Abstract
We reevaluated a previously reported family with an X-linked mental retardation syndrome and attempted to identify the underlying genetic defect. Screening of candidate genes in a 10-Mb region on Xq25 implicated CUL4B as the causative gene. CUL4B encodes a scaffold protein that organizes a cullin-RING (really interesting new gene) ubiquitin ligase (E3) complex in ubiquitylation. A base substitution, c.1564C→T, converted a codon for arginine into a premature termination codon, p.R388X, and rendered the truncated peptide completely devoid of the C-terminal catalytic domain. The nonsense mutation also results in nonsense-mediated mRNA decay in patients. In peripheral leukocytes of obligate carriers, a strong selection against cells expressing the mutant allele results in an extremely skewed X-chromosome inactivation pattern. Our findings point to the functional significance of CUL4B in cognition and in other aspects of human development.
Mental retardation (MR) affects ∼2% of the human population and represents a major health care challenge.1 Although its etiology is very heterogeneous, the largest single category, accounting for ∼10% of all types of MR, is caused by mutations at various loci on the X chromosome and is called “X-linked mental retardation” (XLMR).2,3 XLMR can be divided into syndromic and nonsyndromic forms on the basis of the presence or absence of other symptoms and features. Since the early 1990s, genetic studies of families with XLMR have led to the identification of at least 61 X-linked loci that are responsible for MR.3 The identification of such genetic defects is invaluable in discovering genes that govern brain function and cognition development. It also makes diagnosis more accurate and genetics counseling more informed.
We identified a family with XLMR with five affected males in four sibships during a population survey for common hereditary diseases conducted in Zoucheng, Shandong Province, China, in 1990. Because of the presence of MR, short stature, and some other physical features that were suggestive of Smith-Fineman-Myers syndrome (SFMS), this family was initially reported as having putative SFMS.4 The causative gene was mapped by linkage analysis to an interval of 20 Mb between DXS8064 and DXS8050 on Xq25.5
The initially reported SFMS—together with Carpenter-Waziri syndrome, Chudley-Lowry syndrome, Juberg-Marsidi syndrome, and Holmes-Gang syndrome—was found to be caused by mutations in XH2 (also called ATRX and XNP)3,6 (MIM *300032), a gene that is located at Xq13 and encodes X-linked helicase 2. These syndromes are now collectively called “XLMR-hypotonic facies syndrome” (MIM #309580).3
Employing two polymorphic markers within the XH2 gene, we excluded linkage of the XH2 gene to the disease in the Zoucheng family with XLMR.7 This finding, together with the mapping of the causative gene to Xq25 instead of Xq13, suggests that cases in this family with XLMR probably represent a syndrome different from SFMS. Therefore, we revisited the family and reexamined the patients in 2005. An updated pedigree is shown in figure 1. In addition to the five patients reported elsewhere,4 individual IV-11, aged 8 years, was identified as a new patient. All adult patients showed mild to moderate MR and led their daily lives under the care and supervision of their parents and other relatives. None of the adult patients were able to speak a single word during their lifetimes. Deviating from other patients, IV-11 showed only mild MR and was able to speak several short sentences. No remarkable facial features were noticed. In particular, the small palpebral fissures, flat nasal bridge, micrognathia, and hypertelorism that are characteristic of SFMS were not obvious. However, several other physical features were distinct, including short stature, brachydactyly, large tongue (macroglossia), and a unique gait of walking with the toes pointing inward. Neuroimaging by magnetic resonance imaging and computed axial tomography scanning revealed no structural abnormalities. Interestingly, peripheral blood tests revealed that monocyte counts, ranging from 0.8×109 to 1.4×109 per liter in four patients tested, and their percentages, ranging from 10.9% to 17.9% in total white blood cells (WBC), were remarkably increased, whereas the total WBC counts were in the normal range. An updated summary of phenotypes of patients is listed in table 1. No abnormal phenotypes were noted in obligate carriers. Considering the exclusion of XH2 as the causative gene and the presence of some unique clinical features, we now propose that the disease in this family represents a novel syndrome.
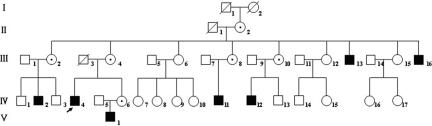
Figure 1. .
Updated pedigree of a family with XLMR. V-1 was aged 1 year at the time of examination and was found to carry the mutation. IV-4 was proband (arrow). Blackened symbols represent affected individuals, and symbols with dots represent carriers.
