Abstract
Understanding patterns of gene-expression variation within and among human populations will provide important insights into the molecular basis of phenotypic diversity and the interpretation of patterns of expression variation in disease. However, little is known about how gene-expression variation is apportioned within and among human populations. Here, we characterize patterns of natural gene-expression variation in 16 individuals of European and African ancestry. We find extensive variation in gene-expression levels and estimate that ∼83% of genes are differentially expressed among individuals and that ∼17% of genes are differentially expressed among populations. By decomposing total gene-expression variation into within- versus among-population components, we find that most expression variation is due to variation among individuals rather than among populations, which parallels observations of extant patterns of human genetic variation. Finally, we performed allele-specific quantitative polymerase chain reaction to demonstrate that cis-regulatory variation in the lymphocyte adaptor protein (SH2B adapter protein 3) contributes to differential expression between European and African samples. These results provide the first insight into how human population structure manifests itself in gene-expression levels and will help guide the search for regulatory quantitative trait loci.
Gene expression is the primary mechanism by which information encoded in the genome is converted into developmental, morphological, and physiological phenotypes.1 Gene expression is also an important source of evolutionary change within and among species,2 and aberrant gene expression has been implicated in the pathogenesis of numerous diseases.3,4 Thus, understanding the amount, structure, and patterns of gene-expression variation is of fundamental importance to both biomedical research and evolutionary biology.5
Although it is well known that 85%–95% of human genetic variation is due to variation among individuals within a population, whereas 5%–15% is attributable to variation among populations,6–9 it remains unclear whether similar levels of within- versus among-population components of variation will extend to higher-level phenotypes such as gene-expression levels. Whereas some prior work on gene-expression differences among human populations has been done in the context of disease studies,10,11 to our knowledge, there have been no systematic and quantitative attempts to apportion natural variation in gene-expression levels into within- and among-population components akin to several excellent studies in model organisms.5,12–14
To begin to address these issues, we used Affymetrix Human Focus Arrays to study gene-expression levels in B lymphoblastoid cells derived from eight unrelated individuals of northern and western European ancestry (CEU) and eight unrelated individuals from the Yoruba of Ibadan, Nigeria (YRI). These samples are a subset of the CEU and YRI individuals used in the International HapMap project,15 and cell lines were obtained from the Coriell Cell Repositories (samples GM06995, GM07029, GM07349, GM10845, GM10851, GM10856, GM10857, GM10860, GM19138, GM18516, GM18859, GM18871, GM18501, GM18504, GM18507, and GM18522). All study individuals were males, to eliminate the potential confounding effects of sex on gene-expression levels.
We performed tissue culture and RNA extraction as described elsewhere16,17 and assessed RNA integrity by measuring the optical density 260/280 ratio and subjecting the sample to analysis with the Agilent Bioanalyzer 2100. Extracted RNA was labeled and hybridized according to the manufacturer’s protocol (Affymetrix). We performed quantile normalization and used the RMA algorithm to combine probe-set intensities into a single measure of expression for each gene.18 Low-intensity probe sets that were deemed absent in ⩾50% of the arrays with use of the algorithms implemented in MAS519 were discarded in subsequent statistical analyses, resulting in 5,194 analyzable probe sets. All reported results were robust to different normalization methods and definitions of low-intensity genes (results not shown). Technical replicates were obtained for each individual, resulting in a total of 32 microarrays.
Of the ∼8,500 genes on the array, 5,194 were expressed in lymphoblastoid cells, which is comparable with previous observations.17 We used a fully nested, mixed-model analysis to identify genes differentially expressed among individuals within populations and genes differentially expressed among populations (see appendix A). This model allows tests of differential expression among individuals to be performed while properly accounting for population effects and technical variation. Similarly, it allows tests of differential expression among populations while properly accounting for individual effects and technical variation.
