Abstract
ChIP coupled with microarray provides a powerful tool to determine in vivo binding profiling of transcription factors to deduce regulatory circuitries in mammalian cells. Aiming at improving the specificity and sensitivity of such analysis, we developed a new technology called ChIP-DSL using the DNA selection and ligation (DSL) strategy, permitting robust analysis with much reduced materials compared with standard procedures. We profiled general and sequence-specific DNA binding transcription factors using a full human genome promoter array based on the ChIP-DSL technology, revealing an unprecedented number of the estrogen receptor (ERα) target genes in MCF-7 cells. Coupled with gene expression profiling, we found that only a fraction of these direct ERα target genes were highly responsive to estrogen and that the expression of those ERα-bound, estrogen-inducible genes was associated with breast cancer progression in humans. This study demonstrates the power of the ChIP-DSL technology in revealing regulatory gene expression programs that have been previously invisible in the human genome.
Keywords: breast cancer, genome-wide, promoter array
The elucidation of genomes for humans and other model organisms has made it possible to conduct analysis of gene expression and regulation at the genome scale. Gene expression is generally accompanied by chromatin remodeling activities and histone modifications. An important conceptual advance has been the “histone code” hypothesis, which suggests that histone modifications reflect a sequential action of enzymes associated with the transcriptional machinery such that one prior activity may influence the next during regulated gene expression (1, 2). Histone acetylation results in charge neutralization of modified lysines, which is generally associated with gene activation (3). In contrast, histone methylation on different residues appears to provide binding sites for specific transcription regulators, thereby positively or negatively affecting gene expression (4). Although histone methylation may modulate gene expression in a gene-specific and context-dependent manner, certain site-specific modifications appear to be generally applicable to most genes. The epigenetic markers thus provide a roadmap to identify and characterize functional DNA elements in the genome.
The nuclear receptor (NR) superfamily of transcriptional regulators plays a central role in many developmental and disease processes, and the system has been extensively studied as a model to learn the mechanism for spatial and temporal control of gene expression (5). Individual NRs have consensus binding sites in promoters and enhancers, which have been characterized in detail, but only in a limited number of NR-regulated genes. In the case of the pS2 gene (also known as TFF1), for example, binding by estrogen receptor α (ERα) initiates sequential recruitment of a large number of transcription factors onto the promoter to start transcription (6). However, despite extensive mechanistic insights in transcriptional initiation in this and other well studied cases, little is known about how many genes are direct targets for an NR. Genome-wide ChIP coupled with microarray, known as ChIP-on-chip, offers a solution to this problem by determining promoters bound directly by transcription factors (7–10). Surprisingly, however, recent promoter and tiling array analyses suggest that ERα binds relatively rarely to gene promoters compared with intergenic regions, suggesting a critical role of long-distance enhancers in regulated gene expression in mammalian cells (11–13).
Here we describe an approach to detecting in vivo DNA–protein interactions by coupling ChIP with a DNA selection and ligation (DSL) strategy, permitting analysis of many fewer cells than required by the conventional ChIP-on-chip method. We constructed a full genome promoter array based on this ChIP-DSL platform, and our analysis revealed that ERα bound to >3% of human genes in promoter-proximal regions in MCF-7 cells, reinforcing the importance of direct binding events in the promoter-proximal regions during regulated gene expression. Results from built-in tiling arrays allowed direct visualization of binding events even without statistical filtering of raw data, and a comprehensive histone modification profile extended the current histone code hypothesis. These results demonstrate the versatility and accuracy of the ChIP-DSL technology in a genome-wide search for direct target genes by specific transcription factors and in comprehensive analysis of regulatory programs within specific genomic loci. Furthermore, comparison between profiles of ERα binding and 17β-estradiol (E2)-induced gene expression in MCF-7 cells revealed a subset of genes whose expression tracks breast cancer progression in humans, which not only suggests the prognostic value of these genes as biomarkers for breast cancer but also illustrates a general strategy for dissecting molecular pathways in cancer.
Results and Discussion
Design and Development of the ChIP-DSL Technology.
Aiming to detect functional DNA elements with high sensitivity and specificity, we devised a multiplex assay by coupling ChIP with a DSL approach (Fig. 1). A signature 40-nt sequence is first computationally identified in a genomic segment 0.5–1 kb in length. For promoter profiling each such probe corresponds to a proximal promoter region from +200 nt to −800 nt relative to the transcription start, which contains ≈95% of known binding sites for transcription factors in humans (8). To construct a tiling array, each probe is used to represent an ≈0.5-kb nonrepetitive genomic block in a path to be tiled. This probe density takes into account the number of probes required for maximal coverage of genomic sequences and the sufficiency in detecting immunoprecipitated DNA, which is generally sheared to an average length of 0.5–1 kb. Amine-modified 40-mers are spotted onto solid support to form an array.