Table 1. .
A Summary of Phenotypes Observed in Patients of a Family with XLMR[Note]
| Measure | III-13 | III-16 | IV-2 | IV-4 | IV-11 | IV-12 |
| Age (years) | 39 | 31 | 30 | 26 | 8 | 18 |
| MR severity | Moderate | Moderate | Moderate | Moderate to mild | Mild | Moderate |
| Height (cm) and percentile | 152, <3rd | 159, <10th | 140, <1st | 160, <10th | 124, <50th | 158, <10th |
| OHCa (cm) and percentile | 53, <5th | 55, <50th | 52, <5th | 55, <50th | 53, <5th | 54, <50th |
| Restlessness | + | + | + | + | − | + |
| Absence of speech | + | + | + | + | − | + |
| Macroglossia | + | + | + | + | + | NE |
| Brachydactyly | + | + | + | + | + | NE |
| Abduction of small fingers | + | + | NE | + | + | NE |
| Gait with toes facing inward | + | + | + | + | + | + |
| Monocyte count (109/liter)b | 1 | 1.4 | NE | 1.3 | .8 | NE |
| Monocyte percentagec | 17.9 | 14.3 | NE | 17.4 | 10.9 | NE |
Note.— NE = not examined.
OHC = occipitofrontal head circumference.
Normal range is 0.3–0.8.
Normal range is 4%–10%.
When additional polymorphic markers became available, the candidate interval was narrowed to a region of 10.26 Mb between XSTR3 and XSTR4.8 More than 60 genes are located in this region (UCSC Genome Browser). The search for the disease gene was performed by sequencing the genes that had been previously associated with MR or that were expressed in brain within the candidate interval (fig. 2A). After excluding the first 13 genes, we found a nonsense mutation in the 14th gene tested, CUL4B (cullin 4B transcript [GenBank accession number NM_003588]). CUL4B is broadly expressed in most human tissues and is highly expressed in brain, testis, prostate, colon, and leukocytes, as determined by northern blot analysis.9 This observation was confirmed by semiquantitative RT-PCR (data not shown). CUL4B consists of 22 exons and encodes 913 aa. With the use of intronic primers (table 2), all exons of the CUL4B gene were amplified and sequenced for two patients of the family. A base substitution at 1564 in exon 9, c.1564C→T, converted a codon for arginine (CGA) to a termination codon (TGA), p.R388X (fig. 2B and 2C). By screening this mutation in family members, by direct sequencing of exon 9, we found that this mutation cosegregated with the disease phenotype in the family. We also detected the mutation in a boy aged 1 year, V-1 (fig. 1), and his mother, IV-6, which suggests that this boy is affected and that his mother is a carrier. The boy, now aged 2 years, appeared to be delayed in mental development and could not speak or walk. To confirm the 1564C→T mutation in the family and to detect it in controls, a modified primer was used to create a restriction endonuclease (BclI) site to distinguish 1564C and 1564T alleles. With the use of PCR-RFLP assay, the sequencing results of all family members were confirmed (fig. 2D and table 3), and no 1564T allele was detected among 312 X chromosomes from 264 unrelated, unaffected Chinese controls (216 males and 48 females) (table 3). Thus, it is unlikely that this mutation represents a polymorphism.
Figure 2. .
Mutation in CUL4B that causes XLMR. A, Minimal candidate region of 10.26 Mb between XSTR3 and XSTR4. The candidate genes screened are shown at the bottom. The location of CUL4B is highlighted. B, Partial sequence chromatograms of exon 9 of CUL4B in a patient and a healthy male from the family. The normal and altered nucleotides at 1564 are labeled with an asterisk (*). C, Genomic structure of CUL4B and schematic representation of CUL4B protein. The position of the introduced stop codon is indicated by a red arrow. The position of serine-rich and cullin domains are indicated. D, Confirmation of the c.1564C→T mutation by PCR-RFLP assay. All patients and carriers exhibited the 172-bp fragment representing the 1564T allele. Part of exon 9 of CUL4B was amplified from genomic DNA and was digested with BclI, to distinguish between the 1564C allele (absence of BclI site) and the 1564T allele (presence of BclI site). Sizes of the resulting restriction fragments are indicated. The primers used in this experiment were 5′-CCAATCATCATGCTTCTCAACT-3′ (forward) and 5′-CGGTTAGTTTCTTCCAATGATC-3′ (reverse).
Table 2. .