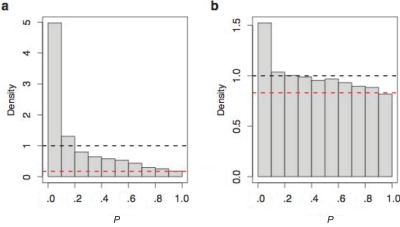
We used methodology described elsewhere22,25 to analyze the complete distribution of P values resulting from tests of differential expression, to estimate the proportion of all genes that are differentially expressed either within or between the CEU and YRI samples. Under the null hypothesis of no differential expression, we expect the P values to be uniformly distributed between 0 and 1. Conversely, if the data set contains differentially expressed genes, the distribution of P values will be skewed toward 0.22 We estimated that ∼83% of genes are differentially expressed among individuals and ∼17% of genes are differentially expressed between the CEU and YRI samples (fig. 1). That these two percentages add up to 100% is a coincidence; we found that the significance of tests for differential expression within and among populations was uncorrelated (see appendix A). The estimated proportion of genes that possess interindividual variation is consistent with previous studies describing pervasive cis-regulatory variation in humans.26–28 To our knowledge, there have been no systematic studies of gene-expression differences among human populations. Thus, these results demonstrate substantial natural variation in gene-expression levels both within and among populations and show that population structure exists in levels of transcript abundance.
Figure 1. .
Estimates of the proportion of genes differentially expressed within and among populations. Histograms of all P values calculated for tests of differential expression among individuals within populations (a) and between the CEU and YRI samples (b) are shown. The Y-axis is drawn to reflect a histogram density, such that the total area of all rectangles is 1. Under the null hypothesis of no differential expression, we expect the P values to be uniformly distributed between 0 and 1, forming a histogram with frequencies following the dashed black line. However, the observed P values in each graph are skewed toward 0, suggesting that these data sets contain differentially expressed genes. Using methodology described elsewhere,22,25 we estimated that 82.6% of genes are differentially expressed among individuals and that 17.4% of genes are differentially expressed between the CEU and YRI samples. The dashed red lines indicate these estimates by showing that the P values close to 1 flatten out at a height of ∼17% (a) and 83% (b).
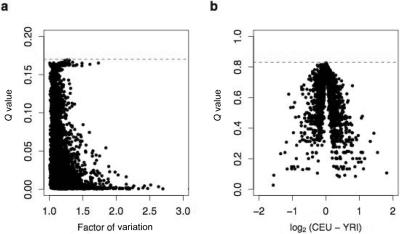
We next investigated the magnitude of expression differences observed within and among populations, which, in general, was relatively small (fig. 2). For example, of the ∼83% of genes estimated to be differentially expressed among individuals, 1,210 were significant at a false-discovery rate (FDR) ⩽1%. These genes varied by an average factor of 1.25 across individuals (see fig. 2), although 20 loci varied by a factor of 2. Of the ∼17% of genes estimated to be differentially expressed between the CEU and YRI samples, 50 were significant at an FDR ⩽20%. The average absolute log2 difference in mean expression levels between samples for these 50 genes was 0.73 (corresponding to a 1.65-fold change). Although the majority of observed gene-expression differences within and between populations are modest, even small perturbations in expression can have significant functional and phenotypic consequences.29,30 The results for all tests of differential expression are presented in a tab-delimited txt file.
Figure 2. .
Magnitude of expression differences within and between populations. a, Magnitude of gene-expression differences among individuals, shown as the factor of variation (X-axis) versus Q value (Y-axis). The Q value is a measure of statistical significance in terms of the FDR.22,25 For each gene, the factor of variation is calculated as the ratio of the maximum:minimum log2 expression level across all individuals.5 b, Magnitude of gene-expression differences between populations, shown by a volcano plot of the average log2-fold change between the CEU and YRI samples (X-axis) versus Q value (Y-axis). In panels a and b, the horizontal dashed lines at 0.17 and 0.83 indicate the estimated proportion of truly null hypotheses in tests of differential expression among individuals and between the CEU and YRI samples, respectively.
To get a broad overview of the types of pathways that differentially expressed genes participate in, we tested whether they were overrepresented among PANTHER biological pathways.31 In this analysis, we considered the top 10% of genes differentially expressed either between individuals or between populations. Only two pathways were nominally significant (P=.05) for genes differentially expressed among individuals, and no pathways remained significant after correction for multiple hypothesis tests (table 1). Thus, gene-expression differences among individuals are found in a wide variety of pathways, which is consistent with our estimate that ∼83% of genes are differentially expressed among individuals. Examples of genes with large interindividual variation in expression include RAGE (MIM *605762) and LRAP (MIM *609497), the expression levels of which correlate with diabetic complications32 and improper antigen processing,33 respectively. Genes differentially expressed between the CEU and YRI samples were strongly enriched in inflammatory pathways, even after a strict Bonferroni correction for multiple-hypothesis tests (table 1). Included in this set of genes are several cytokines and chemokine receptors (CCL22 [MIM *602957], CCL5 [MIM *187011], CCR2 [MIM *601267], CCR7 [MIM *600242], and CXCR3 [MIM *300574]) that have been implicated in numerous cardiovascular, infectious, and immune-related diseases.34,35
Table 1. .
