Abstract
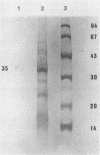
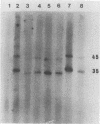
The only major structural protein (35 kDa) of the lactococcal small isometric-headed bacteriophage ul36, a member of the P335 species, was isolated from a preparative sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. Monoclonal antibodies (MAbs) were raised against the denatured 35-kDa protein. Six MAbs were selected and characterized. Western blots (immunoblots) showed that all MAbs recognized the 35 kDa but also a 45 kDa that is in lower concentration in the phage structure. Binding inhibition assays identified five families of MAbs that recognized nonoverlapping epitopes of the 35- and 45-kDa proteins. Immunoelectron microscopy showed that these two proteins are localized within the phage head, therefore indicating that the 35 kDa is a major capsid protein of ul36 and that the 45 kDa is a minor capsid protein. With two MAbs, a sandwich enzyme-linked immunosorbent assay (ELISA) was developed for direct detection of lactococcal phages in whey and milk samples. Whey and milk components, however, interfered with the conduct of the assay. Partial denaturation of milk samples by heat treatment in the presence of SDS and β-mercaptoethanol removed the masking effect and increased the sensitivity of the assay by 100-fold. With the method used here, 107 PFU/ml were detected by the ELISA within 2 h without any steps to enrich or isolate bacteriophages.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alatossava T., Klaenhammer T. R. Molecular Characterization of Three Small Isometric-Headed Bacteriophages Which Vary in Their Sensitivity to the Lactococcal Phage Resistance Plasmid pTR2030. Appl Environ Microbiol. 1991 May;57(5):1346–1353. doi: 10.1128/aem.57.5.1346-1353.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D. K., Kim J. H., Batt C. A. Cloning and nucleotide sequence of the major capsid protein from Lactococcus lactis ssp. cremoris bacteriophage F4-1. Gene. 1991 May 15;101(1):121–125. doi: 10.1016/0378-1119(91)90233-2. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Jarvis A. W., Fitzgerald G. F., Mata M., Mercenier A., Neve H., Powell I. B., Ronda C., Saxelin M., Teuber M. Species and type phages of lactococcal bacteriophages. Intervirology. 1991;32(1):2–9. doi: 10.1159/000150179. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Moineau S., Durmaz E., Pandian S., Klaenhammer T. R. Differentiation of Two Abortive Mechanisms by Using Monoclonal Antibodies Directed toward Lactococcal Bacteriophage Capsid Proteins. Appl Environ Microbiol. 1993 Jan;59(1):208–212. doi: 10.1128/aem.59.1.208-212.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moineau S., Pandian S., Klaenhammer T. R. Restriction/Modification systems and restriction endonucleases are more effective on lactococcal bacteriophages that have emerged recently in the dairy industry. Appl Environ Microbiol. 1993 Jan;59(1):197–202. doi: 10.1128/aem.59.1.197-202.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourad W., Pelletier G., Boulet A., Islam N., Valet J. P., Hébert J. Allergenicity and cross-reactivity of rye grass pollen extracts revealed by monoclonal antibodies. J Immunol Methods. 1986 May 1;89(1):53–59. doi: 10.1016/0022-1759(86)90031-1. [DOI] [PubMed] [Google Scholar]
- Prevots F., Mata M., Ritzenthaler P. Taxonomic differentiation of 101 lactococcal bacteriophages and characterization of bacteriophages with unusually large genomes. Appl Environ Microbiol. 1990 Jul;56(7):2180–2185. doi: 10.1128/aem.56.7.2180-2185.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouler C., Bouet C., Ritzenthaler P., Drouet X., Mata M. Characterization of Lactococcus lactis phage antigens. Appl Environ Microbiol. 1992 Aug;58(8):2479–2484. doi: 10.1128/aem.58.8.2479-2484.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenson L. R., Klaenhammer T. R. Streptococcus cremoris M12R transconjugants carrying the conjugal plasmid pTR2030 are insensitive to attack by lytic bacteriophages. Appl Environ Microbiol. 1985 Oct;50(4):851–858. doi: 10.1128/aem.50.4.851-858.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt R. F., Jr, Phillips D. L., Henderson L. O., Whitfield W., Spierto F. W. Quantitative differences among various proteins as blocking agents for ELISA microtiter plates. J Immunol Methods. 1987 Jul 16;101(1):43–50. doi: 10.1016/0022-1759(87)90214-6. [DOI] [PubMed] [Google Scholar]
- Wernars K., Heuvelman C. J., Chakraborty T., Notermans S. H. Use of the polymerase chain reaction for direct detection of Listeria monocytogenes in soft cheese. J Appl Bacteriol. 1991 Feb;70(2):121–126. doi: 10.1111/j.1365-2672.1991.tb04437.x. [DOI] [PubMed] [Google Scholar]