Primers Used for PCR Amplification of CUL4B Exons
| Primer (5′→3′) |
||||
| Exon | Forward | Reverse | Product Size (bp) |
Annealing Temperature (°C) |
| 1 | GGGAATAACGAAACCTAGCA | CAGACTCACCCTTACCCAAG | 801 | 48 |
| 2 | CATTCTCCTGCCTAAGCCTCCT | CCTGCCAAATCAGCTTGAAATG | 542 | 51 |
| 3 | CCCTCCACCTCATTGATTAT | CTTTCCTTCTCGTCCTCTGT | 829 | 48 |
| 4 | TTAGAAGGGAACCAAAAGTC | GAGAAAAACCTCACCTTCAA | 464 | 48 |
| 5 | GGCTGAAAAGTCAAAATGTG | CAGCCCAAGTAGGATTTTCT | 580 | 51 |
| 6 | AAATGTCACCTGTACCCCAAT | TGCCCTATCAGATTTCCACTT | 560 | 51 |
| 7 | ACTTTAGTCCCCTCGGCTAAC | AATTATATGAGCGTCCCCTTG | 598 | 48 |
| 8 | GAGATGTGCTTCCTTCAAAC | GCAAAATGAGTTTCCTGTGT | 474 | 51 |
| 9/10 | CCCCTAATAATCGAATCCTT | TGAAGGAAGCACATCTCATA | 849 | 48 |
| 11 | CTTGGGATTAGAAGTGCTTG | TTGCCTGTCAGTTCCTTTAT | 751 | 48 |
| 12 | AGCTACCATTCTCAGGTTGA | CAGAAAGATGGTTGGGTTTA | 766 | 51 |
| 13 | TAGTGTCCAACGTACTGTCTTT | TGTCCTCATTTCACTATCAGC | 689 | 48 |
| 14 | CCCCCAATGAATTAAACAC | TTGATCTTCCTGACTTTGGA | 393 | 48 |
| 15/16 | CCTCAGTTGCCATGAATTAC | GGCACAAAAATGGATCTCTA | 933 | 48 |
| 17 | AGCAGTTGGTATTTGGTGTT | TGATCCTTGTATGCCTTTGT | 595 | 48 |
| 18 | GGAAAGACCACACACCTAAAA | ATGTTCAGAGAGAGCAAAAGG | 839 | 48 |
| 19 | CCTGAGCCATGATTGTACC | GATTTCTTGCTGCTTGTGAT | 600 | 51 |
| 20 | TACAACCACTCTCCCAAACA | CCATTGGCTAGAACAAAGAA | 498 | 48 |
| 21 | AGGGTTTGGCTATCTACCTC | AGGATTTTTCTCTGTCAGCA | 920 | 51 |
| 22 | CAAGTCAAGGGTGGTTTTTC | GCCGAAACTGTTGTCTTTTT | 653 | 51 |
Table 3. .
1564C→T Mutation in Patients, Carriers, and Controls[Note]
| Group | No. of Subjects | Nucleotide at 1564a |
| Pedigree with XLMR: | ||
| Patients | 7 | T |
| Carriers | 6 | T/C |
| Other members | 9 | C |
| Controls: | ||
| Males | 216 | C |
| Females | 48 | C |
mRNAs that contain premature termination codons are usually eliminated by a process called “nonsense-mediated decay” (NMD).10 To determine whether NMD also occurs in individuals carrying the 1564C→T mutation, we measured the mRNA levels of CUL4B in peripheral leukocytes of patients, carriers, and controls. As shown in table 4, only ∼30% of the mRNA, a 70% reduction, was retained in the patients when compared with the controls, suggesting that the mRNA was readily eliminated in those patients, possibly via the NMD mechanism. To check whether the reduced mRNA level was due to an altered splicing pattern, we performed RT-PCR, with different primer sets flanking the mutation site, and sequenced the PCR products. No altered splicing sites were detected in the mRNA extracted from peripheral leukocytes of patients (data not shown). Thus, most likely, the CUL4B mRNA in patients was being degraded via NMD. The reduced CUL4B mRNA level in patients substantiates the above notion that 1564C→T is a mutation with functional consequence, instead of a polymorphism.
Table 4. .