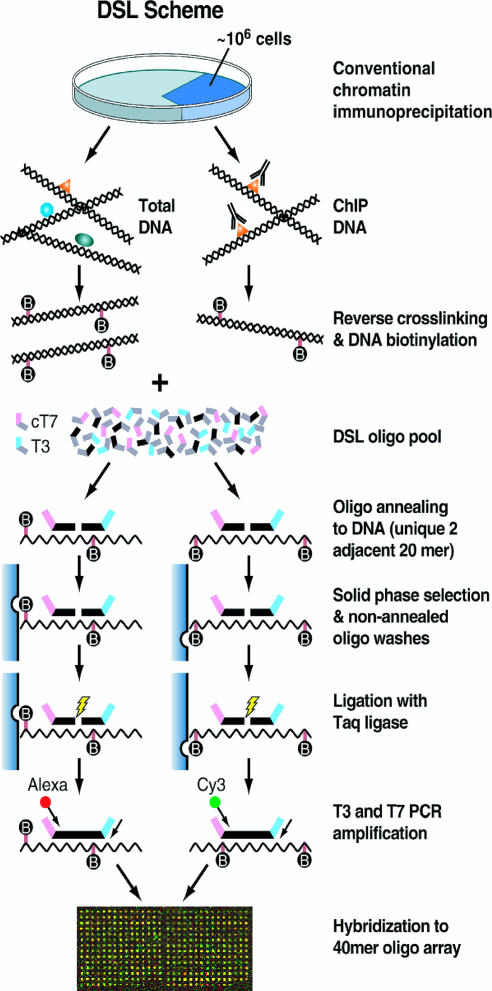
Fig. 1.
The ChIP-DSL scheme. A key feature of the technology is oligonucleotide ligation templated by chromatin immunoprecipitated (ChIP) DNA followed by DSL. This permits high-throughput analysis of target genes with much improved specificity and sensitivity.
Corresponding to each 40-mer, a pair of assay oligonucleotides are synthesized, each consisting of the two 20-mer halves in the 40-mer and flanked by a universal primer landing site. Multiple oligonucleotide pairs are mixed to form a pool. The assay begins with standard ChIP, and the isolated DNA is randomly biotinylated followed by annealing to the oligonucleotide pool. Annealed oligonucleotides are selected on streptavidin-conjugated magnetic beads, and unannealed oligonucleotides are washed away. This selection strategy allows the use of an excessive amount of oligonucleotides to achieve maximal annealing that follows the pseudo first-order kinetics and prevents interference of later steps by excess free oligonucleotides in solution. All selected oligonucleotides are immobilized, and those paired by specific target DNA are ligated, thereby converting only correctly targeted oligonucleotides to full amplicons for PCR amplification. One of the PCR primers is end-labeled with a fluorescent dye so that the PCR products can be directly hybridized to the 40-mer array.
This technology is distinct from the conventional ChIP-on-chip assay in several key aspects. First, chromatin immunoprecipitated DNA was used to template oligonucleotide ligation, instead of being directly amplified for hybridization. This step can tolerate incomplete decross-linking because cross-linking adducts should have less effect on oligonucleotide hybridization than signal amplification by a polymerase. Second, we targeted only unique signature sequences in the human genome, thereby avoiding potential interference of repetitive and related sequences during hybridization. Third, the sensitivity is significantly elevated by PCR amplification of ligated oligonucleotides in an unbiased fashion because all amplicons contain the same pair of specific primer landing sites and are uniform in length as previously documented (14).
We progressively enlarged the multiplicity of the assay to eventually cover most annotated gene promoters in the human genome. Titration experiments indicate that the ChIP-DSL technology could routinely operate with cells from one-third of a single 100-mm culture dish, which corresponds to 1–5 × 106 cells, depending on the cell type under investigation. Despite the fact that each promoter is targeted by one oligonucleotide pair, high-quality data generated as reported in this and other studies (15) demonstrate the reproducibility and robustness of the ChIP-DSL technology.
