Abstract
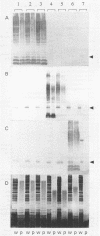
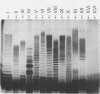
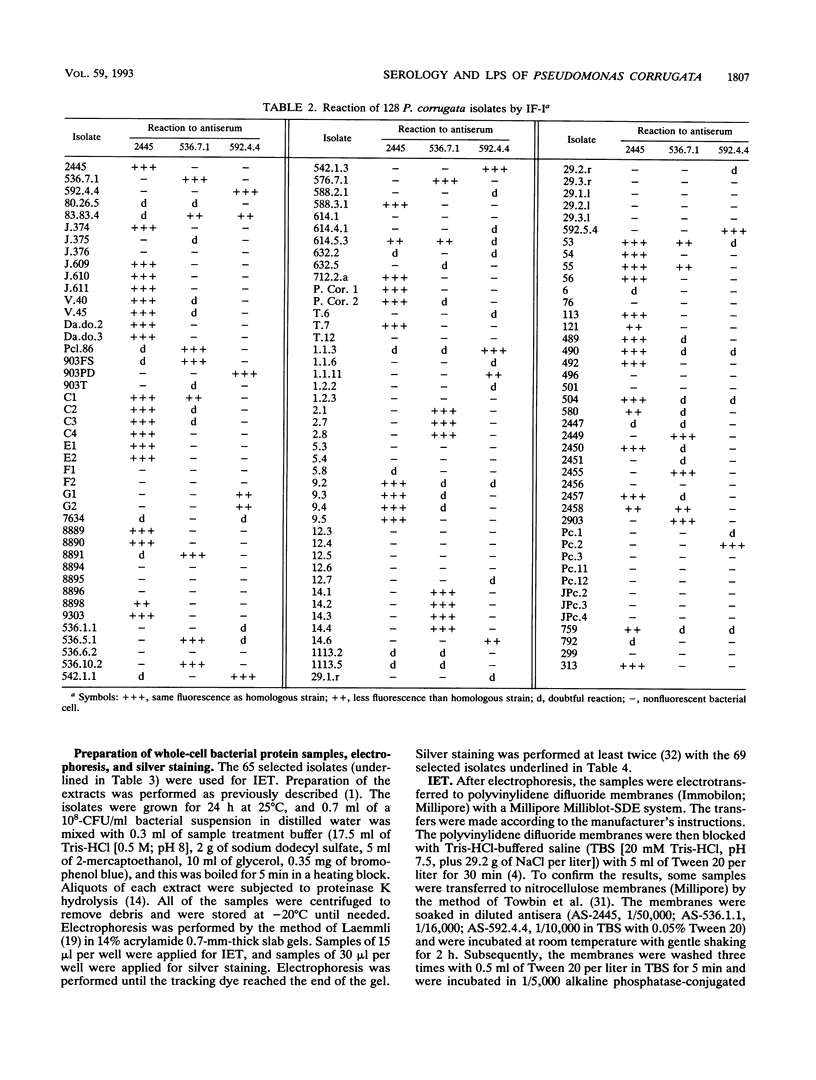
The variation in biochemical and serological features of 128 isolates of Pseudomonas corrugata has been studied with 56 isolates from Spain and 72 isolates from other countries. Isolates were analyzed with common diagnostic tests and with the AP150CHE system. Variability among isolates for some standard tests usually listed as positive or negative for this species, such as arginine dihydrolase and gelatin hydrolysis, lipase and lecithinase activities, pigment production, and wrinkled colony morphology, was observed. Three antisera were raised against the type strain and two Spanish isolates from tomato and pepper plants. Serological reactions were studied by indirect immunofluorescence and indirect enzyme-linked immunosorbent assay. Eighty-three isolates reacted with a single antiserum, 6 reacted with two antisera, and none reacted with three antisera. Thirty-nine isolates did not react with any of the three antisera. These results suggest that serology will not be a useful method for routine diagnosis of P. corrugata unless common antigens can be identified. Electrophoresis and immunoelectrotransfer were used to study the antigens involved. Each antiserum reacted with whole-cell lysates, giving two common bands for P. corrugata isolates and other Pseudomonas species and a ladder-like pattern characteristic of lipopolysaccharides (LPS). Common bands were not observed after proteinase K treatment. More than 10 LPS patterns were distinguished in 98 isolates after silver staining of polyacrylamide gels. There was no correlation between the geographical origin or host of the isolates and the LPS patterns. A correlation between LPS groups and serological reaction was observed.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker C. J., Neilson M. J., Sequeira L., Keegstra K. G. Chemical Characterization of the Lipopolysaccharide of Pseudomonas solanacearum. Appl Environ Microbiol. 1984 May;47(5):1096–1100. doi: 10.1128/aem.47.5.1096-1100.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batteiger B., Newhall W. J., 5th, Jones R. B. The use of Tween 20 as a blocking agent in the immunological detection of proteins transferred to nitrocellulose membranes. J Immunol Methods. 1982 Dec 30;55(3):297–307. doi: 10.1016/0022-1759(82)90089-8. [DOI] [PubMed] [Google Scholar]
- Blake M. S., Johnston K. H., Russell-Jones G. J., Gotschlich E. C. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem. 1984 Jan;136(1):175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- Goldman R. C., Hunt F. Mechanism of O-antigen distribution in lipopolysaccharide. J Bacteriol. 1990 Sep;172(9):5352–5359. doi: 10.1128/jb.172.9.5352-5359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUGH R., LEIFSON E. The taxonomic significance of fermentative versus oxidative metabolism of carbohydrates by various gram negative bacteria. J Bacteriol. 1953 Jul;66(1):24–26. doi: 10.1128/jb.66.1.24-26.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrick C. A., Sequeira L. Lipopolysaccharide-Defective Mutants of the Wilt Pathogen Pseudomonas solanacearum. Appl Environ Microbiol. 1984 Jul;48(1):94–101. doi: 10.1128/aem.48.1.94-101.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock P. J., Brown T. M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983 Apr;154(1):269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KING E. O., WARD M. K., RANEY D. E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954 Aug;44(2):301–307. [PubMed] [Google Scholar]
- KOVACS N. Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature. 1956 Sep 29;178(4535):703–703. doi: 10.1038/178703a0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. M., Frasch C. E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982 Jan 1;119(1):115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- de Weger L. A., Jann B., Jann K., Lugtenberg B. Lipopolysaccharides of Pseudomonas spp. that stimulate plant growth: composition and use for strain identification. J Bacteriol. 1987 Apr;169(4):1441–1446. doi: 10.1128/jb.169.4.1441-1446.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]