Reduced CUL4B mRNA Levels in Patients, but Not in Carriers, with the 1564C→T Mutation[Note]
| Group | No. of Subjects | Average ΔCt (CUL4B−18S rRNA) |
ΔCt−ΔCt ofUnaffected Females | CUL4B mRNA Ratioa |
| Patients | 5 | 12.539 ± .315 | 1.732 ± .315 | .303 |
| Carriers | 4 | 10.680 ± .397 | −.137 ± .397 | 1.099 |
| Unaffected females | 3 | 10.816 ± .572 | .000 ± .572 | 1.000 |
| Unaffected males | 3 | 10.669 ± .326 | −.147 ± .326 | 1.107 |
Note.— Total RNA, extracted from peripheral blood cells, was reverse transcribed into cDNA. Real-time PCR was performed using an ABI PRISM 7500 Sequence Detection System (Applied Biosystems). 18S rRNA was used as endogenous control. All reactions were run in triplicates. TaqMan probe and primer oligonucleotide sequences are as follows: CUL4B forward primer 5′-TGCTGCTCAGGAGGTCAGATC-3′, reverse primer 5′-TGGAATCAAAGTCTTCTCTCTCGTT-3′, probe 5′-FAM-TAATACCAGCACCACTCCGCCCACCT-TAMRA-3′; 18S rRNA forward primer 5′-CGGCTACCACATCCAAGGAA-3′, reverse primer 5′-GCTGGAATTACCGCGGCT-3′, probe 5′-FAM-TGCTGGCACCAGACTTGCCCTC-TAMRA-3′. Results were obtained as threshold cycle (Ct) values ± SD, which are inversely proportional to the starting template copy number.
mRNA ratio of each group to unaffected females.
Interestingly, the CUL4B mRNA levels in carriers were as high as those in unaffected individuals (table 4). This is in contrast to a 35% reduction expected in carriers if X chromosomes were randomly inactivated and cells expressing either the 1564C allele or the 1564T allele had equal proliferation capacity. We speculated that the maintenance of a high CUL4B mRNA level in carriers was probably achieved by a selection against cells expressing the 1564T allele, the allele associated with NMD. To test this hypothesis, we evaluated the X-chromosome inactivation pattern in carriers and unaffected females, by characterizing the methylation status of a HpaII site near the highly polymorphic CAG repeats in the androgen receptor gene (AR [MIM *313700]).11 The CAG polymorphism distinguishes between the maternal and paternal alleles. We found that X-chromosome inactivation was extremely skewed in all the informative carriers tested (fig. 3A). In addition, only the wild-type allele was detected in leukocyte cDNA from those carriers (fig. 3B), indicating that the mutant allele is completely silenced. This finding suggests that leukocytes lacking CUL4B are strongly selected against in vivo. Therefore, CUL4B probably plays a critical role in normal hematopoiesis. The elevation in the monocyte counts may represent one consequence of CUL4B deficiency.
Figure 3. .
X-chromosome inactivation that was extremely skewed in leukocytes of the carriers with the CUL4B mutation. A, Methylation status of an HpaII site near the CAG repeat of AR was analyzed. U = undigested; D = digested with HpaII. Methylated DNA on the inactive X chromosome is resistant to HpaII digestion and thus can be PCR amplified with primers flanking the restriction site. DNA on the active X chromosome, as in patient III-16, is cleavable by HpaII and gives no PCR products. X-chromosome inactivation pattern was measured by densitometry as the ratio between the PCR products from two X chromosomes and was scored as “random” (ratios 50:50–80:20) or “skewed” (ratios >80:20–95:5). When only one band was visible, the pattern was scored as “extremely skewed” (ratio >95:5). Random X inactivation was observed in unaffected females III-6 (ratio 51:49), III-12 (ratio 52:48), and III-15 (59:41). X inactivation was extremely skewed toward one chromosome in CUL4B carriers III-2, III-4, III-8, and III-10, in which only one allele was visible. B, Chromatograms of the partial sequences from genomic DNA (gDNA) and leukocyte cDNA in the carriers. Although heterozygous status was observed in genomic DNA from the carriers (left panel), only the wild-type allele was detected in their leukocyte cDNA (right panel). The normal and altered nucleotides at 1564 are labeled with an asterisk (*).