| Sample Comparison and PANTHER Biological Pathway | P |
| Individuals: | |
| Inflammation mediated by chemokine and cytokine | 1.91×10−2 |
| T-cell activation | 3.01×10−2 |
| Populations: | |
| Inflammation mediated by chemokine and cytokine | 2.91×10−4 |
| Histamine H1 receptor–mediated signaling pathway | 3.90×10−3 |
| Toll-receptor signaling pathway | 1.02×10−2 |
| Fibroblast growth factor–signaling pathway | 1.11×10−2 |
| Vascular endothelial growth factor–signaling pathway | 1.14×10−2 |
| T-cell activation | 1.32×10−2 |
| EGF receptor–signaling pathway | 1.53×10−2 |
| B-cell activation | 2.70×10−2 |
| Notch-signaling pathway | 2.99×10−2 |
| Enkephalin release | 2.99×10−2 |
| 5HT2 type receptor–mediated signaling pathway | 4.24×10−2 |
Note.— All pathways nominally enriched at P=.05 are shown; bold type indicates significance after a Bonferroni correction for multiple hypothesis tests.
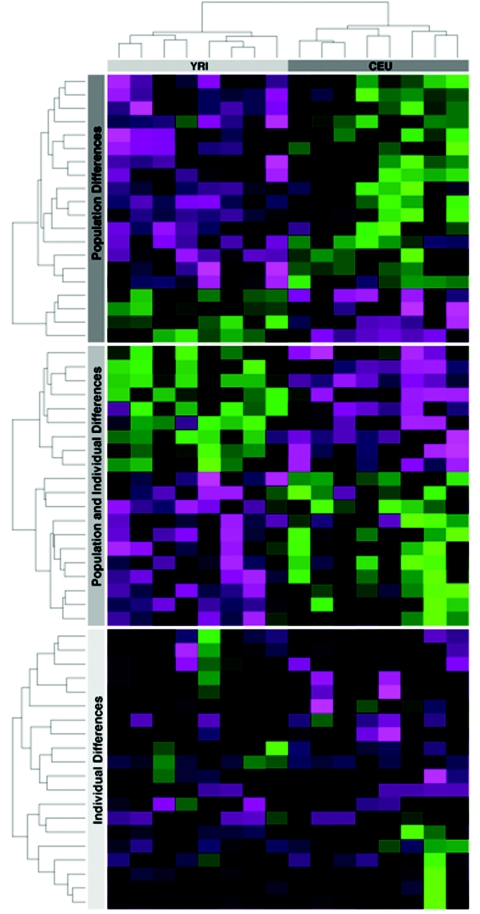
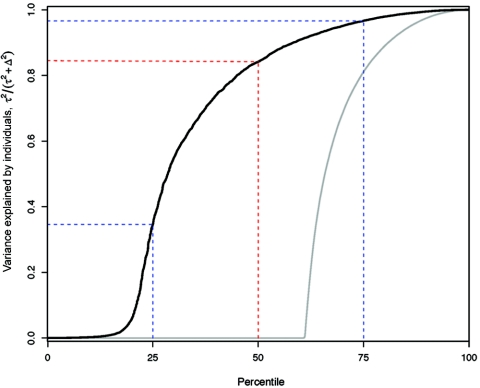
Simply identifying genes differentially expressed within or among populations may provide an incomplete view of the quantitative details of gene-expression variation. For instance, we found examples where expression variation was observed primarily between populations but not individuals, both among individuals and between populations, or among individuals but not between populations (fig. 3). Classifying genes by differential versus no differential expression fails to accurately reflect the quantitative patterns of how expression variation is apportioned into within- and among-population components; therefore, it is important to investigate how much of total gene-expression variation is explained by individual and population effects. To this end, for each gene, we estimated the proportion of total gene-expression variation due to either differences among individuals or differences between populations, while properly taking into account technical variation (see appendix A). The median proportion of variation due to interindividual variation is 0.85 (fig. 4), which is nearly identical to levels of population structure observed in extant patterns of human genetic variation.6–9 In addition, similar to estimates of genetic structure at individual loci,7,36 the distribution of population structure in expression levels across genes varies considerably (fig. 4).
Figure 3. .
