mu) transposable element line. An active Mu line was established in the inbred W64A, because ig1-O homozygotes are male-sterile as W64A inbreds and W64A/W23 hybrids. Crosses were made between ig1-O/ig1-O W23 females and Mu W64A males. Rare male-steriles from an F1 population of 60,000 individuals were crossed as females with standard W64A inbred plants. The progeny of the selections were tested for ig1 mutant phenotypes. Specifically, these plants were tested for seed phenotypes similar to those of ig1-O (i.e., miniature seeds, twins, and aborted seeds) and retested for their failure to complement the male-sterile phenotype of ig1-O. By these criteria, one of these selections was shown to carry a new mutant allele of ig1, named ig1-mum. In addition to causing a male-sterile phenotype in transheterozygotes with ig1-O or as homozygotes, ig1-mum causes the same abnormal seed types as ig1-O, suggesting that the same embryo sac defects are present in ig1-mum mutants as in ig1-O. Most of the mutant classes occur at a lower frequency in ig1-mum than in ig1-O, suggesting that ig1-mum is a weaker allele in the embryo sac (Figure 1, Table 1).
Fine Mapping and Cloning of ig1
Fine mapping of ig1 was initiated in a population segregating ig1-O from a cross between ig1-Ow23/+Mo17 females and +/+Mo17 males. Fine mapping of ig1 against available simple sequence repeat markers placed ig1 between umc1311 and umc1973 on chromosome 3 of maize. The rice orthologs of the markers around ig1 were identified by BLAST search for each sequence against the rice genome using the Gramene database for comparative grass genomics (www.gramene.org) (Ware et al., 2002). The orthologs of umc1311 and umc1973 on rice chromosome 1 are ∼845 kb apart (Figure 2). By generating mapping populations among multiple inbred lines (between ig1-O in a W23 inbred background and wild-type Mo17, W64A, A158, or M14), polymorphisms were found with the umc1539 simple sequence repeat marker that further reduced this interval in the rice map to 378 kb covered by three rice BAC clones.
Figure 2.
Comparative Mapping between Rice and Maize around ig1.
BAC clones and maize BAC contigs are stippled. Arrows indicate the physical distance between markers on rice chromosome 1. Vertical lines show the positions of the closest sequence matches in the rice genome for maize clones in the ig1 region. Maize markers shown above the maize BACs had been placed on the BAC clones but not on the genetic map. Underlines indicate PCR-based markers designed using rice genome information.
The Gramene database, in which cDNA and genomic clones of many grasses, including maize, are aligned with their predicted rice orthologs, was used to develop more maize mapping markers. Maize ESTs and methyl-filtered library clones that were orthologs of the rice genes in this interval were used to design PCR primers to generate additional mapping markers. These markers reduced the interval carrying the potential rice ortholog of ig1 to a 65-kb region containing 12 annotated genes.
A Mu insertion in the ig1-mum allele in a maize gene orthologous to the rice gene Os01g66590, a gene within the 65-kb interval of the rice genome described above, was identified by PCR amplification using a primer for the Mu terminal inverted repeat and a gene-specific primer. Additionally, a small population of 24 individuals segregating for the ig1-mum allele was used to test for a cosegregating Mu band using a modified amplification of insertion mutagenized sites protocol (Frey et al., 1998). One band that cosegregated with ig1-mum contained a Mu insertion in the assembled Zea mays contig, AZM4_49905, orthologous to Os01g66590 that was used to design primers for the directed Mu search described above. This gene encodes a member of the LATERAL ORGAN BOUNDARIES (LOB) protein family, a plant-specific gene family with 43 members in Arabidopsis (Iwakawa et al., 2002; Shuai et al., 2002).
Both ig1-mum and ig1-O were compared along with their progenitors by DNA gel blot hybridization of a probe for the LOB domain of the ig1 gene (probe 2-4 in Figure 3) with the DNA of heterozygous mutant and homozygous wild-type plants (data not shown). Both alleles have a novel band not present in their progenitors. The ig1-mum allele contains a Mu8 insertion within the first intron, 86 nucleotides upstream of the start codon, and the ig1-O allele contains a Hopscotch retrotransposon insertion within codon 120 of ig1, 14 residues before the end of the LOB domain (Figure 3). Hopscotch is one of a group of low-copy retrotransposons that are more commonly inserted into genes than the high-copy retroelements that make up the bulk of the maize genome(White et al., 1994). The ig1 gene contains four exons that constitute an mRNA 1264 bases in length (Figure 3). The structures of ig1, Oryza sativa ig1 (Osig1), and the closest maize homolog to ig1, ig1/as2-like1 (ial1), are very similar, with three introns in the same positions.
Figure 3.
ig1 Gene Structure and Mutations.
(A) Structures of the ig1 gene and its rice ortholog. Positions of the Mu insertion in ig1-mum and the Hopscotch insertion in ig1-O are indicated. The LOB domain is indicated by dark gray. The first exon contains most of the 5′ untranslated region (UTR); the second exon contains the LOB domain; the third exon contains the C-terminal domain; and the fourth exon contains most of the 3′ untranslated region. The third intron is 1.6 kb in length and extends beyond the end of AZM4_49905. The last exon is part of a separate AZM contig, AZM4_116957.
(B) Effect of ig1 mutations on ig1 RNA levels. RT-PCR was performed on RNA from whole ear primordia ∼5 cm in length using primers for both ig1 around the first intron and for ubiquitin (ubi). Lane 1, wild type; lane 2, ig1-mum homozygote; lane 3, ig1-O homozygote. Homozygous mutants were identified on the basis of male sterility.
(C) Relative ig1 message levels in ig1-O and ig1-mum ear primordia normalized to ubi. The left graph shows PCR results using primers around the first intron of ig1 (around the insertion in ig1-mum and upstream of the insertion in ig1-O), and the right graph shows PCR results using primers around the second intron of ig1 (downstream of the insertion in ig1-mum and around the insertion in ig1-O). Error bars indicate se. Five replicate assays were performed for each pair of PCR primers per sample.
To test the effect of both ig1-O and ig1-mum on ig1 mRNA levels, RT-PCR was performed on whole ear primordia of homozygous ig1-O and ig1-mum and the wild type (Figure 3). Primers flanking the insertion in either ig1-mum or ig1-O were used for the RT-PCR experiments. Both ig1-mum and ig1-O cause a significant decrease in the levels of normal ig1 mRNA, as measured by amplification around the first intron and the ig1-mum Mu8 insertion site. There is a 10-fold decrease in the amount of ig1 message in ig1-O and in ig1-mum, as measured by quantitative real-time PCR around intron 1. In the ig1-mum samples, melting curve and gel analyses demonstrate that most of this PCR product is of abnormal size. Quantitative RT-PCR downstream of the ig1-mum insertion site using primers flanking intron 2 and the ig1-O Hopscotch insertion site detects a twofold reduction in ig1 RNA in ig1-mum and a 100-fold reduction in ig1-O.
Presumably, the discrepancy between the findings for the two pairs of primers in ig1-mum results from the presence of many mRNAs in ig1-mum ears that have 5′ untranslated regions that are too long to amplify under the conditions used. Sequencing of the RT-PCR products from ig1-mum ears obtained with longer extension times revealed that mutant transcripts retained a variable amount of both the first intron and the Mu8 element. This alteration in the 5′ untranslated region could have affects on the translation efficiency of these messages, even though the abundance of ig1 message 3′ of the Mu insertion is only mildly affected.