CUL4B belongs to a family of proteins that are characterized by the presence of a highly conserved globular C-terminal domain and a variable N-terminal helical domain.12 Each member of the cullin family can function as a scaffold in organizing a cullin-RING (really interesting new gene) ubiquitin ligase complex that recruits and ubiquitylates proteins for proteosomal degradation. The C-terminal domain, together with a RING protein, forms a catalytic core that covalently attaches ubiquitin to the targeted protein. The N-terminal domain, together with proteins that function as adaptors and receptors, recognizes and recruits protein substrates. The cullin family consists of at least seven members in mammals.12 Although Cul4 presents as a single gene in Schizosaccharomyces pombe, Caenorhabditis elegans, and Drosophila melanogaster, it has two closely related paralogs, CUL4A and CUL4B, in mammals. A recent study of human DDB1-CUL4A-ROC1 machinery confirmed that CUL4A adopts a structure similar to those of other cullins. As an adaptor, DDB1 binds cullin scaffold through one β-propeller domain and uses a separate double-β-propeller fold for substrate presentation.13 WD40-repeat proteins can bind the double-β-propeller fold and serve as a substrate-recruiting module of E3. The C-terminus of CUL4A, like those of other cullins, forms a catalytic core. Because of the high degree of homology between CUL4A and CUL4B, it can be assumed that CUL4B would adopt a similar structure. As a consequence of the 1564C→T mutation, the truncated peptide would retain only less than half of the whole protein length at the N-terminus, leaving the C-terminal domain completely abolished (fig. 2C). Since the C-terminus is required for the formation of catalytic core, the truncated peptide, if it is to be synthesized, will have no function. As a matter of fact, the mRNA level of the mutant CUL4B allele was greatly reduced in the patients (table 4), which would result in a very limited production, if any, of the truncated peptides. In addition, no transcripts due to alternative splicing were detected (data not shown). In light of these observations, we propose that the 1564C→T mutation represents a null mutation.
Our study linked a mutation in CUL4B to a human genetic disorder. Patients lacking CUL4B exhibit structural or functional abnormalities in multiple systems, notably in CNS, skeleton, and hematopoiesis. This indicates that CUL4B plays a critical role in many aspects of human development. Indeed, CUL4B was shown to be expressed in several human tissues examined.9
Unlike the presence in mammals of two closely related paralogs, CUL4A and CUL4B, only one gene, Cul4, is present in lower organisms. Also, CUL4A and CUL4B were recently identified in the same CUL4-DDB1-ROC1 complex that ubiquitylates histones H3 and H4.14 Such findings would suggest that CUL4A and CUL4B may have overlapping functions and compensate for each other. However, in contrast to the survival of patients with the CUL4B mutation into adulthood, knockout mice for Cul4a die in the embryonic stage,15 indicating that Cul4b is incapable of compensating for the lack of Cul4a in the mouse. The tissue-specific expression pattern also differs between CUL4A and CUL4B.9 More importantly, CUL4B has a much longer N-terminus than CUL4A, which may enable them each to bind unique adaptors and receptors in protein recognition and recruitment. A human genetic disorder caused by mutation in CUL4A has not been reported. A Cul4b-knockout mouse model, which is not yet available, will definitely help to achieve more insight into the functional difference between CUL4A and CUL4B during mammalian development and will answer the question of whether the phenotypes exhibited by the patients are species specific.
Protein ubiquitination regulates the functions of a broad spectrum of protein substrates in diverse cellular pathways.16 Many human genetic disorders have been found to be caused by errors in ubiquitination and proteasomal degradation.17 For example, lack of expression of UBE3A, a gene that encodes an E3 ubiquitin-protein ligase, causes Angelman syndrome, which is characterized by MR, ataxia, absence of speech, and other features.18 Opitz syndrome, which is associated with midline abnormalities—such as cleft lip, laryngeal cleft, heart defects, and hypospadias—and with MR, is caused by an impairment of the E3 ubiquitin ligase activity of the MID1 protein.19,20 Recently, UBE2A, a gene encoding ubiquitin-conjugating enzyme E2, has also been identified to be the cause of a type of syndromic XLMR.21 These findings suggest that ubiquitylation is critical in cognition as well as other aspects of human development.
Acknowledgments
We thank the patients and their families for their participation in this study. This work was supported by State Key Basic Research and Development Plan Program grant 2001CB510303, National Science Fund for Distinguished Young Scholars grant 30225020, and National Science Fund grant 30671163.