Patterns of gene-expression variation within and between the CEU and YRI samples. The log2 expression levels (averaged across replicates) of each individual are shown for 20 genes that are differentially expressed between the CEU and YRI samples but exhibit little within-population variation (top), for 20 genes that are differentially expressed both within and between populations (middle), and for 20 genes that are differentially expressed among individuals but not between populations (bottom). For each gene, the expression of each individual is expressed as the deviation from the overall mean across all 16 individuals. Values range in color from magenta to green, indicating expression levels relatively smaller or larger, respectively, compared with the overall mean. Black values indicate expression levels close to the overall mean across individuals. The dendrograms on the X- and Y-axes correspond to individuals and genes, respectively. The topology of the dendrogram is based on the 50 genes differentially expressed between the CEU and YRI samples.
Figure 4. .
Distribution of the proportion of total gene-expression variation explained by variation among individuals. The percentile of the proportion of all genes (X-axis) versus the proportion of total expression variation explained by interindividual differences (Y-axis) is shown for the observed (solid black line) and randomized (solid gray line) data. For example, the median (50th percentile) (dashed red line) proportion of variation explained by differences among individuals is 85%, leaving 15% explained by differences between populations. The dashed blue lines indicate the interquartile range. Note that the magnitude of total gene-expression variation attributable to interindividual differences in the observed data is considerably greater compared with the randomized data.
To fully understand the genetic architecture of gene-expression levels and how population structure influences patterns of gene-expression variation, it will be necessary both to confirm predictions from microarray data and to delineate the molecular mechanisms governing regulatory variation. To begin to address these issues, we performed quantitative allele-specific PCR (qPCR) on SH2B3 (MIM *605093), which encodes for an adaptor protein that regulates growth factor and cytokine receptor-mediated pathways37 and was in the top 1% of genes differentially expressed between the CEU and YRI samples. We generated double-stranded cDNA and used TaqMan (Applied Biosystems) allelic discrimination assays to interrogate the expression level of each allele for a SNP (rs1107853) in the coding region of SH2B3, as described elsewhere.38 We constructed a dilution series of heterozygous genomic DNA to estimate dye effects and differences in allele-specific hybridization efficiency, as described elsewhere.38 To obtain the overall expression level for each individual, we summed the expression levels of the two alleles after adjusting for dye effects and hybridization effects. All qPCR experiments were performed in triplicate.
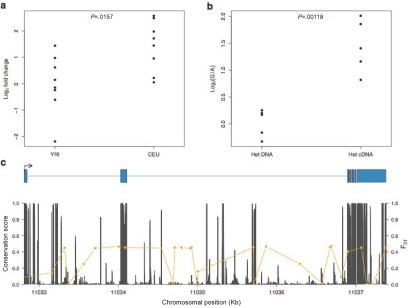
In our microarray experiments, SH2B3 showed an average log2-fold change between the CEU and YRI samples of 0.52 (P=6.5×10-4; FDR=0.134). Consistent with the microarray data, the qPCR results also demonstrate that SH2B3 is differentially expressed between the CEU and YRI samples (P=.0157) (fig. 5a). To better understand the molecular basis for the observed difference in expression, we asked whether the expression level of one allele was different from the other in heterozygous individuals. If so, this provides evidence of cis-regulatory effects.26 There was a significant difference (P=1.18×10-3) in expression between alleles in heterozygous cDNA versus genomic DNA, strongly suggesting cis-regulatory effects (fig. 5b).
Figure 5. .
Allele-specific qPCR analysis of SH2B3. a, Log2-fold change of SH2B3 expression for all CEU and YRI individuals, relative to the average expression level in the YRI sample obtained from allele-specific qPCR. The distribution of SH2B3 expression is significantly different between samples (t-test, P=.0157), which confirms the microarray results. b, Allele-specific qPCR of a coding polymorphism (rs1107853), which demonstrates that the log2-fold change of the G allele relative to the A allele is significantly different between heterozygous DNA (Het DNA) and heterozygous cDNA (Het cDNA) samples (t-test, P=.00118). c, The gene structure of SH2B3, shown in blue with rectangles denoting exons. The arrow indicates transcriptional orientation. The graph below shows the distribution of conservation scores (dark gray) (University of California–Santa Cruz Genome Browser) across the SH2B3 gene and pairwise FST values (yellow) between the CEU and YRI samples for each SNP in this region (derived from HapMap phase II release 21 data).