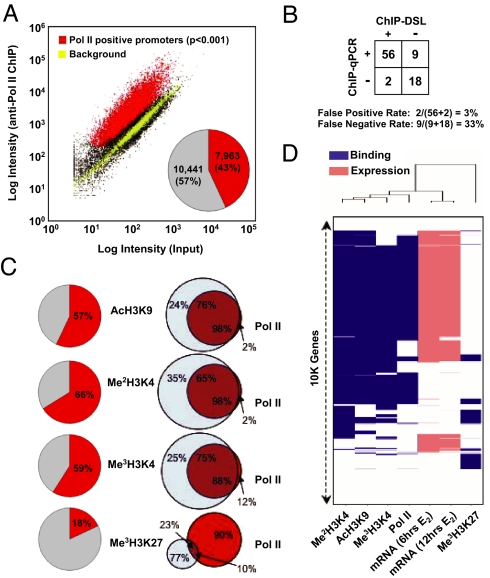
We initially assessed promoters potentially active in transcription based on their association with RNA polymerase (Pol) II in E2-treated MCF-7 cells (Fig. 2A). Anti-Pol II-enriched (Pol II+) promoters (red) were clearly segregated from background marked by built-in tiling array controls (yellow), finding that 43% of total promoters were Pol II+ at a standard P value < 0.001. Quantitative ChIP/quantitative PCR (qPCR) analysis of randomly selected promoters suggests a false positive rate of ≈3% and a false negative rate of ≈33% [Fig. 2B and supporting information (SI) Fig. 6]. A similar false positive rate was observed by using an irrelevant IgG (SI Fig. 7A). The false negative rate is quite similar to that reported in published ChIP-chip studies (16). Pol II+ promoters were also marked by AcH3K9 (98%), Me2H3K4 (98%), and Me3H3K4 (88%), although a significant fraction of promoters were associated only with these gene activation marks, but not with Pol II (Fig. 2C). In contrast, the “repressive” histone mark Me3H3K27 was detected in only a small fraction (10%) of Pol II+ promoters. Indeed, this repressive histone mark has been shown to associate with some active genes (17). An RNA profiling experiment in the same E2-stimulated MCF-7 cells showed that most Pol II+ promoters were actively transcribing (Fig. 2D). Collectively, these robust and highly consistent data testify to the utility and sensitivity of the ChIP-DSL technology.
Fig. 2.
Global analysis of promoter occupancy by ChIP-DSL. (A) Global analysis of Pol II-bound promoters in E2-stimulated MCF-7 cells. A set of tiled genomic loci (yellow) served as internal negative controls because most genomic sequences are not expected to interact with general and sequence-specific transcription factors. Pol II-positive (red) and -negative (black) promoters were identified based on the single-array error model at P < 0.001, and the percentages of Pol II-positive and -negative promoters are shown in Inset. (B) ChIP/qPCR validation of the ChIP-DSL results. (C) Promoter profiling of modified histones in E2-treated MCF-7 cells. (Left) Percentages of positive promoters. (Right) Overlap of positive promoters with Pol II. The overlap between Pol II binding and individual histone modification events is shown in individual Venn diagrams. (D) Correlation of gene expression with promoter occupancy by Pol II and histone modification markers. Gene expression profiling in E2-induced MCF-7 cells was carried out on Illumina gene expression arrays. Approximately 10,000 genes common to both promoter and expression profiling arrays and reliably scored in all measurements were used to construct the binary map by unsupervised hierarchical clustering analysis.
Identification of ERα-Occupied Gene Promoters in the Human Genome.
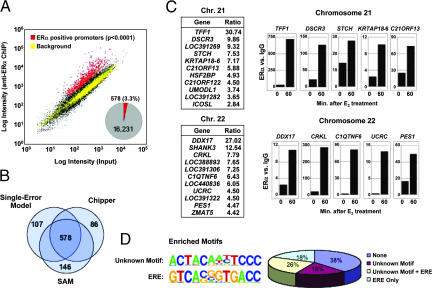
We next applied the ChIP-DSL technology to identify target genes for sequence-specific DNA-binding transcription factors. ERα plays an important role in human reproduction and breast cancer. Recent promoter profiling analysis using 1-kb promoter regions detected 153 ERα-bound promoters (13). Further, tiling analysis of ERα binding suggests that ERα binds prevalently to intergenic regions in the human genome, suggesting a new paradigm that estrogen-regulated gene expression may be largely driven by long-distance enhancers (11, 12). Complimentary to these recent genomic analyses, we scored ≈1,300 anti-ERα-enriched (ERα+) promoters in E2-stimulated MCF-7 cells based on the single-array error model (9) at the standard cutoff of P < 0.001, and ≈700 at a more stringent cutoff of P < 0.0001 (Fig. 3A). A significant number of ERα+ promoters were also identified in vehicle-treated MCF-7 cells, suggesting a class of hormone-independent recruitment events (SI Fig. 7B). To identify ERα+ promoters with high statistical confidence, we analyzed the data from multiple biological repeats using three statistical methods that are based on distinct mathematical principles, revealing an overlapping set of 578 highest confidence ERα+ promoters, which represents 3.3% of all reliably scored promoters (Fig. 3B and SI Data Set 1).