ig1 Encodes a LOB Domain Protein Similar to AS2
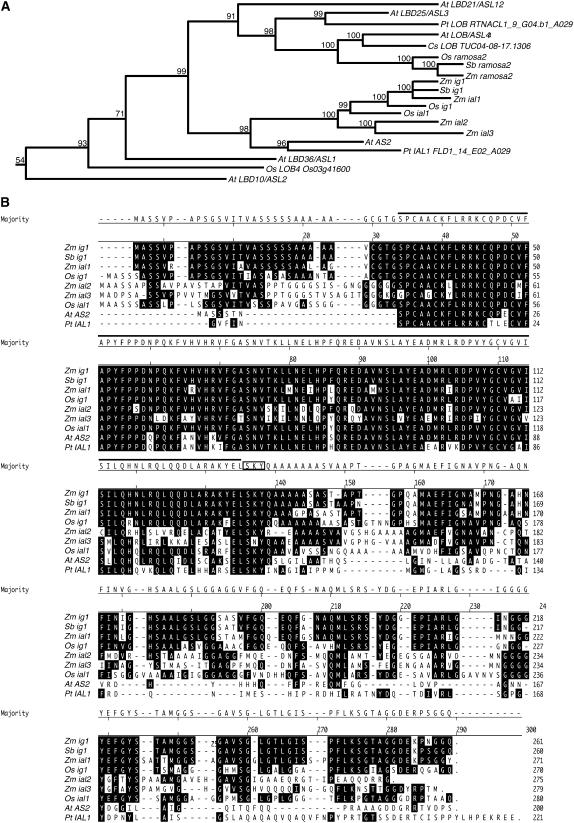
The LOB domain protein encoded by ig1 has high similarity to that of ASYMMETRIC LEAVES2 (AS2) of Arabidopsis (Figure 4). An alignment was made between the proteins encoded by ig1, its nearest maize homologs, the entire families of LOB DOMAIN/AS2-LIKE (LBD/ASL) genes of rice (36 genes) and Arabidopsis (43 genes), and available LBD genes most similar to AS2 from other plants (Iwakawa et al., 2002; Shuai et al., 2002). Figure 4 shows the portion of this tree encompassing AS2, its five closest homologs in Arabidopsis (LBD36/ASL1, LBD10/ASL2, LBD25/ASL3, LOB/ASL4, and LBD21/ASL12), and ial genes from other species. This subclade includes the ramosa2 (ra2) gene of maize, Sbra2 of Sorghum bicolor, and Osra2 of rice, which are most closely related to LOB/ASL4 of Arabidopsis (Bortiri et al., 2006). ig1 is one of four genes, along with ial1, ial2, and ial3, in maize whose proteins align most closely with that of AS2, suggesting substantial redundancy for AS2 function in maize. There are two rice genes in this clade for the four maize genes (Os LOB2 and Os LOB3 [Bortiri et al., 2006], named here Os ig1 [Os01g66590] and Os ial1 [Os05g34450] to reflect their inclusion in the ig1/AS2 subclade of LOB genes), as one would expect based on comparative mapping of grasses and the allotetraploid origin of maize (Gaut and Doebley, 1997). To date, the AS2 subgroup includes genes from several grasses, including maize, rice, and sorghum (as well as partial sequences of genes from Hordeum vulgare, Triticum aestivum, and Saccharum officinarum; data not shown), one dicot in Arabidopsis, and one gymnosperm in Pinus taeda.
Figure 4.
The ig1 Gene Family.
(A) Relationship of IG1 with other closely related LOB domain proteins. Only the portion of the LOB domain protein family including AS2 and its five most closely related homologs from Arabidopsis is shown. Of the rice and maize genes and the full-length LOB domain genes from a few other species, only those that fall into this same LOB domain subfamily are included. The number above each branch corresponds to the posterior probability for that node.
(B) Alignment of proteins within the IG1/AS2 subgroup. Residues identical to IG1 are highlighted. The LOB domain is indicated by a black line over the residues, and the SKY motif is boxed. The translation start of the P. taeda gene is unknown. Zm, Zea mays; Sb, Sorghum bicolor; Os, Oryza sativa; At, Arabidopsis thaliana; Pt, Pinus taeda; Cs, Citrus sinensis.
The members of this clade are distinct from the other LOB domain genes in possessing a SKY motif (often SKYQ) immediately after the LOB domain that is not present in other LOB domain genes in either Arabidopsis or rice, even the ASL1, -2, -3, and -4 proteins (Figure 4). These residues immediately follow the last Leu of the predicted Leu zipper coiled-coil domain of the LOB domain. Additionally, the Tyr and Gln residues of the SKYQ motif occupy the g and a positions, respectively, in the Leu zipper after this last Leu. The a, e, and g positions in the helix of Leu zippers determine the specificity of dimerization between Leu zippers (Acharya et al., 2002; Deppmann et al., 2004). Additionally, the presence of a positively charged amino acid in the a position of one of the heptads—Lys in ig1, ial1, Sbig1, Osig1, and AS2 and Arg in ial2, ial3, and Osial1—suggests that these proteins do not homodimerize. This conserved structure suggests that the LOB domain proteins in this clade interact with a conserved partner. The IAL proteins from grass species are distinct from AS2 in Arabidopsis in having a longer Ser-rich N-terminal leader upstream of the LOB domain and an Ala-rich sequence downstream of the SKY motif. Whether these are peculiarities of grasses/monocots or are found in other plant LOB proteins cannot be determined until these genes are cloned from more species. Within the LOB domain, IG1 and AS2 are 84% identical; however, the C-terminal domain is not conserved, with only 17% identity between IG1 and AS2. Interestingly, the divergent portion of the protein is in a separate exon from the conserved portion—the LOB domain plus the SKYQ motif—in ig1 and all grass IAL genes for which there are genome sequence data. There is also one gene in the rice genome, Os02g21390, that lacks a LOB domain but whose C-terminal half is 60% identical to the C-terminal domain of ig1 and 76% identical to that of Osig1. Based on the protein alignments, there is redundancy of AS2 gene function in both rice and maize not present in Arabidopsis. However, there are also genes in Arabidopsis that have no clear ortholog in maize or rice, such as LBD10/ASL2 and LBD25/ASL3. Assignment of functional redundancy and orthology, therefore, are tentative without more detailed expression or mutant analysis of these genes.
Expression of ig1 and Closely Related Genes
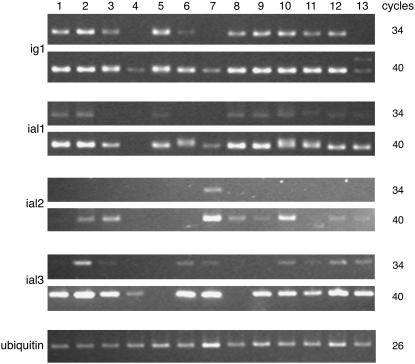
ig1 and the ial genes are broadly expressed in a variety of tissues in wild-type plants (Figure 5). ig1 transcript can be detected by RT-PCR in leaves, leaf primordia, immature ears (before and during floret development), immature tassels, whole ovules, silk, and husk leaves. ig1 has lower expression in endosperm and much lower expression in roots and mature pollen grains. ial1 is expressed in many of the same tissues but generally at lower levels than ig1. ial2, unlike ig1 and ial1, has its highest expression in pollen and has very low expression elsewhere. ial2 RNA levels are too low to be detected in mature leaves, roots, silk, endosperm, or ovules, but it has detectable expression in developing leaves, husks, ears, and tassel primordia. ial3 is more broadly expressed at higher levels overall than ial2. Unlike ial2, ial3 is expressed in ovules and endosperm but is not expressed in young ear primordia. The low expression of ig1 in pollen grains is consistent with the absence of an effect of ig1 mutations on the male gametophyte. ial2 may perform a similar function in the male gametophyte to that of ig1 in the female gametophyte.
Figure 5.
Expression of ig1 and ial Genes in Various Tissues.
RT-PCR was performed on total RNA using ubi primers or ig1 or ial primers. Note the larger ig1 transcript seen after 40 cycles in ig1-mum mutant leaves (lane 13). Lane 1, fully expanded leaf; lane 2, immature leaf; lane 3, husk leaf; lane 4, root; lane 5, silk; lane 6, endosperm at 9 d after pollination; lane 7, mature pollen; lane 8, 1-cm-long ear primordium; lane 9, 5-cm-long ear primordium; lane 10, 2-cm-long tassel primordium; lane 11, ovule; lane 12, ligular region of wild-type seedling leaf; lane 13, ligular region of ig1-mum flag leaf.
To gain a better understanding of the expression pattern of ig1 in flowers, in situ hybridization was performed on sections of female flowers from the ear. A probe for the last 33 codons and the 3′ untranslated region was used to reduce the amount of cross-hybridization to ial1. Flowers at several stages were examined to detect ig1 in embryo sacs and floral organs. ig1 is expressed in the adaxial domain of all organs examined (Figure 6). ig1 mRNA is found on the adaxial side of lemmas and paleas (sepals), glumes (bracts), and silk (fused carpels) surrounding the single ovule. The expression of ig1 in the adaxial walls of the carpels that make up the silk forms a ring on the inner wall of the ovary surrounding the ovule. This expression is stronger in the lateral regions of the ovary wall than in the medial domain (Figure 6C). Additionally, ig1 is expressed at the boundary between the integuments and the nucellus of the ovule (Figures 6B and 6C). The expression on the adaxial side of floral organs becomes restricted to the basal portion of this domain and eventually fades, becoming more difficult to detect as the flowers age. This decrease in ig1 message is evident in the difference between the adaxial sides of the paleas of the upper and lower florets, because the lower floret is arrested at an earlier stage of development than the upper floret (Figure 6E).