Interestingly, these observations coincide with patterns of genetic variation at SH2B3, since there are 13 SNPs with large allele-frequency differences (FST⩾0.45) between the CEU and YRI samples (fig. 5c). Five of these highly differentiated SNPs occur in conserved regions, as determined by alignment of 17 vertebrate genomes, making them strong candidates for future functional studies. We calculated the empirical probability of observing a SNP with a pairwise FST⩾0.45 between the CEU and YRI samples, on the basis of all autosomal markers contained in HapMap release 21, to be ∼0.05, and this magnitude of allele-frequency difference is consistent with a signature of local adaptation.7,39 SH2B3 also possesses unusually large levels of linkage disequilibrium compared with the rest of the genome,40 which provides additional support for the hypothesis that this locus has been subject to adaptive evolution, although additional studies will be necessary to make more-definitive inferences about its evolutionary history.
In summary, consistent with previous studies of model organisms,5,12–14 our results demonstrate that considerable natural variation in gene-expression levels exists within and among human populations. Genes differentially expressed among populations may be particularly relevant to explore as candidate susceptibility loci for diseases whose prevalence varies as a function of ethnicity and may be amenable to genetic dissection by admixture linkage-disequilibrium mapping.41 Importantly, we also show that simply focusing on differentially expressed genes can lead to an incomplete understanding of how gene-expression variation is apportioned within and among human populations. By decomposing expression variation into its component sources, we find that, similar to that observed for genetic variation, the majority of gene-expression variation is due to differences among individuals rather than among populations.
These observations are subject to several caveats, including the fact that a relatively small number of individuals and populations were studied and that expression levels were measured only in a single (transformed) cell type. For example, of the ∼17% of genes expressed in B lymphoblasts that we estimate to be differentially expressed between the CEU and YRI samples, 50 could be identified at an FDR <20%. Thus, to fully catalog the specific genes that are differentially expressed, it will be necessary to increase the sample size. In addition, it is plausible that probes that overlap SNPs could lead to biased estimates of gene-expression levels42 and confound our interpretation of gene-expression variation within and among populations. Although the algorithms we used to normalize the raw expression data and to combine individual probe sets into an overall measure of gene expression should be relatively robust to low levels of sequence divergence, it remains a formal possibility that probes interrogating sequences with SNPs contribute to the observed patterns of gene-expression variation. However, recent work suggests that this is unlikely to have a large influence on estimates of gene-expression levels among closely related populations.43–45 Despite these limitations, our results and methodology provide the foundation for building a more principled understanding of natural variation in gene-expression levels that will be useful for testing hypotheses of regulatory evolution and interpreting patterns of expression variation in disease.
Supplementary Material
Acknowledgments
We thank the Center for Expression Arrays core facility at the University of Washington for technical assistance in microarray hybridizations, and we thank Dayna Akey and Bruce Weir for helpful discussions. This work was supported by National Science Foundation starter grant DEB 0512279 (to J.M.A.).
Appendix A
Tests of Differential Expression Within and Among Populations
We used the recently developed optimal discovery procedure20 (ODP), available in the EDGE (J.D.S.'s Web site) software package,21 to test for differential expression between the CEU and YRI samples. We averaged technical replicates for each individual and tested for a difference in mean expression between samples, as detailed in the ODP method and EDGE manual (J.D.S.'s Web site). This produced an FDR Q value (J.D.S.'s Web site)22 for differential expression between samples for each gene, as well as a conservative estimate of the total proportion of population–differentially expressed genes (∼17%). The ODP is derived from the same principles yielding more-traditional methods, such as a t test and its popular microarray extensions.23 However, the ODP is aimed at optimizing a more relevant balance between true positive and false positive results, to yield substantially greater power to identify genes, as has been shown elsewhere.20 Although the global estimate of the proportion of differentially expressed genes, on the basis of a t test, was not significantly different from the ∼17% estimate produced by the ODP, the power of the ODP to identify genes as significant was substantially better here as well.
We formulated a new model of gene-expression variation from individuals in structured populations, to estimate the proportion of expression variation due to differences among individuals and among populations and to identify genes showing differential expression within populations. Specifically, we employed a mixed model in which population effects were treated as fixed and individual effects were treated as random. The model for each gene can be written as: expression=baseline+population+individual+error, where “baseline” is the fixed baseline expression level, “population” is the fixed effect due to potential differences between populations, “individual” is the random effect representing each individual’s potentially different average level of expression, and “error” represents the remaining random fluctuations in expression due to technical and measurement variation. Treating individual effects as fixed5 attributes too much variation to interindividual differences, which leads to substantial underbias in estimating the proportion of variation due to population effects. The mixed model allows for unbiased estimates of both population and interindividual differential expression effects and allows us to separate the technical and measurement errors from the biological signals of interest.
