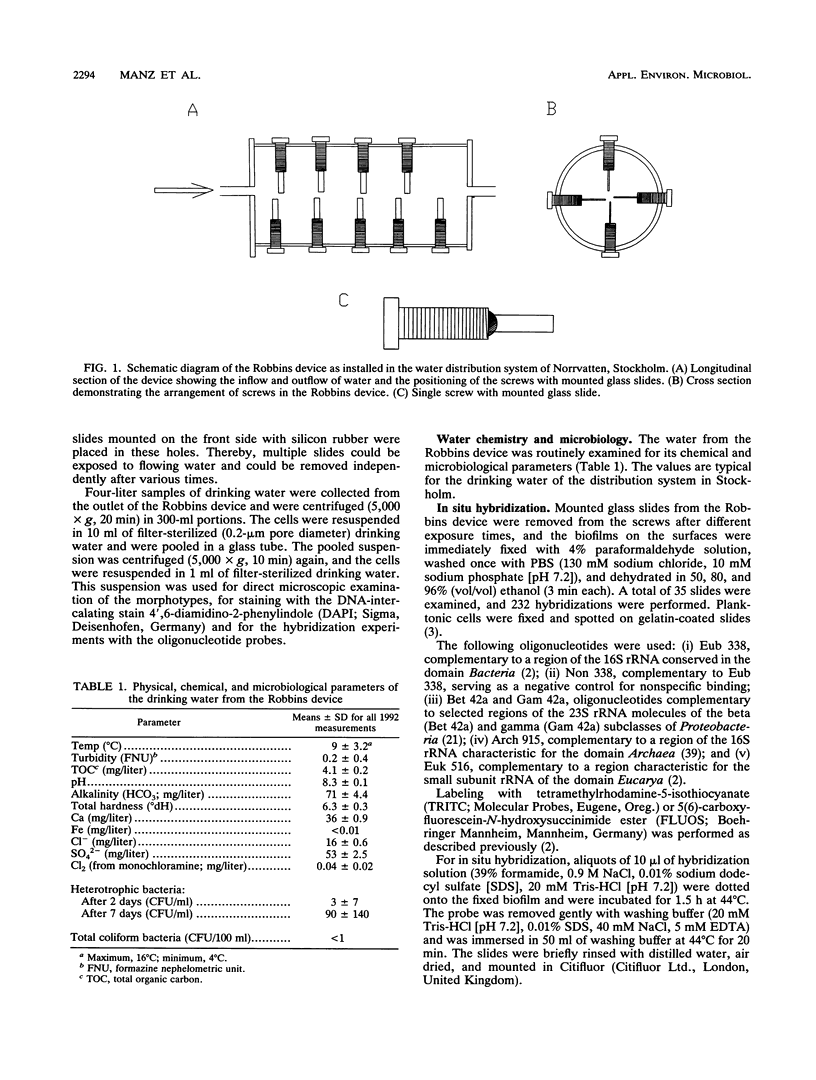
Abstract
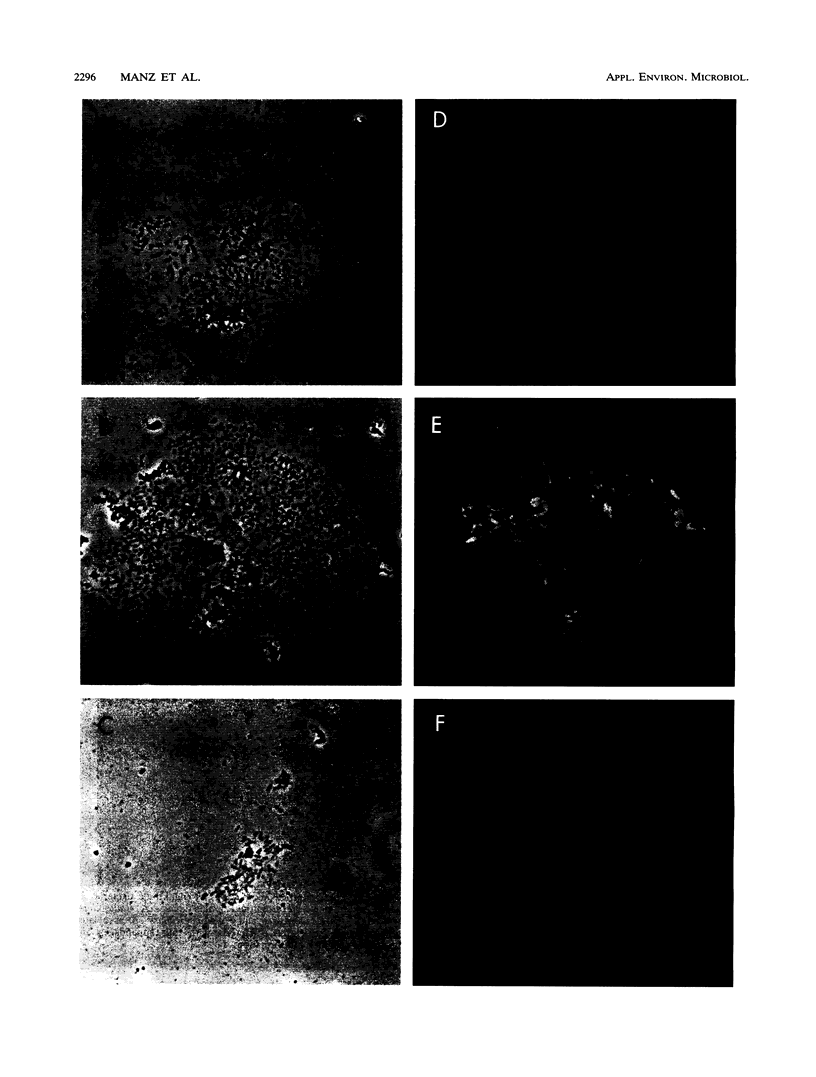
Free-water-phase and surface-associated microorganisms from drinking water were detected and roughly identified by hybridization with fluorescence-labeled oligonucleotide probes complementary to regions of 16S and 23S rRNA characteristic for the domains Bacteria, Archaea, and Eucarya and the beta and gamma subclasses of Proteobacteria. Samples of glass-attached biofilms and plankton were taken from a Robbins device installed in a water distribution system. More than 70% of the surface-associated cells and less than 40% of the planktonic cells visualized by 4',6-diamidino-2-phenylindole staining bound detectable amounts of rRNA-targeted probes. These findings are an indication for higher average rRNA content and consequently higher physiological activity of the attached microbial cells compared with the free-living cells. All detectable cells hybridized with the bacterial probe, whereas no Archaea and no Eucarya cells could be detected. Simultaneous hybridization with probes specific for the beta and gamma subclasses of Proteobacteria revealed that microcolonies already consisted of mixed populations in early stages with fewer than 50 cells. These observations provide further evidence that the coexistence and interaction of bacteria in drinking water biofilms may be an integral part of their growth and survival strategies.