Fig. 3.
Promoter profiling of ERα in E2-induced MCF-7 cells. (A and B) ERα-bound promoters were identified (red) at P < 0.0001. The percentage of ERα-bound promoters scored positively by all three analytical methods is shown in A Inset, and additional promoters scored positively by one or two methods are indicated in the peripheries of the Venn diagram in B. (C) Listed are the newly identified ERα-positive promoters on chromosomes 21 and 22. Ratios were deduced from array measurements. Selected promoters were validated by ChIP/qPCR (Right). (D) Motif analysis of anti-ERα-enriched promoters. The first motif appears common to gene promoters in general, but the protein(s) recognizing this motif is unknown. When this motif is masked, the most enriched motif corresponds to full- or half-consensus estrogen responsive element. Allowing one base mismatch, the percentage of promoters containing a full- or half-consensus estrogen responsive element among total ERα-bound promoters was calculated and shown in Right.
ChIP/qPCR analysis confirmed all ERα+ promoters examined, including those residing in chromosomes 21 and 22 (Fig. 3C) and 20 additional promoters in other chromosomes (data not shown), indicating a negligible false positive rate for anti-ERα+-enriched promoters supported by stringent statistical tests. Estimation of the false negative rate proved to be challenging when the majority of the probes are in the “negative” population (8). We used ChIP/qPCR-confirmed promoters recently reported (13) to objectively estimate our false negative rate. Among 27 validated promoters common between the two array platforms, 20 were scored positive in our array at P < 0.0001 and 24 at P < 0.001, indicating that our false negative rate is ≈26% and 11% at the two P value cutoffs, respectively, which is probably an overestimate, because three promoters (CYP4F3, PROP1, and ABCG2) not detected were enriched only <2-fold in previous ChIP/qPCR experiments (13). Together, these results demonstrate the accuracy of the ChIP-DSL data and conservatively identify ≈4-fold as many ERα target promoters as were detected in previous genome-wide location analysis, suggesting that the promoter array based on the ChIP-DSL technology is a useful resource for the general research community.
We next conducted motif analysis using a newly refined algorithm, which compares ChIP-enriched promoters against normalized nucleotide frequencies in all promoters (C.B. and C.K.G., unpublished data), revealing a highly enriched, but uncharacterized, motif associated with ERα-bound promoters (Fig. 3D). When this motif was masked, the next most enriched motifs were the classic ERα-binding consensus sequences (18), which are present in 44% of the total ERα+ promoters (Fig. 3D). Interestingly, whereas the algorithm confirmed the presence of FoxA1 recognition motifs surrounding a fraction of intergenic ERα binding sites (11), it did not detect extensive association of the FoxA1 binding site with the ERα+ promoters identified by ChIP-DSL. In light of the finding that FoxA1 is critical for ERα binding to several target genes examined (11, 13), it will be interesting to determine in future studies whether FoxA1 is selectively or universally required for ERα targeting.
Locus-Specific Tiling Array Analysis of ERα Binding and Histone Modifications.