Figure 6.
Expression of ig1 in Floral Organs.
(A) Diagram of a maize female spikelet with a fully developed upper floret and an arrested lower floret (yellow). In a median section, the lemma and palea (the first-whorl organs), one of the arrested stamens, and the silk (gynoecium) with a single ovule can be seen. The area in box 1 is shown in (B), (C), and (D). The area in box 2 is shown in (E). The area in box 3 is shown in (F) and (G). The area in box 4 is shown in Figures 7A to 7C. The area in box 5 is shown in Figures 7D to 7H.
(B) and (D) Median longitudinal sections of wild-type ovules.
(C) Longitudinal section through the margin of a wild-type ovule. ig1 is expressed in the adaxial epidermis at the base of the carpels that make up the silk as well as at the boundary between the integuments and the nucellus.
(E) The lower floret and the palea of the upper floret.
(F) and (G) The lemma of the upper floret and the inner glume (bract).
Probes were as follows: (B), (C), (E), and (F), ig1 antisense probe; (D) and (G), ig1 sense probe. Arrowheads point to adaxial expression of ig1 in floral organs and bracts. Arrows point to embryo sacs. g, glume; i, integument; l, lemma; n, nucellus; p, palea; si, silk; st, stamen. Bar = 100 μm.
In embryo sacs, ig1 message is detectable as early as the one-nucleus stage (Figure 7A) but is not detectable in the nonfunctional megaspores. In older embryo sacs, ig1 message is highest in antipodal cells (Figure 7D). In older embryo sacs, the background staining becomes more intense even with a sense probe for ig1 (Figure 7H). Consequently, it is difficult to determine whether ig1 expression continues at these later stages. In particular, it is difficult to determine whether ig1 expression decreases as embryo sacs age, as it does in lateral organs. Expression in most of the micropylar cells is frequently not above background. However, ig1 appears to be expressed in the developing egg cell, and staining around the polar nuclei is also detected occasionally. ig1 expression is not detected in the developing synergids, however. Therefore, ig1 may have asymmetric expression within the embryo sac, as it does in lateral organs.
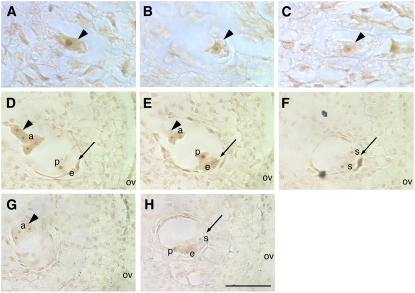
Figure 7.
Expression of ig1 in Embryo Sacs.
(A) to (C) Ovules with stage 1 (one-nucleus) embryo sacs.
(A) and (C) Wild-type stage 1 embryo sacs (arrowheads). The ig1 antisense probe detects message in stage 1 embryo sacs.
(B) Nonfunctional megaspore (arrowhead) lacking ig1 expression from the same ovule as the embryo sac in (A).
(D) to (H) Ovules with immature cellularized embryo sacs. Arrowheads point to chalazal nuclei and cells of embryo sacs, and arrows point to micropylar nuclei and cells of embryo sacs.
Probes were as follows: (A), (B), (D), (E), and (F), ig1 antisense probe; (C), (G), and (H), ig1 sense probe. Signal with the ig1 antisense probe is higher than that with the sense probe control in the antipodal cells. In the cells at the micropylar end of the embryo sac, the signal is more variable and the background staining with the sense probe is higher, making it difficult to determine whether there is any ig1 signal. a, antipodal cell; e, egg; ov, ovary wall; p, polar nucleus; s, synergid. Bar = 25 μm for (A) to (C) and 50 μm for (D) to (H).
ig1 Controls Leaf Development
The AS2 gene has been shown to repress the expression of the knotted1-like homeobox (knox) genes KNAT1/BREVIPEDICELLUS1 (BP1), KNAT2, and KNAT6 in leaf primordia (Ori et al., 2000; Semiarti et al., 2001). During vegetative development, the initiation of lateral organ primordia from an undifferentiated stem cell population is associated with the downregulation of knox genes (Jackson et al., 1994; Long et al., 1996). knox genes are normally expressed in the shoot apical meristem and excluded from leaf primordia. In maize leaves with ectopic knox gene expression, differentiation fails to progress properly and regions of the leaf overproliferate, producing outgrowths and abnormal boundaries between leaf domains (Muehlbauer et al., 1997). In severe cases of knox misexpression, leaf cells revert to a stem cell identity and shoot apical meristems develop on the adaxial surfaces of leaves (Sinha et al., 1993).
In Arabidopsis, knox genes are ectopically expressed in leaf primordia in as2 mutants (Ori et al., 2000; Semiarti et al., 2001; Iwakawa et al., 2002). Ectopic knox gene expression in leaf primordia is also caused by mutations in rough sheath2 (rs2) in maize and AS1 in Arabidopsis, which have been shown to be functionally orthologous, myb domain–containing ARP (for AS1 RS2 Phantastica) genes (Schneeberger et al., 1998; Timmermans et al., 1999; Tsiantis et al., 1999; Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001; Iwakawa et al., 2002; Theodoris et al., 2003). No effect of a mutation in a maize LOB domain gene on leaf development has been reported previously.
To analyze the effects of ig1-mum on sporophyte development, ig1-mum heterozygotes were self-pollinated after having been backcrossed three generations to inactive Mu W64A and one generation to inactive Mu Mo17 inbreds. Interestingly, plants with abnormal morphology of the flag leaf (the last vegetative leaf) were segregating in families segregating ig1-mum (Figure 8). The plants in these families were tested for the ig1-mum mutation by PCR. All of the plants with leaf abnormalities were ig1-mum homozygotes (19 of 19). However, not all ig1-mum homozygotes had abnormal flag leaves (i.e., the phenotype was not completely penetrant). In the W64A/Mo17 hybrid background examined, only 19 of 28 homozygotes had abnormal leaf morphology. The incomplete penetrance may reflect a segregation of modifiers that were introduced from either the W64A or Mo17 inbred. Because of this phenotype in ig1-mum homozygotes, ig1-O homozygotes in a W64A inbred background were examined for leaf defects and discovered to have a similar flag leaf phenotype as that of ig1-mum. ig1-O W64A homozygotes have leaf flaps on the adaxial side of the midrib of the leaf blade, a phenotype not previously reported in homozygous ig1-O lines in other inbred backgrounds. Additionally, closer examination of ig1-O homozygotes in the original W23 inbred line revealed that most but not all homozygotes (10 of 14) had flag leaves with leaf flaps on the adaxial side of the leaf sheath but not the leaf blade, with occasional disruption of the ligular region (Figure 8E).
Figure 8.
Phenotype of the Flag Leaf (Last Vegetative Leaf) in ig1/ig1 Homozygotes.
(A) and (C) Wild type.
(B), (D), (F), and (G) ig1-mum/ig1-mum.
(E) ig1-O/ig1-O W23.
(A) and (B) Adaxial side of the middle of the leaf blade.
(C) to (E) Adaxial ligular region. The arrow points to the leaf flap along the midrib. Arrowheads point to the ligular region.
The leaf in (B) shows the most common phenotype in ig1-mum plants. The leaf in (D) shows the most severe ig1-mum leaf phenotype. The leaf in (E) shows the most common ig1-O flag leaf phenotype in a W23 inbred background, with leaf flaps on the sheath and mild ligule distortion.
(F) Close-up of leaf flaps on the adaxial surface of an ig1-mum leaf. Note that there are epidermal hairs on the outer epidermis of the flaps but not the inner surfaces of the flaps. The edges of the flaps also have hairs, like normal leaf margins.
(G) Knot protruding from the abaxial surface of an ig1-mum flag leaf, with anthocyanin and abaxial epidermal hairs typical of sheath tissue.
(H) Expression of knox genes in ig1-mum and rs2. PCR was performed on cDNA from the ligular region of a wild-type flag leaf (lane 1), the ligular region of an ig1-mum flag leaf (lane 2), the ligular region of wild-type seedling leaves (lane 3), the ligular region of rs2 seedling leaves (lane 4), ear primordia (lane 5), or genomic DNA (lane 6).