We fit the above model to each gene, by maximum likelihood under the assumption that the individual random effects and error terms are normally distributed, using the statistical software package R.24 From this, we obtained point estimates of the fixed population effect (Δ), the variance of the individual random effect (τ2), and the variance of the error term (σ2) for each gene. We can show that the total variance for a gene’s expression is equal to Δ2+τ2+σ2. We calculated the proportion of variance explained by population differences as the ratio of the variance due to population differences to the sum of the variances due to population differences and interindividual differences: Δ2/(Δ2+τ2). The proportion of variance explained by interindividual differences is equal to one minus this quantity: τ2/(Δ2+τ2). Note that the mixed model allowed us to remove the nonbiological variance component when partitioning the variance into within- and among-population components.
To test for differential expression among individuals, we performed a hypothesis test for each gene, to determine whether the variance corresponding to the individual random effect (τ2) is zero, where a nonzero variance indicates the presence of interindividual differences in expression. For each gene, the full model was fit by maximum likelihood, as described above, as well as the analogous model with no individual random-effect term. The two models were compared by a generalized likelihood-ratio statistic. We simulated >500,000 statistics from the null distribution by permuting the individual labels within each population and recomputing the generalized likelihood-ratio statistics on these permuted data. The observed and null statistics were then used to estimate an FDR Q value (J.D.S.'s Web site) for each gene as described elsewhere.22 This also provides a conservative estimate of the total proportion (83%) of interindividual differentially expressed genes.
Assessing Data Quality
We performed several diagnostic procedures to make sure that the results derived from tests of differential expression within and among populations were genuine and not confounded by technical artifacts. First, we observed that the significance of these two types of differential expression appeared to be independent. Genes showed both types of differential expression, only one type, or neither type in proportions expected by chance, given the fact that the two types of differential expression occur independently. Second, the correlation of log[p/(1-p)] of the two sets of P values was only 5%, which is well within the range observed under random permutations of the P values. Third, the error-variance estimates did not show any functional relationships with the estimated proportion of variation due to interindividual or population differences, indicating that our model successfully separated the biological signal of interest from the technical and measurement errors.
Web Resources
The URLs for data presented herein are as follows:
- Coriell Cell Repositories, http://ccr.coriell.org/
- HapMap, http://www.hapmap.org/
- J.D.S.'s Web site, http://faculty.washington.edu/~jstorey/ (for the EDGE and QVALUE software)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for RAGE, LRAP, CCL22, CCL5, CCR2, CCR7, CXCR3, and SH2B3)
- PANTHER, http://www.pantherdb.org/ (to test for overrepresentation of differentially expressed genes in biological pathways)
- University of California–Santa Cruz Genome Browser, http://genome.ucsc.edu/ (for conservation scores)
References
- 1.Rifkin SA, Kim J, White KP (2003) Evolution of gene expression in the Drosophila melanogaster subgroup. Nat Genet 33:138–144 10.1038/ng1086 [DOI] [PubMed] [Google Scholar]
- 2.King MC, Wilson AC (1975) Evolution at two levels in humans and chimpanzees. Science 188:107–116 10.1126/science.1090005 [DOI] [PubMed] [Google Scholar]
- 3.Knight JC (2005) Regulatory polymorphisms underlying complex disease traits. J Mol Med 83:97–109 10.1007/s00109-004-0603-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan H, Zhou W (2004) Allelic variations in gene expression. Curr Opin Oncol 16:39–43 10.1097/00001622-200401000-00008 [DOI] [PubMed] [Google Scholar]
- 5.Oleksiak MF, Churchill GA, Crawford DL (2002) Variation in gene expression within and among natural populations. Nat Genet 32:261–266 10.1038/ng983 [DOI] [PubMed] [Google Scholar]