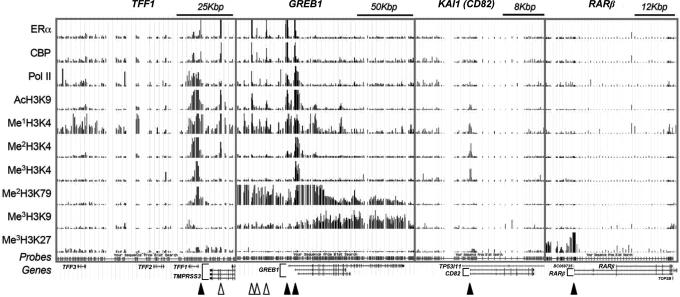
To facilitate data analysis of promoter arrays, we built in a number of tiled loci to serve as internal negative controls because not all genomic regions are expected to be occupied by general and sequence-specific DNA binding transcription factors. The data in turn illustrate the usefulness of the ChIP-DSL technology in revealing specific molecular recognition events that constitute the regulatory programs in individual genomic loci. As illustrated in Fig. 4, we found that ERα bound to the promoter (filled arrow) and a putative enhancer (open arrow) of the TFF1 gene, as previously reported (11). The transcriptional coactivator CBP similarly interacted with both promoter and enhancer, whereas Pol II covered the body of this relatively small gene. In the case of the GREB1 gene, we observed a similar pattern with ERα, CBP, and Pol II present on two of the three promoters that were previously characterized (19). Interestingly, we found that all three factors interacted with three distinct loci upstream of the GREB1 promoters, suggesting that these sites may function as enhancers. These observations are consistent with a large body of literature that gene promoters and enhancers are recognized by sequence-specific DNA-binding transcription factors, which in turn recruit transcription coactivators.
Fig. 4.
Locus-specific tiling array analysis of ERα binding and histone modifications in E2-induced MCF-7 cells. Individual genes and scales are shown at the top, and probe positions and gene structure are indicated at the bottom. Individual transcription factors and chromatin remodeling markers profiled are indicated on the left. Transcription starts and known or putative enhancers are designated by filled and open arrowheads, respectively, at the bottom.
Acetylated histone (AcH3K9) was observed in both promoters and enhancers in TFF1 and GREB1 as expected. Histone 3 lysine-4 methylation is generally associated with active genes, but the profile of individual modifications is significantly distinct: Me1H3K4 seems to associate broadly with active genes, but, in contrast to the situation in yeast, this modification is not preferentially linked to the 3′ end of active genes (2, 20, 21). Me2H3K4 marks both promoters and enhancers with a clear preference for promoters over enhancers. Again, in contrast to events in yeast, we did not detect substantial Me2H3K4 in the transcribed regions of GREB1 and other tiled genes. Me3H3K4 was found exclusively in promoters, which agrees with most mapping studies in yeast and mammalian cells (20, 22, 23).
Interestingly, AcH3K9 and methylated H3K4 marks were present in a number of gene promoters, including KAI1 (Fig. 4), where no RNA transcripts were detected in MCF-7 cells. These observations suggest either that some histone modifications take place before the recruitment of the general transcriptional machinery or that these genes may be transcribing at an undetectable basal level. Although one cannot formally distinguish between these possibilities, the histone modification pattern is clearly different from “silent” genes such as RARβ (Fig. 4). The RARβ promoter was specifically marked by Me3H3K27, which is generally associated with silent genes in heterochromatin (24). Thus, there is heterogeneity in marks of nonexpressed genes exemplified by the observation that the KAI1 promoter is accessible to transcription factors, whereas the RARβ promoter is actively repressed in MCF-7 cells.
To further characterize histone modifications associated with gene repression, we mapped Me2H3K79 and Me3H3K9 (Fig. 4). Me2H3K79 has been previously implicated in interactions with Sir proteins during gene silencing (25, 26), although a more recent study suggested a link of this modification to gene activation (27). We found that Me2H3K79 was indeed associated with active genes, but in a distinct, gene-specific manner. In the case of TFF1, this modification took place in the transcribing region near the promoter, whereas in the case of GREB1, Me2H3K79 was spread in the entire transcription unit, including both coding and promoter/enhancer regions. Me3H3K9 has also been previously linked to gene repression by serving as the binding site for HP1 to facilitate the assembly of heterochromatin (25, 26, 28–30). Here we found that Me3H3K9 decorated most 3′ transcribed regions of both TFF1 and GREB1, consistent with a role of this specific histone modification in transcription elongation as recently suggested in yeast (31). These findings illustrate that it is still quite precipitous to generalize the significance of most histone modification events with respect to gene activation or repression, as the same histone modification may reflect or influence transcription positively or negatively in a highly gene-specific and locus-dependent manner, consistent with a combinatorial histone code.
Expression of Direct ERα Target Genes in Breast Cancer Cells and Tissues.