Web Resources
The accession number and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for CUL4B transcript [accession number NM_003588])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.gov/Omim/ (for XH2, XLMR-hypotonic facies syndrome, and AR)
- UCSC Genome Browser, http://genome.ucsc.edu/
References
- 1.Leonard H, Wen X (2002) The epidemiology of mental retardation: challenges and opportunities in the new millennium. Ment Retard Dev Disabil Res Rev 8:117–134 10.1002/mrdd.10031 [DOI] [PubMed] [Google Scholar]
- 2.Ropers HH, Hamel BC (2005) X-linked mental retardation. Nat Rev Genet 6:46–57 10.1038/nrg1501 [DOI] [PubMed] [Google Scholar]
- 3.Ropers HH (2006) X-linked mental retardation: many genes for a complex disorder. Curr Opin Genet Dev 16:260–269 10.1016/j.gde.2006.04.017 [DOI] [PubMed] [Google Scholar]
- 4.Wei J, Chen B, Jiang Y, Yang Y, Guo Y (1993) Smith-Fineman-Myers syndrome: report on a large family. Am J Med Genet 47:307–311 10.1002/ajmg.1320470302 [DOI] [PubMed] [Google Scholar]
- 5.Gong Y, Wei J, Shao C, Guo Y, Chen B, Guo C, Warman M (1999) [Mapping the gene responsible for Smith-Fineman-Myers syndrome to Xq25]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 16:277–280 [PubMed] [Google Scholar]
- 6.Villard L, Fontes M, Ades LC, Gecz J (2000) Identification of a mutation in the XNP/ATR-X gene in a family reported as Smith-Fineman-Myers syndrome. Am J Med Genet 91:83–85 [DOI] [PubMed] [Google Scholar]
- 7.Liu Q, Gong Y, Chen B, Guo C, Li J, Guo Y (2002) [Linkage analysis of X-linked nuclear protein gene in Smith-Fineman-Myers syndrome]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 19:22–25 [PubMed] [Google Scholar]
- 8.Liu QJ, Gong YQ, Li JX, Zhang XY, Gao GM, Guo YS (2004) [Fine mapping of Smith-Fineman-Myers syndrome and exclusion of GPC3, GPCR2 MST4 and GLUD2 as candidate genes]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 21:198–202 [PubMed] [Google Scholar]
- 9.Hori T, Osaka F, Chiba T, Miyamoto C, Okabayashi K, Shimbara N, Kato S, Tanaka K (1999) Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene 18:6829–6834 10.1038/sj.onc.1203093 [DOI] [PubMed] [Google Scholar]
- 10.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE (2004) Nonsense-mediated decay approaches the clinic. Nat Genet 36:801–808 10.1038/ng1403 [DOI] [PubMed] [Google Scholar]
- 11.Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW (1992) Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet 51:1229–1239 [PMC free article] [PubMed] [Google Scholar]
- 12.Petroski MD, Deshaies RJ (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6:9–20 10.1038/nrm1547 [DOI] [PubMed] [Google Scholar]
- 13.Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N (2006) Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443:590–593 [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y (2006) Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell 22:383–394 10.1016/j.molcel.2006.03.035 [DOI] [PubMed] [Google Scholar]
- 15.Li B, Ruiz JC, Chun KT (2002) CUL-4A is critical for early embryonic development. Mol Cell Biol 22:4997–5005 10.1128/MCB.22.14.4997-5005.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67:425–479 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- 17.Jiang YH, Beaudet AL (2004) Human disorders of ubiquitination and proteasomal degradation. Curr Opin Pediatr 16:419–426 10.1097/01.mop.0000133634.79661.cd [DOI] [PubMed] [Google Scholar]
- 18.Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL (1997) De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet 15:74–77 10.1038/ng0197-74 [DOI] [PubMed] [Google Scholar]
- 19.Quaderi NA, Schweiger S, Gaudenz K, Franco B, Rugarli EI, Berger W, Feldman GJ, Volta M, Andolfi G, Gilgenkrantz S, et al (1997) Opitz G/BBB syndrome, a defect of midline development, is due to mutations in a new RING finger gene on Xp22. Nat Genet 17:285–291 10.1038/ng1197-285 [DOI] [PubMed] [Google Scholar]
- 20.Trockenbacher A, Suckow V, Foerster J, Winter J, Krauss S, Ropers HH, Schneider R, Schweiger S (2001) MID1, mutated in Opitz syndrome, encodes an ubiquitin ligase that targets phosphatase 2A for degradation. Nat Genet 29:287–294 10.1038/ng762 [DOI] [PubMed] [Google Scholar]
- 21.Nascimento RM, Otto PA, de Brouwer AP, Vianna-Morgante AM (2006) UBE2A, which encodes a ubiquitin-conjugating enzyme, is mutated in a novel X-linked mental retardation syndrome. Am J Hum Genet 79:549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]