- 6.Barbujani G, Magagni A, Minch E, Cavalli-Sforza LL (1997) An apportionment of human DNA diversity. Proc Natl Acad Sci USA 94:4516–4519 10.1073/pnas.94.9.4516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akey JM, Zhang G, Zhang K, Jin L, Shriver MD (2002) Interrogating a high-density SNP map for signatures of natural selection. Genome Res 12:1805–1814 10.1101/gr.631202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW (2002) Genetic structure of human populations. Science 298:2381–2385 10.1126/science.1078311 [DOI] [PubMed] [Google Scholar]
- 9.Excoffier L, Hamilton G (2003) Comment on “Genetic structure of human populations.” Science 300:1877 [DOI] [PubMed] [Google Scholar]
- 10.He XS, Ji X, Hale MB, Cheung R, Ahmed A, Guo Y, Nolan GP, Pfeffer LM, Wright TL, Risch N, et al (2006) Global transcriptional response to interferon is a determinant of HCV treatment outcome and is modified by race. Hepatology 44:352–359 10.1002/hep.21267 [DOI] [PubMed] [Google Scholar]
- 11.Dysvik B, Vasstrand EN, Lovlie R, Elgindi OA, Kross KW, Aarstad HJ, Johannessen AC, Jonassen I, Ibrahim SO (2006) Gene expression profiles of head and neck carcinomas from Sudanese and Norwegian patients reveal common biological pathways regardless of race and lifestyle. Clin Cancer Res 12:1109–1120 10.1158/1078-0432.CCR-05-0115 [DOI] [PubMed] [Google Scholar]
- 12.Oleksiak MF, Roach JL, Crawford DL (2005) Natural variation in cardiac metabolism and gene expression in fundulus heteroclitus. Nat Genet 37:67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Townsend JP, Cavalieri D, Hartl DL (2003) Population genetic variation in genome-wide gene expression. Mol Biol Evol 20:955–963 10.1093/molbev/msg106 [DOI] [PubMed] [Google Scholar]
- 14.Jin W, Riley RM, Wolfinger RD, White KP, Passador-Gurgel G, Gibson G (2001) The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat Genet 29:389–395 10.1038/ng766 [DOI] [PubMed] [Google Scholar]
- 15.The International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437:1299–1320 10.1038/nature04226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monks SA, Leonardson A, Zhu H, Cundiff P, Pietrusiak P, Edwards S, Phillips JW, Sachs A, Schadt EE (2004) Genetic inheritance of gene expression in human cell lines. Am J Hum Genet 75:1094–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen KY, Morley M, Spielman RS (2003) Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet 33:422–425 10.1038/ng1094 [DOI] [PubMed] [Google Scholar]
- 18.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31:e15 10.1093/nar/gng015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubbell E, Liu WM, Mei R (2002) Robust estimators for expression analysis. Bioinformatics 18:1585–1592 10.1093/bioinformatics/18.12.1585 [DOI] [PubMed] [Google Scholar]
- 20.Storey JD, Dai JY, Leek JT (2006) The optimal discovery procedure for large-scale significance testing, with applications to comparative microarray experiments. Biostatistics (http://biostatistics.oxfordjournals.org/cgi/reprint/kxl019v1) (electronically published August 23, 2006; accessed January 10, 2007) [DOI] [PubMed] [Google Scholar]
- 21.Leek JT, Monsen E, Dabney AR, Storey JD (2006) EDGE: extraction and analysis of differential gene expression. Bioinformatics 22:507–508 10.1093/bioinformatics/btk005 [DOI] [PubMed] [Google Scholar]
- 22.Storey JD, Tibshirani R (2003) Statistical significance for genome-wide studies. Proc Natl Acad Sci USA 100:9440–9445 10.1073/pnas.1530509100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98:5116–5121 10.1073/pnas.091062498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storey JD (2002) A direct approach to false discovery rates. J R Stat Soc Ser B 64:479–498 10.1111/1467-9868.00346 [DOI] [Google Scholar]
- 26.Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW (2002) Allelic variation in human gene expression. Science 297:1143 10.1126/science.1072545 [DOI] [PubMed] [Google Scholar]
- 27.Lo HS, Wang Z, Hu Y, Yang HH, Gere S, Buetow KH, Lee MP (2003) Allelic variation in gene expression is common in the human genome. Genome Res 13:1855–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bray NJ, Buckland PR, Owens MJ, O’Donovan MC (2003) Cis-acting variation in the expression of high proportion of genes in human brain. Hum Genet 113:149–153 [DOI] [PubMed] [Google Scholar]