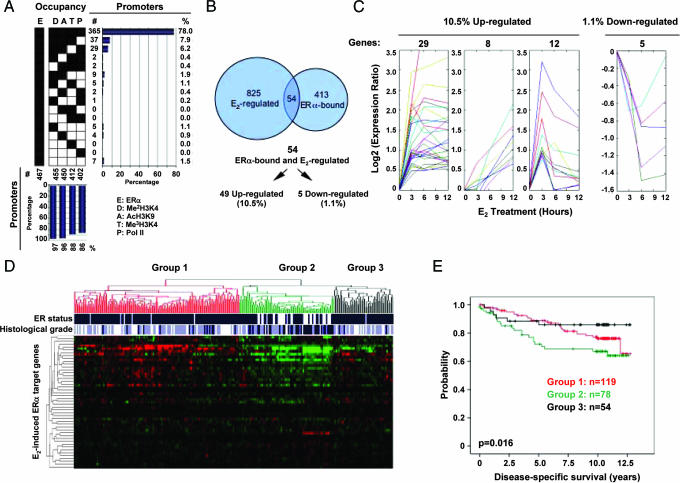
While the majority of ERα-bound promoters were also marked by Pol II and epigenetic markers associated with gene activation (Fig. 5A), we evaluated the time course of regulated gene expression by RNA profiling and identified 879 genes that responded to E2-induction in MCF-7 cells, which generally agrees with other published gene expression profiling studies (32–34). Strikingly, only 54 of these 879 E2-affected genes were bound by ERα in the promoter-proximal region (Fig. 5B), indicating that the majority of E2-induced genes might be indirectly affected or regulated by ER-responsive elements located away from the promoter-proximal region. Among these 879 E2-regulated genes, 562 were up-regulated and 317 were down-regulated. Contrary to the expectation that a similar percentage of genes in these two categories would be targeted by ERα in the promoter-proximal region, we found that 49 (10.5%) of ERα-bound promoters were up-regulated by E2, whereas only 5 (1.1%) were down-regulated (Fig. 5 B and C). These observations suggest that many genes in both up- and down-regulated categories might be indirectly affected, with more down-regulated genes influenced by indirect mechanisms than up-regulated ones (35).
Fig. 5.
E2-induced gene expression and the biological relevance of direct ERα target genes. (A) Relationship between ERα binding and histone modifications. To directly compare ERα binding and E2-induced gene expression, 467 of 578 ERα-bound promoters common between our promoter array and the Illumina gene expression array were analyzed. The majority of ERα-positive promoters was also marked by Pol II and modified histones associated with gene activation. (B) Venn diagram showing the overlap between ERα-bound promoters and E2-induced genes. (C) Gene expression profiling in response to E2 treatment. ERα-bound and E2-regulated genes are grouped into four distinct classes. Among up-regulated genes, 29 were rapidly induced, and the level remained relatively constant afterward; eight were induced in a time-dependent manner; and 12 were induced followed by a rapid decay. E2-induced, genes represent 10.5% of total ERα-bound genes in the promoter-proximal region. Only five ERα-bound genes were down-regulated by E2, which represent 1.1% of total ERα-bound genes in the promoter-proximal region. (D) Segregation of ER expression and breast tumor grade (both indicated at the top by blue bars) based on ERα-bound and E2-induced genes in MCF-7 cells. (E) Kaplan–Meier plots of patient survival in different groups segregated based on ERα-bound and E2-inducted genes in MCF-7 cells. Statistical significance was determined by the χ2 test.
Conversely, the fact that only 54 ERα-bound promoters responded to E2 with rapid changes in mRNA levels suggests that most ERα-bound promoters may require additional cofactors for E2-dependent gene expression, as has been previously documented (36–39). Consequently, we predicted that different sets of ERα-occupied promoters might respond to E2 stimulation in different cell types. Indeed, we have found that a subset of ERα-bound, but E2-insensitive, promoters in MCF-7 cells could be directly targeted by ERα and induced by E2 in U2OS cells stably expressing ERα (data not shown). This observation suggests that, at least for some promoters, they represent bona fide estrogen target genes under different circumstances.
To further investigate the biological relevance of ERα-binding and estrogen-regulated gene expression, we asked how the newly identified 54 E2-responsive ERα target genes might be differentially regulated in breast cancer tissues using a comprehensive set of gene expression profiling data from 251 breast cancer patients (40). We found a direct correlation between gene expression and tumor progression by unsupervised hierarchical clustering (Fig. 5D). Patients were clustered into three groups. About half of the genes were strongly suppressed in group 2, which displayed an ER-negative status and advanced tumor grade (Fig. 5D). Significantly, this patient group exhibited a much reduced survival rate compared with the two other groups (Fig. 5E). These results illustrate a general strategy for disease etiology studies by combining gene expression profiling with location analysis of key transcriptional regulators altered in specific diseases.