Although incompletely penetrant, the most common leaf phenotype of ig1 mutants is ectopic outgrowths of leaf lamina on the adaxial side of the midrib. The dominant maize mutant Lax midrib1-O (Lxm1-O) has similar flaps of tissue on either side of the midrib (Schichnes et al., 1997; Schichnes and Freeling, 1998). In contrast with ig1, the flaps in Lxm1-O are on the abaxial side of the leaf and have adaxial tissue between them, the opposite polarity of that in ig1. Some ig1-mum mutants have distortions of the ligular region at the boundary between the proximal leaf sheath and the distal leaf blade, a phenotype that more closely resembles that of rs2 mutants, although these distortions are often associated with leaf flaps on the adaxial sheath (Figure 8). The leaf defects also include rare knots of sheath tissue in the leaf blade (Figure 8G).
To test whether these leaf phenotypes are associated with ectopic knox gene expression, RT-PCR was performed on RNA extracted from the ligular region of ig1-mum mutant leaves with abnormal leaf morphology as well as rs2 mutant leaves and wild-type ear primordia as positive controls (Figure 8H). The effect of ig1-mum was tested on all class I knox genes, knotted1 (kn1), roughsheath1 (rs1), liguleless3 (lg3), lg4a/b, knox3, knox8, knox10, and gnarley1 (gn1), as well as on a class II knox gene, knox6. Ectopic expression of kn1, rs1, lg3, lg4a/b, gn1, and knox3 was detected in ig1-mum and rs2 leaves; knox6, knox10, and possibly knox8 were unaffected by ig1-mum. No ectopic knox gene expression was detected in ig1-mum seedling leaves, which are normal in appearance (data not shown). These results demonstrate that ig1-mum affects many but not all class I knox genes and expand the number of knox genes known to be affected by rs2 to include lg4 and knox3. Additionally, they show that not all class I knox genes are repressed in leaf primordia; knox10 has equivalent levels of expression in wild-type leaf and ear primordia, and knox8 nearly so.
ig1-mum mutant leaves also have defects in abaxial–adaxial polarity. The leaf flaps along the midrib are usually found in pairs and have hairs and sclerenchymal cells on their margins typical of normal leaf margins (Figures 8 and 9). The epidermis of these flaps lacks macrohairs (i.e., is abaxial) on the inner surfaces toward the midrib and has macrohairs (i.e., is adaxial) on the outer surfaces that are continuous with the adaxial surface of the normal leaf lamina (Figure 8F). The switch in leaf polarity includes the internal layers of the leaf as well (Figure 9). In the wild type, the midrib contains large clear cells on the adaxial side of the vasculature and small sclerenchyma cells on the abaxial side. In ig1-mum flag leaves, the cells immediately adaxial to the midvein between the two ectopic leaf flaps are sclerenchyma cells typical of the abaxial side of the wild-type leaf, and the adaxial clear cells are absent. The polarity of the vascular bundle in the midrib is not affected. In wild-type leaves, the phloem is located on the abaxial side and the xylem is located on the adaxial side of the veins, and the polarity of cell types within the midvein is normal in ig1-mum. Just outside of the two leaf flaps, polarity is normal, with the clear cells present on the adaxial side. Therefore, the leaf flaps appear to be bordered by adaxial tissue on the marginal side and abaxial tissue on the midrib side. The vascular bundles in the leaf flaps also show the same polarity as the macrohairs on the surface. In the ig1-mum leaf flaps, the abaxial phloem cells are on the midrib side and the adaxial xylem cells are on the margin side.
Figure 9.
Polarity Defects in ig1 Flag Leaves.
(A) Midrib of a wild-type flag leaf. The arrowhead points to abaxial sclerenchyma.
(B) Midrib and flaps of an ig1-mum flag leaf. Arrowheads point to abaxial and adaxial sclerenchyma.
(C) Normal leaf margin. Arrowhead points to marginal sclerenchyma.
(D) Margin of an ig1-mum leaf flap. Arrowhead points to marginal sclerenchyma.
(E) Model of adaxial and abaxial domains in wild-type and ig1 leaves.
cc, clear cells; ph, phloem; x, xylem. Bars = 150 μm.
This morphology has been seen in other leaf polarity mutants, such as phantastica in snapdragon (Antirrhinum majus), in which ectopic leaf flaps form at the juxtaposition of adaxial and abaxial tissues (Waites and Hudson, 1995). as2 loss-of-function mutants in Arabidopsis have also been shown to have polarity defects, similar to ig1-mum, with abaxial tissue on the adaxial surfaces of leaves, particularly in double mutants with either erecta or rna-dependent rna polymerase6 (Xu et al., 2003; Li et al., 2005). In approximately half of the ig1-mum leaves that have these leaf flaps, the abaxial side of the midrib opposite the flaps has a file of epidermal hairs (i.e., is adaxial in character), demonstrating a switch of leaf epidermal identity rather than the duplication of abaxial identity in as2 mutants or of adaxial identity in dominant phabulosa1-d mutants (McConnell et al., 2001). Although this is different from the affect of as2 in Arabidopsis, a similar difference was seen between dominant polarity mutants in the maize rolled leaf1 gene and those in its homologs in Arabidopsis, REVOLUTA and PHABULOSA (Nelson et al., 2002; Juarez et al., 2004).
Expression of knox Genes in Embryo Sacs
The molecular identity of ig1 and the phenotype of ig1-mum leaves suggested that the ig1 embryo sac phenotype may be caused by ectopic expression of knox genes. To test this model, embryo sacs were isolated from homozygous wild-type W23 plants and homozygous ig1-O W23 mutant plants. The W23 inbred background was chosen because of its strong expression of the ig1-O mutant embryo sac phenotype. Ovules were hand-dissected from developing ears and subjected to digestion with cell wall–degrading enzymes. Embryo sacs were dissected out of ovules with some nucellus cells still attached.
In the ovules used, wild-type embryo sacs had recently cellularized and antipodal cells were not finished proliferating (similar in stage to the embryo sacs shown in Figures 7D to 7H). The ig1-O embryo sacs from similarly staged ovules were much more variable in size and developmental stage. Three replicates were isolated for each genotype, and each replicate contained 13 to 15 embryo sacs. Because of the small amount of RNA isolated from each sample, linear amplification of RNA was performed before quantitative real-time RT-PCR. As a positive control for the ability to detect RNA after linear amplification for each of the primer combinations used, RNA from rs2 leaves and 5-cm-long wild-type ears used previously and shown in Figure 8 was also subjected to linear amplification and RT-PCR in the same manner as the embryo sac samples. PCRs were repeated at least three times for each pair of PCR primers for each sample. Quantitative real-time PCR was terminated after 61 cycles.
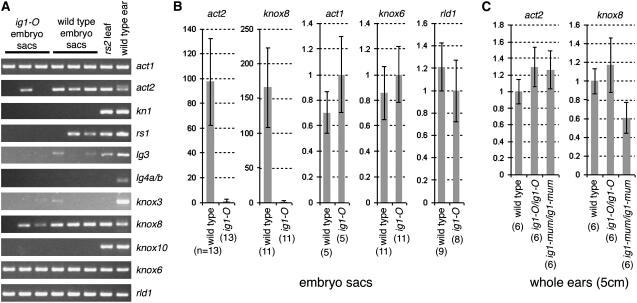
By the end of each run, cDNAs of ubi (data not shown) as well as actin1 (act1) and knox6 (a class II knox gene) (Figure 10A) were detected in all embryo sac samples. A second actin gene termed act2 was consistently expressed in wild-type embryo sacs but was rarely detected in ig1-O mutant embryo sacs. Most class I knox genes were not consistently amplified from either wild-type or ig1-O embryo sacs. kn1, gn1, lg4a/b, and knox10 were never detected in any embryo sac samples. rs1, lg3, and knox3 were seen in only a few PCR samples (2 of the 9 to 12 replicates) after 61 cycles, and for rs1 and lg3 only in the wild type. The only class I knox gene consistently detected in either wild-type or ig1-O embryo sacs was knox8. Interestingly, knox8 RNA levels were greatly reduced in ig1-O embryo sacs compared with wild-type sacs. No knox genes were overexpressed in ig1-O embryo sacs. Additionally, the organ polarity gene rolled leaf1 was examined in these samples and detected in all mutant and wild-type samples. Expression of kn1 and lg3 was also examined in sections of wild-type and ig1-O embryo sacs using immunolocalization and in situ hybridization, respectively. Neither kn1 nor lg3 showed any difference in expression pattern between mutant and wild-type embryo sacs before or after cellularization (data not shown).
Figure 10.
Expression of Genes from Whole Embryo Sacs.
RNA was subjected to linear amplification and then used for quantitative real-time PCR.
(A) Products after the completion of real-time RT-PCR.