- 29.Yan H, Dobbie Z, Gruber SB, Markowitz S, Romans K, Giardiello FM, Kinzler KW, Vogelstein B (2002) Small changes in expression affect predisposition to tumorigenesis. Nat Genet 30:25–26 10.1038/ng799 [DOI] [PubMed] [Google Scholar]
- 30.Bray NJ, Buckland PR, Williams NM, Williams HJ, Norton N, Owen MJ, O’Donovan MC (2003) A haplotype implicated in schizophrenia susceptibility is associated with reduced COMT expression in human brain. Am J Hum Genet 73:152–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, Ladunga I, Ulitsky-Lazareva B, Muruganujan A, Rabkin S, Vandergriff JA, Doremieux O (2003) PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucl Acids Res 31:334–341 10.1093/nar/gkg115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.dos Santos KG, Canani LH, Gross JL, Tschiedel B, Pires Souto KE, Roisenberg I (2005) The -374A allele of the receptor for advanced glycation end products gene is associated with a decreased risk of ischemic heart disease in African-Brazilians with type 2 diabetes. Mol Genet Metab 85:149–156 10.1016/j.ymgme.2005.02.010 [DOI] [PubMed] [Google Scholar]
- 33.Fruci D, Ferracuti S, Limongi MZ, Cunsolo V, Giorda E, Fraioli R, Sibilio L, Carroll O, Hattori A, van Endert PM, et al (2006) Expression of endoplasmic reticulum aminopeptidases in EBV-B cell lines from healthy donors and in leukemia/lymphoma, carcinoma, and melanoma cell lines. J Immunol 176:4869–4879 [DOI] [PubMed] [Google Scholar]
- 34.Charo IF, Peters W (2003) Chemokine receptor 2 (CCR2) in atherosclerosis, infectious diseases, and regulation of T-cell polarization. Microcirculation 10:259–264 10.1038/sj.mn.7800191 [DOI] [PubMed] [Google Scholar]
- 35.Leung TF, Tang NL, Lam CW, Li AM, Fung SL, Chan IH, Wong GW (2005) RANTES G-401A polymorphism is associated with allergen sensitization and FEV1 in Chinese children. Respir Med 99:216–219 10.1016/j.rmed.2004.06.004 [DOI] [PubMed] [Google Scholar]
- 36.Weir BS, Cardon LR, Anderson AD, Nielsen DM, Hill WG (2005) Measures of human population structure show heterogeneity among genomic regions. Genome Res 15:1468–1476 10.1101/gr.4398405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fitau J, Boulday G, Coulon F, Quillard T, Charreau B (2006) The adaptor molecule Lnk negatively regulates tumor necrosis factor-α-dependent VCAM-1 expression in endothelial cells through inhibition of the ERK1 and -2 pathways. J Biol Chem 281:20148–20159 10.1074/jbc.M510997200 [DOI] [PubMed] [Google Scholar]
- 38.Ronald J, Brem RB, Whittle J, Kruglyak L (2005) Local regulatory variation in Saccharomyces cerevisiae. PLoS Genet 1:e25 10.1371/journal.pgen.0010025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akey JM, Eberle MA, Rieder MJ, Carlson CS, Shriver MD, Nickerson DA, Kruglyak L (2004) Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol 2:e286 10.1371/journal.pbio.0020286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang ET, Kodama G, Baldi P, Moyzis RK (2006) Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc Natl Acad Sci USA 103:135–140 10.1073/pnas.0509691102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halder I, Shriver MD (2003) Measuring and using admixture to study the genetics of complex diseases. Hum Genomics 1:52–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilad Y, Rifkin SA, Bertone P, Gerstein M, White KP (2005) Multi-species microarrays reveal the effect of sequence divergence on gene expression profiles. Genome Res 15:674–680 10.1101/gr.3335705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stranger BE, Forrest MS, Clark AG, Minichiello MJ, Deutsch S, Lyle R, Hunt S, Kahl B, Antonarakis SE, Tavare S, et al (2005) Genome-wide associations of gene expression variation in humans. PLoS Genet 1:e78 10.1371/journal.pgen.0010078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doss S, Schadt EE, Drake TA, Lusis AJ (2005) Cis-acting expression quantitative trait loci in mice. Genome Res 15:681–691 10.1101/gr.3216905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.GuhaThakurta D, Xie T, Anand M, Edwards SW, Li G, Wang SS, Schadt EE (2006) Cis-regulatory variations: a study of SNPs around genes showing cis-linkage in segregating mouse populations. BMC Genomics 7:235 10.1186/1471-2164-7-235 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.