Materials and Methods
Cell Culture and Antibodies.
MCF-7 cells were cultured in MEM supplemented with 10% FBS. Before induction, cells were hormone-deprived for 4 days in phenol-free MEM plus charcoal-depleted FBS and then treated with 100 nM E2 (Sigma–Aldrich, St. Louis, MO) for 1 h for ChIP or various periods of time for RNA profiling as indicated. Antibodies used for ChIP analyses were anti-RNAP (8WG16) (MMS-126R; Covance, Princeton, NJ), anti-ERα (HC-20 and H-184 combined; Santa Cruz Biotechnology, Santa Cruz, CA), anti-CBP (C-20 and A22 combined; Santa Cruz Biotechnology). All anti-modified histone antibodies are from Upstate Biotechnology (Lake Placid, NY), including anti-AcH3K9 (07-352), anti-Me1H3K4 (07-436), anti-Me2H3K4 (07-030), anti-Me3H3K4 (07-473), anti-Me3H3K9 (07-442), anti-Me3H3K27 (07-449), and anti-Me2H3K79 (07-366).
Array Fabrication and the ChIP-DSL Assay.
Human promoters were annotated by aligning Refseq mRNAs against the human genome and extended by using existing ESTs. A sequence from +200 to −800 bp relative to each transcription start was used to determine the most unique 40-mer to represent that promoter. All 40-mer oligonucleotides were amino-derived during oligo synthesis and printed on the 3D-CodeLink slides according to the manufacturer's instructions (Amersham Biosciences). Corresponding to each 40-mer, a pair of assay oligonucleotides were synthesized, each containing a half of the 40-mer sequence, flanked by a universal primer binding site. The built-in tiling paths for internal controls were based on sequences from multiple human genes, and oligonucleotides probes were selected at the ≈0.5-kb interval across each gene unit. The genomic coordinates for the annotated human gene promoters and the array data have been submitted to ArrayExpress (www.ebi.ac.uk/aerep).
Cells were cross-linked by formaldehyde and subjected to standard ChIP as previously described (41). Cells in one 100-mm dish were used for each ChIP-DSL experiment. Both input (≈5% of total DNA) and antibody-enriched DNA were randomly biotinylated by using a kit (Vector Laboratories) according to the manufacturer's instructions. All T7-linked assay oligonucleotides were kinased and then mixed with all T3-linked oligonucleotides. For each reaction, we used 0.1 pmol per oligonucleotide in a pool suspended in 10 μl of TE buffer. The procedure for oligonucleotide annealing, solid phase selection, ligation, and PCR amplification was as described (42), except Taq ligase was used in place of T4 ligase to improve ligation specificity. Input DNA was labeled with Alexa Fluor 647 and chromatin immunoprecipitated DNA with Cy3. The PCR products were mixed, denatured, and hybridized to the 40-mer Hu20K array. Slides were scanned on the GenPix 4000B scanner (Axon Instruments). The Hu20K array and the associated assay kit with detailed instruction are commercially available from Aviva Systems Biology.
Data Analysis.
The single-array error model was previously described (43, 44). The SAM analysis package (www-stat.stanford.edu/∼tibs/SAM) was previously described (45). Chipper (http://llama.med.harvard.edu/cgi/Chipper/chip3.py?id=725676) was described (46). After conducting analysis with these methods, we first obtained genes at P < 0.0001 according to the single-error model and selected the same number of genes from the top list of the other two methods to identify genes that were scored significantly by all three methods. Clearly, genes identified by only one or two of the methods may still be highly significant.
Supplementary Material
Acknowledgments
We are indebted to Bing Ren for generous help during the course of the technology development. We thank colleagues in our laboratories, especially V. Lunyak for insightful discussion and advice and C. Nelson for cell culture. M.G.R. is an Investigator of the Howard Hughes Medical Institute. This work was supported by grants from the National Institutes of Health and the Vitamin Cases Consumer Settlement Fund (to M.G.R.), and by National Cancer Institute Grant CA114184 and National Human Genome Research Institute Grant HG003119 (to X.-D.F.).
Abbreviations
- DSL
DNA selection and ligation
- E2
17β-estradiol
- Pol
polymerase
- qPCR
quantitative PCR
- NR
nuclear receptor.
Footnotes
Data deposition: The genomic coordinates for the annotated human gene promoters and the array data have been deposited in the ArrayExpress database, www.ebi.ac.uk/aerep (accession nos. E-MEXP-984 and E-TABM-231).