(B) Comparison of RNA levels of act2, knox8, act1, knox6, and rld1 in wild-type and ig1-O embryo sacs. Expression of ubi was used to normalize RNA levels between samples. Expression is given relative to that of ig1-O. Because of the dramatic difference between ig1-O and wild-type embryo sacs for act2 and knox8, these genes are compared using a log scale, whereas all others are presented on a linear scale.
(C) Relative levels of RNA of act2 and knox8 in developing wild-type, ig1-O, and ig1-mum ear primordia.
Error bars in (B) and (C) represent se. The number of replicates is given in parentheses below each column.
Quantitative analysis was performed for all of the genes—act1, knox6, rld1, act2, and knox8—that were consistently detected in all wild-type samples. Multiple PCR replicates were performed for each gene. All PCR replicates and biological samples of the same genotype were averaged together and normalized to ubi (Figure 10B). act1, knox6, and rld1 were expressed at similar levels in ig1-O and wild-type embryo sacs. By contrast, there was at least a 100-fold reduction in detectable act2 and knox8 message in ig1-O embryo sacs compared with wild-type sacs at this stage. The effect of ig1 mutations on act2 and knox8 did not occur in all tissues; mRNA levels of both genes were not significantly different in whole 5-cm-long ear primordia of ig1-O and ig1-mum homozygotes compared with wild-type primordia (Figure 10C). Additionally, act2 expression was not altered in ig1-mum flag leaves (Figure 8H).
DISCUSSION
The failure to limit proliferation in ig1 embryo sacs leads to a variety of structural defects, including the production of extra gametes and synergids. Additionally, the fertilization process is frequently abnormal, producing seeds with haploid embryos and embryos and endosperms derived from fertilization by different pollen tubes. These late defects have often been interpreted as resulting from the abnormal structure of ig1 embryo sacs rather than from a requirement for ig1 in the female gametes per se. The synergids are known to attract pollen tubes (Higashiyama et al., 2001; Huck et al., 2003; Rotman et al., 2003; Marton et al., 2005), and the presence of extra synergids may promote the entry of extra pollen tubes into the embryo sac, leading to heterofertilization. Alternatively, ig1 may directly prevent these abnormal events by inhibiting embryogenesis in the egg cell until fertilization and karyogamy occur.
Although the leaf phenotype of ig1 is incompletely penetrant, the molecular identity of the gene, its expression pattern in lateral organs of the flower, and the presence of the phenotype in two independent alleles demonstrate that this phenotype is a consequence of mutation in ig1. Genetic background differences, coupled with the subtle phenotype in W23, could explain why this phenotype had not been reported previously, although it may also be dependent on certain growing conditions. The incomplete penetrance of the leaf phenotype in a pure W23 line demonstrates that this variability need not be caused by segregation of modifiers but can reflect either an inherent variability in the development of ig1 leaves or a dependence of the phenotype on particular environmental conditions.
Both alleles affect only the last one or two vegetative leaves of the plant in all backgrounds tested. The absence of an effect of ig1-mum on most leaves of the plant may reflect redundancy with ial1, ial2, or ial3 or incomplete loss of ig1 function in ig1-mum. However, ial1, ial2, and ial3 are all expressed in flag leaves that have the phenotypes of ig1-mum, suggesting that the absence of expression of one of these genes is not responsible for the phenotype in ig1-mum flag leaves. Alternatively, there may be qualitative differences between these leaves that make the flag leaves more sensitive to a partial reduction in ig1/ial function. For example, the level of expression of lg4a/b is higher in wild-type flag leaves than in wild-type seedling leaves (Figure 8H).
AS2 is a nuclear protein that interacts physically with AS1 protein, suggesting a role for LOB domain genes in regulating transcription (in the case of AS2 in combination with AS1) (Iwakawa et al., 2002; Xu et al., 2003). IG1 protein (referred to as maize AS2) was recently shown to interact with RS2 in vitro, supporting the model that it also acts as a transcriptional regulator and is orthologous with AS2 (Phelps-Durr et al., 2005). It is not yet known whether AS1 and AS2 repress the transcription of knox genes directly or indirectly. Interestingly, regarding the role of ig1 in restricting the embryogenic potential of cells lacking a maternal or paternal genome, leaf sections from as2 mutants have an enhanced ability to develop autonomous shoots in vitro compared with wild-type leaves (Semiarti et al., 2001). To date, no effect of as2 on the Arabidopsis embryo sac has been reported.
AS2 mRNA is polarly localized to the adaxial domains of developing cotyledons (Iwakawa et al., 2002). Similarly, ig1 is expressed in the adaxial domains of lateral organs. This expression is strongest throughout the adaxial domain in early primordia. Later in organ development, this expression is restricted to the basal region and eventually is undetectable in older primordia. Loss of function of ig1 causes some of the cells in the adaxial domain of the midrib to adopt an abaxial fate. The subsequent juxtaposition of abaxial and adaxial domains leads to the outgrowth of ectopic flaps of leaf lamina flanking the midrib on the upper surface of leaves. These flaps are more commonly found in the basal portion of the leaf and are often associated with disruption of the proximal–distal axis and with distortion of the ligular region as well. Like as2 mutant leaves, ig1 flag leaves also have ectopic expression of knox genes.
In embryo sacs, ig1 is expressed at low levels as early as the one-nucleus stage. The early time of ig1 expression in embryo sacs is consistent with an early role for ig1 in embryo sac development. After studying the effects of ig1-O in an embryonic marker line, Enaleeva et al. (1998) concluded that ig1 acts as early as the first division of the embryo sac and affects the establishment of polarity and the subsequent progression of nuclear divisions and cytokinesis (Enaleeva et al., 1998). Later in embryo sac development, expression is even lower and is near the threshold for detection, but it appears to be higher at the chalazal end. The ig1 loss-of-function phenotype, like the leaf phenotype, may be caused by a partial loss of polarity in the embryo sac, and the later defects, including prolonged proliferation, are consequences of this defect.
However, not all aspects of the leaf and embryo sac phenotypes in ig1 are similar. There are no knox genes overexpressed in ig1-O mutant embryo sacs after cellularization, although ectopic expression of one or more knox genes in a brief period of embryo sac development cannot be completely ruled out. Two genes, knox8 and act2, have been identified, however, with reduced expression in ig1-O embryo sacs. The fact that ig1 does not affect these genes in the same way in other tissues suggests that they are not direct targets of ig1 but instead are associated with the morphological defects of ig1 embryo sacs. One explanation is that these genes are normally expressed in maturing embryo sacs and ig1-O delays the switch from proliferation to maturation programs, thereby suppressing their expression. Alternatively, knox8 and act2 may be expressed asymmetrically in the embryo sac, and this pattern is disrupted in ig1 mutants.
The molecular identity of ig1 and the phenotype of ig1 mutant leaves suggested that common mechanisms had been used in switching from proliferation to differentiation in gametophytic (embryo sac) and sporophytic (lateral organ primordia) tissues. ig1 function may reflect ancestral homology between gametophyte and sporophyte shoot development. In this case, an ig1-like LOB gene in conjunction with an ARP gene imposes determinacy by negatively regulating knox genes in both gametophyte and sporophyte development. In primitive land plants, such as Physcomitrella patens, the gametophyte is the dominant phase of the life cycle and contains a leafy-shoot phase called the gametophore with at least superficial similarity to the vegetative shoots of seed plants (Cove and Knight, 1993). However, there is currently no evidence that ig1 interacts with the same partners or affects the expression levels of the same genes in embryo sacs and lateral organ primordia.
Another possible explanation is that ig1 function in the female gametophyte is a later adaptation and is not derived from ancestral gametophyte function. This adaptation may have occurred at any of several points in plant gametophyte evolution. For example, the adoption of ig1 for gametophyte development may have occurred in basal angiosperm lineages to limit the growth of the gametophyte, or it may have occurred in only a subset of angiosperm lineages. Basal angiosperms are thought to have been four-nuclei-type, and the eight-nuclei Polygonum type arose from an early duplication event along the micropylar–chalazal axis, making a domain of nuclei at the chalazal end as well as the micropylar end of the embryo sac. ig1 may function to restrict this to a single duplication event, and in ig1 mutants the process is reiterated. In basal angiosperms, ig1 genes may act to prevent this duplication entirely, and a slight modification in ig1 function or timing led to this duplication in plants with the Polygonum type of megagametogenesis. Additionally, ig1 function in the embryo sac may have been adopted to restrict proliferation in the postcellularization phase to the antipodal cells in angiosperms, such as maize, in which the antipodals proliferate. knox genes do not seem to be repressed by ig1 in embryo sacs as they are in leaves. Instead, ig1 likely controls the expression of other downstream genes that promote proliferation in the embryo sac. Whether these genes are also affected in mutant leaves is unknown.
Alternatively, the C-terminal domain of the IG1 protein may be critical for ig1 function in the embryo sac, whereas the LOB domain is critical for lateral organ function. The C-terminal domain of AS2 is different from that of IG1 and perhaps does not confer a function in the embryo sac, which would explain the lack of any reported effect of as2 mutations on embryo sac development. Interestingly, there appears to be a gene in rice and putatively also in maize that has this C-terminal domain but lacks an N-terminal LOB domain, suggesting that the C-terminal domain may have a separate molecular function. By contrast, there are no genes in Arabidopsis with high similarity to this domain.
The analysis of these processes in lower plant gametophytes with more extensive development will help elucidate whether ig1 function in leaves and embryo sacs arose through conservation of an ancestral mechanism in gametophytes and sporophytes or through the convergence of the mechanisms regulating sporophyte and gametophyte development. Finally, changes in the timing and expression pattern of ig1 orthologs within the gametophyte may have led to changes in the duration of the haploid phase of the plant life cycle in higher plants.
METHODS
Genetics
Maize (Zea mays) plants were grown in summer field conditions or in greenhouses in 16-h-light/8-h-dark conditions. To identify a new allele of ig1, active Mu; r1; W64A plants were crossed as males onto male-sterile ig1-O/ig1-O; R1-navajo (R1-nj) females in either a W23 inbred or a W23/W64A hybrid genetic background. The ig1-O mutation is on a W23 chromosome. A total of 60,000 F1 individuals were screened for male sterility (failure to extrude anthers). Male-sterile individuals were pollinated as females by standard r1; W64A males. Maternal haploids were distinguished based on their shorter stature, and maternal diploids and ig1-O sibling pollen contaminants were distinguished by homozygosity for R1-nj. Eight male-sterile individuals heterozygous for R1-nj were identified. The progeny of these plants were tested for markers linked to ig1 to identify individuals homozygous for W64A alleles (i.e., not carrying ig1-Ow23). Only one line had ig1 seed phenotypes when pollinated by standard W64A pollen, demonstrating the presence of a new ig1 mutant allele designated ig1-mum.
ig1 fine mapping was performed simultaneously in four different populations. ig1-Ow23 was crossed with one of the following inbred lines: Mo17, W64A, A158, and M14. The F1 plants were backcrossed as females by the appropriate inbred line. ig1-O individuals were identified as seeds with miniature endosperm or twin embryos. Recombinants were identified as individuals homozygous for the allele of the backcross parent (Mo17, W64A, A158, or M14). Rare androgenetic progeny that arise in ig1-O resemble recombinants, as they also have lost the W23 allele for that particular marker, but are distinguished by their apparent homozygosity at all loci. PCR conditions were as described previously (Evans and Kermicle, 2001).
Cloning of ig1
Identification of a Mu insertion in ig1-mum was initially identified by PCR using 5′-GTTGTGCTGGAGGATGGAGATGAC-3′ for the ig1 gene, 5′-TGGCGTTGGCTTCTMTG-3′ as a Mu primer, and 5′-GCCTCCATTTCGTCGAATCCC-3′ as a nested Mu primer. Amplification of insertion-mutagenized sites was performed using a protocol modified from that of Frey et al. (1998). Approximately 500 ng of DNA was digested per sample with 5 units of HinP1I (New England Biolabs), and then adaptors were ligated overnight using 1 unit of T4 DNA ligase (Fermentas). Sequences of the two strands of the adaptors were 5′-GACCACGCGTATCGATGTCGACGAGATGAGTCCTGAG-3′ and 5′-CGCTCAGGATCCACTCAT-3′. Ligation reactions were diluted 1:1, and 15 μL was used for a first round of PCR in a final volume of 25 μL. The 25-μL reaction mix consisted of 10 μM primers, 2.0 mM MgCl2, 250 μM each deoxynucleotide triphosphate, 10 mM Tris, pH 8.3, 50 mM KCl, 3.0% DMSO, and 0.5 units of Platinum Taq (Invitrogen). First-round PCR was performed with a primer for the adaptor, 5′-GACCACGCGTATCGATGTCGAC-3′, and a primer for the ends of the Mu elements, 5′-GAGAAGCCAACGCCAWCGCCTCC-3′, for 30 cycles of 1 min at 94°C, 30 s at 65°C, and 1 min at 72°C. PCRs were performed on a PTC-200 Thermal Cycler (MJ Research). The PCR products were diluted 1:10, and 2.8 μL was used in a 28-μL reaction with the same conditions as the first round except that amplification was performed with 10 cycles of 94°C for 30 s, 66°C for 30 s, and 72°C for 1 min followed by 30 cycles of 94°C for 30 s, 61°C for 30 s, and 72°C for 1 min. Nested primers for the second round of PCR were 5′-GACCACGCGTATCGATGTCGACGAG-3′ for the adaptor and a 5′ Hex-labeled Mu primer, 5′-GCCTCCATTTCGTCGAATCCC-3′. PCR products were separated on a 6% denaturing polyacrylamide gel, and the fluorescent bands were visualized on a Typhoon 8600 laser scanner/imager using 532-nm excitation and 580-nm emission. Bands of interest were excised from the gel, macerated, and allowed to elute overnight in TE (10 mM Tris and 1 mM EDTA, pH 8.0). PCR products were reamplified using Mu and adaptor primers and cloned into pGEM T-easy (Promega). Multiple clones were sequenced for each band selected.
To identify the lesion in the ig1-O allele, genomic DNA of ig1-O/+ heterozygote and its wild-type W23 progenitor was digested with EcoRI and probed with a fragment of the ig1 gene encompassing most of the LOB domain. The ig1-O mutant had a novel band of ∼1.6 kb in length. This band was excised from agarose gels and purified using the Qiagen gel extraction kit. The ig1-O fragment was amplified using inverse PCR according to the method of Cowperthwaite et al. (2002) except that the annealing temperature for the PCR was 67°C. Primers used were 5′-GCGGCCGCGGCTGCTGAGGAAGAAGA-3′ and 5′-CCCTTCCAGCGCGAGGACGCCGTGAACT-3′ for the first round of amplification and 5′-GCTGCTGAGGAAGAAGACGACGCCACGGTGATCA-3′ and 5′-CGGCTGCGTCGGCGTCATCTCCATCCT-3′ for the second round. The PCR product was cloned into pGEM T-easy and sequenced. The Hopscotch insertion was verified by PCR using a primer for the Hopscotch long terminal repeat, 5′-AAATCAAGGAGATCCTGTGCTATCTACGT-3′, and an ig1 primer upstream of the element, 5′-GTCATCTCCATCCTCCAGCACAAC-3′, and the complement of the Hopscotch primer and an ig1 downstream primer, 5′-AACTCTGCCATCGCTTGCGG-3′, to verify the 3′ end of the insertion.