This article contains supporting information online at www.pnas.org/cgi/content/full/0700715104/DC1.
References
- 1.Strahl BD, Allis CD. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Fischle W, Wang Y, Allis CD. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 3.Turner BM. BioEssays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 4.Sims RJ, III, Nishioka K, Reinberg D. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Rosenfeld MG, Lunyak VV, Glass CK. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 6.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 7.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 10.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 12.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 13.Laganiere J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguere V. Proc Natl Acad Sci USA. 2005;102:11651–11656. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan JB, Yeakley JM, Bibikova M, Chudin E, Wickham E, Chen J, Doucet D, Rigault P, Zhang B, Shen R, et al. Genome Res. 2004;14:878–885. doi: 10.1101/gr.2167504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Bassets I, Kwon Y-S, Telese F, Perfontaine GG, Hutt KR, Cheng CS, Ju B-G, Ohgi KA, Wang J, Escoubet-Lozach L, et al. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 18.Klinge CM. Nucleic Acids Res. 2001;29:2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh MG, Thompson DA, Weigel RJ. Cancer Res. 2000;60:6367–6375. [PubMed] [Google Scholar]
- 20.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 21.Schubeler D, Turner BM. Cell. 2005;122:489–492. doi: 10.1016/j.cell.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 22.Ng HH, Robert F, Young RA, Struhl K. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 23.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, III, Gingeras TR, et al. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 25.Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Curr Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 26.Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kouskouti A, Talianidis I. EMBO J. 2005;24:347–357. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maison C, Almouzni G. Nat Rev Mol Cell Biol. 2004;5:296–304. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- 29.Melcher M, Schmid M, Aagaard L, Selenko P, Laible G, Jenuwein T. Mol Cell Biol. 2000;20:3728–3741. doi: 10.1128/mcb.20.10.3728-3741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 31.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 32.Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Endocrinology. 2003;144:4562–4574. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- 33.Coser KR, Chesnes J, Hur J, Ray S, Isselbacher KJ, Shioda T. Proc Natl Acad Sci USA. 2003;100:13994–13999. doi: 10.1073/pnas.2235866100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rae JM, Johnson MD, Scheys JO, Cordero KE, Larios JM, Lippman ME. Breast Cancer Res Treat. 2005;92:141–149. doi: 10.1007/s10549-005-1483-4. [DOI] [PubMed] [Google Scholar]
- 35.Zhu P, Baek SH, Bourk EM, Ohgi KA, Garcia-Bassets I, Sanjo H, Akira S, Kotol PF, Glass CK, Rosenfeld MG, Rose DW. Cell. 2006;124:615–629. doi: 10.1016/j.cell.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 36.Saville B, Wormke M, Wang F, Nguyen T, Enmark E, Kuiper G, Gustafsson JA, Safe S. J Biol Chem. 2000;275:5379–5387. doi: 10.1074/jbc.275.8.5379. [DOI] [PubMed] [Google Scholar]
- 37.Stein B, Yang MX. Mol Cell Biol. 1995;15:4971–4979. doi: 10.1128/mcb.15.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeNardo DG, Kim HT, Hilsenbeck S, Cuba V, Tsimelzon A, Brown PH. Mol Endocrinol. 2005;19:362–378. doi: 10.1210/me.2004-0267. [DOI] [PubMed] [Google Scholar]
- 39.Cheng AS, Jin VX, Fan M, Smith LT, Liyanarachchi S, Yan PS, Leu YW, Chan MW, Plass C, Nephew KP, et al. Mol Cell. 2006;21:393–404. doi: 10.1016/j.molcel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 40.Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu ET, Bergh J. Proc Natl Acad Sci USA. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 42.Yeakley JM, Fan JB, Doucet D, Luo L, Wickham E, Ye Z, Chee MS, Fu XD. Nat Biotechnol. 2002;20:353–358. doi: 10.1038/nbt0402-353. [DOI] [PubMed] [Google Scholar]
- 43.Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B. Proc Natl Acad Sci USA. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren B, Dynlacht BD. Methods Enzymol. 2004;376:304–315. doi: 10.1016/S0076-6879(03)76020-0. [DOI] [PubMed] [Google Scholar]
- 45.Tusher VG, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibbons FD, Proft M, Struhl K, Roth FP. Genome Biol. 2005;6:R96. doi: 10.1186/gb-2005-6-11-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.