Gene Expression Analysis
To determine the transcription start and stop sites as well as the intron–exon structure, 5′ and 3′ rapid amplification of cDNA ends was performed using the GeneRacer kit (Invitrogen) according to the manufacturer's directions. For the amplification of the 5′ end of the ig1 transcript, the first primer was 5′-GGCGCATGTCGGCCTCGTAGGCGAGGGAGTTCA-3′ and the nested primer was 5′-GCGGCCGCGGCTGCTGAGGAAGAAGA-3′. For the amplification of the 3′ end, the first primer was 5′-CGGCTGCGTCGGCGTCATCTCCATCCT-3′ and the nested primer was 5′-TCGGCGTCATCTCCATCCTCCAGCACAACCTACGA-3′. RNA was isolated from tissues using the Trizol reagent (Invitrogen). Genomic DNA was removed using the DNA-free kit (Ambion), and cDNA was synthesized using the Thermoscript RT-PCR system and priming with oligo(dT) (Invitrogen). For RT-PCR of ig1 cDNA, the primers used were 5′-GCTACGCCAAGGCCCAAGTGT-3′ for exon 1 and 5′-GCACCGACGAAGCCATCC-3′ for exon 2. RT-PCR of ial1 was performed with the same exon 2 primer as ig1 and 5′-CGAGTGCCCAGGCATTCTTCAG-3′ for exon 1. RT-PCR primers for ial2 were 5′-GAACTCGCCTGCGCCACCTAC-3′ and 5′-AGCTCCACTGCCTTCACCTTCGTAG-3′, and primers for ial3 were 5′-CGCCGTCAATTCGCTCGTTTAC-3′ and 5′-TCGTGGAGTACCCGGCATTGATAA-3′. Primers for rs1, lg3, and ubi were the same as those described by Schneeberger et al. (1998). Primers for kn1 were 5′-TGGGAACAGCGGCGGTAGC-3′ for exon 1 and 5′-ATCTCCGTCAGCCTCGCCGA-3′ for exon 2. Primers for lg4a and lg4b were 5′-GCCGCAAGGTGGGMGCG-3′ and 5′-GGCAGTAMGYSTCCATGAAYTCGTCG-3′. Primers for knox3 were 5′-GCTGCAGCTGCAGATAAAACTGGAGT-3′ and 5′-TCTTCTCCATCTCCGAGGGGTAGG-3′. Primers for knox6 were 5′-GGCCATACCCAACTGAAGACGACA-3′ and 5′-TGCTCTGGATCAATATCACCTTTTCTTCT-3′. Primers for knox8 were 5′-GACCCGGAGCTCGACCAGTTCAT-3′ and 5′-ACGACCCGGCGACCTCACAGTTAC-3′. Primers for knox10 were 5′-CCCGCCAGAAGCTCCTCCACT-3′ and 5′-CCGTGCTGGCCCATCCTGTAAT-3′. Primers for act2 were 5′-CCTTTCGAAAGTCCTTGCGTCACA-3′ and 5′-GCGAAACCGGCCTTGACCAT-3′. For some genes, at least one of the primers has a binding site that spans the junction between two exons, so it cannot anneal to genomic DNA, thus preventing genomic DNA amplification. For PCR, 40 ng of cDNA was amplified for 26 to 36 cycles of 94°C for 30 s, 65°C for 30 s, and 72°C for 30 s using 1 unit of Taq DNA polymerase (Promega), 200 μM deoxynucleotide triphosphate, and 0.2 μM of each primer.
Identification of Other LOB Genes and Phylogenetic Analysis
ial and other LOB domain genes from maize and other species were identified by searching EST and Genome Survey Sequence databases at Gramene (www.gramene.org) for rice (Oryza sativa); The Institute for Genomic Research (http://maize.tigr.org), Iowa State University (magi.plantgenomics.iastate.edu), and PlantGDB (www.plantgdb.org) for maize; and PlantGDB for other species. The alignment of the full-length proteins of ig1 and related genes was done using ClustalW. Bayesian phylogenetic analysis was conducted on a Clustal-aligned subset of the full LOB domain gene data set (see Supplemental Figure 1 online) using MrBayes version 3.1.2 (Huelsenbeck and Ronquist, 2001). Default settings were used and allowed to run for 100,000 generations. Trees were summarized after discarding the first 25,000 generations.
Embryo Sac Isolation and Gene Expression
Ovules from homozygous wild-type W23 and homozygous ig1-O W23 plants were dissected from flowers with silk of ∼2 cm in length. Three replicates were isolated for each sample. Thirteen to 15 ovules were collected from a single row of flowers to encompass a few different stages of ovule development. Ovules were placed in an enzyme mix of 0.75% pectinase, 0.5% cellulase, and 0.25% pectolyase in 1× PBS for 1 h. Embryo sacs with some attached nucellus cells were liberated from ovules with tungsten needles and, using a Pasteur pipette, placed immediately in extraction buffer RLT (Plant RNeasy kit; Qiagen). Tissue was homogenized on a Mixer Mill300 (Qiagen) with one tungsten-carbide bead in each tube. Nine to 30 ng of total RNA was then subjected to linear amplification using the RiboAmp kit (Arcturus) according to the manufacturer's instructions. After amplification, cDNA was made from 150 ng of amplified antisense RNA using random primers and the Thermoscript RT-PCR system (Invitrogen). cDNA products were brought to a final volume of 100 μL, and 1 μL of cDNA was amplified for 61 cycles of 94°C for 15 s, 65°C for 30 s, and 72°C for 30 s. PCR was performed using the DyNAmo enzyme mix (New England Biolabs) on an Opticon2 thermocycler (MJ Research) to allow detection of amplicons in real time. Primers were primarily the same as those used for leaf tissue except that those for kn1, rs1, lg3, lg4, and knox3 were replaced with primers near the 3′ end of transcripts because the RiboAmp products are biased against long fragments and 5′ ends. New primers for embryo sac expression are as follows: for kn1, 5′-TCAGAAGGTGGCACTGGCTGAGTCTAC-3′ and 5′-GGTACAGCCCGCCGTCGTTGAT-3′; for rs1, 5′-CACTACAAGTGGCCGTACCCTTCAGAGAC-3′ and 5′-ACGGCCCGTCCATGTACAGAGCA-3′; for lg3, 5′-GTCCAATTGTGCCTGTATCGAAGTAGAGT-3′ and 5′-CTACGGAGGAAGACAAGGTGAGGCT-3′; for lg4, 5′-GGCTGCGGTCCGAGTTCCTG-3′ and 5′-TGTCCTCCGACGGCTTCCAAT-3′; for knox3, 5′-CGGAGTCGACGGGGCTGGA-3′ and 5′-ATCCGGCCGGGCGTAGAACA-3′; for rld1, 5′-CTGTGCTGCTCCTTTAAGGAGAAACCTA-3′ and 5′-AGGTACGCGTAGCCCTGTTCCATT-3′; and for act1, 5′-GAAGTGCGACGTCGATATCAGGAAGGA-3′ and 5′-TGAGATCCACATCTGTTGGAAGGTGCTC-3′. Quantitative analysis of PCR results was performed using the qBase analysis package for Microsoft Excel.
Histology
Rows of developing florets of different stages were dissected from ears, fixed, embedded in paraffin, and sectioned according to McSteen and Hake (2001). ig1 cDNA from codon 228 to the end of the 3′ untranslated region was cloned in both orientations in pGEM T-easy (Promega) to get both sense and antisense transcripts driven by the T7 promoter. For both sense and antisense probes, plasmids were digested with SalI, and in vitro transcription reactions performed using the MAXIscript T7 kit (Ambion) according to the manufacturer's instructions with digoxigenin-11-UTP to label probes. In situ hybridizations were performed according to Long and Barton (1998). For histology of leaves, samples were fixed, embedded in paraffin, sectioned, and rehydrated as done for in situ hybridization of florets; sections were then stained for 1 min in 0.2% Toluidine Blue-O.
Accession Numbers
Sequence data from this article can be found in the GenBank data library under accession numbers EF081454 (ig1) and EF081455 (ial1).
Supplemental Data
The following material is available in the online version of this article
Supplemental Figure 1. Alignment of Proteins Used to Produce the Phylogeny of ig1 and Closely Related LOB Genes.
Supplementary Material
Acknowledgments
I thank Jerry L. Kermicle for assistance initiating this project and for valuable discussions. I thank Sarah Hake for anti-KNOTTED1 antibody and M. Kathryn Barton for help with in situ hybridizations. This project was supported by a grant from the National Science Foundation to M.M.S.E.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Matthew M.S. Evans (mmsevans@stanford.edu).
Online version contains Web-only data.
References
- Acharya, A., Ruvinov, S.B., Gal, J., Moll, J.R., and Vinson, C. (2002). A heterodimerizing leucine zipper coiled coil system for examining the specificity of a position interactions: amino acids I, V, L, N, A, and K. Biochemistry 41 14122–14131. [DOI] [PubMed] [Google Scholar]
- Banks, J.A. (1999). Gametophyte development in ferns. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 163–186. [DOI] [PubMed] [Google Scholar]
- Bortiri, E., Chuck, G., Vollbrecht, E., Rocheford, T., Martienssen, R., and Hake, S. (2006). ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971. [DOI] [PubMed] [Google Scholar]
- Cove, D.J., and Knight, C.D. (1993). The moss Physcomitrella patens, a model system with potential for the study of plant reproduction. Plant Cell 5 1483–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowperthwaite, M., Park, W., Xu, Z., Yan, X., Maurais, S.C., and Dooner, H.K. (2002). Use of the transposon Ac as a gene-searching engine in the maize genome. Plant Cell 14 713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deppmann, C.D., Acharya, A., Rishi, V., Wobbes, B., Smeekens, S., Taparowsky, E.J., and Vinson, C. (2004). Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: A comparison to Homo sapiens B-ZIP motifs. Nucleic Acids Res. 32 3435–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews, G.N., and Yadegari, R. (2002). Development and function of the angiosperm female gametophyte. Annu. Rev. Genet. 36 99–124. [DOI] [PubMed] [Google Scholar]
- Ebel, C., Mariconti, L., and Gruissem, W. (2004). Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429 776–780. [DOI] [PubMed] [Google Scholar]
- Enaleeva, N.K., Ot'kalo, O.V., and Tyrnov, V.S. (1998). Phenotypic expression of the ig mutation in megagametophyte of the maize line Embryonic marker. Genetika 34 259–265. [Google Scholar]
- Evans, M.M.S., and Kermicle, J.L. (2001). Interaction between maternal effect and zygotic effect mutations during maize seed development. Genetics 159 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey, M., Stettner, C., and Gierl, A. (1998). A general method for gene isolation in tagging approaches: Amplification of insertion mutagenised sites (AIMS). Plant J. 13 717–721. [Google Scholar]
- Gaut, B.S., and Doebley, J.F. (1997). DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimanelli, D., Leblanc, O., Perotti, E., and Grossniklaus, U. (2001). Developmental genetics of gametophytic apomixis. Trends Genet. 17 597–604. [DOI] [PubMed] [Google Scholar]
- Guo, F., Huang, B.Q., Han, Y., and Zee, S.Y. (2004). Fertilization in maize indeterminate gametophyte1 mutant. Protoplasma 223 111–120. [DOI] [PubMed] [Google Scholar]
- Higashiyama, T., Yabe, S., Sasaki, N., Nishimura, Y., Miyagishima, S., Kuroiwa, H., and Kuroiwa, T. (2001). Pollen tube attraction by the synergid cell. Science 293 1480–1483. [DOI] [PubMed] [Google Scholar]
- Huang, B.Q., and Sheridan, W.F. (1996). Embryo sac development in the maize indeterminate gametophyte1 mutant: Abnormal nuclear behavior and defective microtubule organization. Plant Cell 8 1391–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck, N., Moore, J.M., Federer, M., and Grossniklaus, U. (2003). The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130 2149–2159. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck, J.P., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17 754–755. [DOI] [PubMed] [Google Scholar]
- Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H., Hasebe, M., Soma, T., Ikezaki, M., Machida, C., and Machida, Y. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43 467–478. [DOI] [PubMed] [Google Scholar]
- Jackson, D., Veit, B., and Hake, S. (1994). Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120 405–413. [Google Scholar]
- Johnson, M.A., von Besser, K., Zhou, Q., Smith, E., Aux, G., Patton, D., Levin, J.Z., and Preuss, D. (2004). Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez, M.T., Kui, J.S., Thomas, J., Heller, B.A., and Timmermans, M.C. (2004). microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428 84–88. [DOI] [PubMed] [Google Scholar]
- Kermicle, J.L. (1969). Androgenesis conditioned by a mutation in maize. Science 166 1422–1424. [DOI] [PubMed] [Google Scholar]
- Kermicle, J.L. (1971). Pleiotropic effects on seed development of the indeterminate gametophyte gene in maize. Am. J. Bot. 58 1–7. [Google Scholar]
- Kermicle, J.L. (1994). Indeterminate gametophyte (ig): biology and use. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 388–393.
- Kindiger, B., and Hamann, S. (1993). Generation of haploids in maize: A modification of the indeterminate gametophyte (ig) system. Crop Sci. 33 342–344. [Google Scholar]
- Li, H., Xu, L., Wang, H., Yuan, Z., Cao, X., Yang, Z., Zhang, D., Xu, Y., and Huang, H. (2005). The putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and MicroRNA165/166 in Arabidopsis leaf development. Plant Cell 17 2157–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, B.-Y. (1978). Structural modifications of the female gametophyte associated with the indeterminate gametophyte (ig) mutant in maize. Can. J. Genet. Cytol. 20 249–257. [Google Scholar]
- Lin, B.-Y. (1981). Megagametogenetic alterations associated with the indeterminate gametophyte (ig) mutation in maize. Rev. Bras. Biol. 41 557–563. [Google Scholar]
- Lin, B.-Y. (1984). Ploidy barrier to endosperm development in maize. Genetics 107 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.A., and Barton, M.K. (1998). The development of apical embryonic pattern in Arabidopsis. Development 125 3027–3035. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69. [DOI] [PubMed] [Google Scholar]
- Marton, M.L., Cordts, S., Broadhvest, J., and Dresselhaus, T. (2005). Micropylar pollen tube guidance by egg apparatus 1 of maize. Science 307 573–576. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411 709–713. [DOI] [PubMed] [Google Scholar]
- McSteen, P., and Hake, S. (2001). barren inflorescence2 regulates axillary meristem development in the maize inflorescence. Development 128 2881–2891. [DOI] [PubMed] [Google Scholar]
- Moore, G., Devos, K.M., Wang, Z., and Gale, M.D. (1995). Cereal genome evolution. Grasses, line up and form a circle. Curr. Biol. 5 737–739. [DOI] [PubMed] [Google Scholar]
- Muehlbauer, G.J., Fowler, J.E., and Freeling, M. (1997). Sectors expressing the homeobox gene liguleless3 implicate a time-dependent mechanism for cell fate acquisition along the proximal-distal axis of the maize leaf. Development 124 5097–5106. [DOI] [PubMed] [Google Scholar]
- Murgia, M., Huang, B.-Q., Tucker, S.C., and Musgrave, M.E. (1993). Embryo sac lacking antipodal cells in Arabidopsis thaliana (Brassicaceae). Am. J. Bot. 80 824–838. [Google Scholar]
- Nelson, J.M., Lane, B., and Freeling, M. (2002). Expression of a mutant maize gene in the ventral epidermis is sufficient to signal a switch of the leaf's dorsoventral axis. Development 129 4581–4589. [DOI] [PubMed] [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127 5523–5532. [DOI] [PubMed] [Google Scholar]
- Pagnussat, G.C., Yu, H.J., Ngo, Q.A., Rajani, S., Mayalagu, S., Johnson, C.S., Capron, A., Xie, L.F., Ye, D., and Sundaresan, V. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132 603–614. [DOI] [PubMed] [Google Scholar]
- Phelps-Durr, T.L., Thomas, J., Vahab, P., and Timmermans, M.C. (2005). Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17 2886–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotman, N., Rozier, F., Boavida, L., Dumas, C., Berger, F., and Faure, J.E. (2003). Female control of male gamete delivery during fertilization in Arabidopsis thaliana. Curr. Biol. 13 432–436. [DOI] [PubMed] [Google Scholar]
- Schichnes, D., Schneeberger, R., and Freeling, M. (1997). Induction of leaves directly from leaves in the maize mutant Lax midrib1-O. Dev. Biol. 186 36–45. [DOI] [PubMed] [Google Scholar]
- Schichnes, D.E., and Freeling, M. (1998). Lax midrib1-O, a systemic, heterochronic mutant of maize. Am. J. Bot. 85 481–491. [PubMed] [Google Scholar]
- Schneeberger, R., Tsiantis, M., Freeling, M., and Langdale, J.A. (1998). The rough sheath2 gene negatively regulates homeobox gene expression during maize leaf development. Development 125 2857–2865. [DOI] [PubMed] [Google Scholar]
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128 1771–1783. [DOI] [PubMed] [Google Scholar]
- Shuai, B., Reynaga-Pena, C.G., and Springer, P.S. (2002). The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 129 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, N.R., Williams, R.E., and Hake, S. (1993). Overexpression of the maize homeo box gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fates. Genes Dev. 7 787–795. [DOI] [PubMed] [Google Scholar]
- Theodoris, G., Inada, N., and Freeling, M. (2003). Conservation and molecular dissection of ROUGH SHEATH2 and ASYMMETRIC LEAVES1 function in leaf development. Proc. Natl. Acad. Sci. USA 100 6837–6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans, M.C., Hudson, A., Becraft, P.W., and Nelson, T. (1999). ROUGH SHEATH2: A Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284 151–153. [DOI] [PubMed] [Google Scholar]
- Tsiantis, M., Schneeberger, R., Golz, J.F., Freeling, M., and Langdale, J.A. (1999). The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 284 154–156. [DOI] [PubMed] [Google Scholar]
- Waites, R., and Hudson, A. (1995). phantastica: A gene required for dorsiventrality of leaves in Antirrhinum majus. Development 121 2143–2154. [Google Scholar]
- Ware, D.H., et al. (2002). Gramene, a tool for grass genomics. Plant Physiol. 130 1606–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, S.E., Habera, L.F., and Wessler, S.R. (1994). Retrotransposons in the flanking regions of normal plant genes: A role for copia-like elements in the evolution of gene structure and expression. Proc. Natl. Acad. Sci. USA 91 11792–11796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L., Xu, Y., Dong, A., Sun, Y., Pi, L., and Huang, H. (2003). Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130 4097–4107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.