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann R. I., Binder B. J., Olson R. J., Chisholm S. W., Devereux R., Stahl D. A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990 Jun;56(6):1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Krumholz L., Stahl D. A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990 Feb;172(2):762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Stromley J., Devereux R., Key R., Stahl D. A. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl Environ Microbiol. 1992 Feb;58(2):614–623. doi: 10.1128/aem.58.2.614-623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R., Springer N., Ludwig W., Görtz H. D., Schleifer K. H. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature. 1991 May 9;351(6322):161–164. doi: 10.1038/351161a0. [DOI] [PubMed] [Google Scholar]
- Bright J. J., Fletcher M. Amino Acid assimilation and electron transport system activity in attached and free-living marine bacteria. Appl Environ Microbiol. 1983 Mar;45(3):818–825. doi: 10.1128/aem.45.3.818-825.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong E. F., Wickham G. S., Pace N. R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989 Mar 10;243(4896):1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- Dott W., Schoenen D. Qualitative und quantitative Bestimmung von Bakterienpopulationen aus aquatischen Biotopen. 7. Mitteilung: Entwicklung der Aufwuchsflora auf Werkstoffen im Trinkwasser. Zentralbl Bakteriol Mikrobiol Hyg B. 1985 May;180(5-6):436–447. [PubMed] [Google Scholar]
- Fletcher M. Measurement of Glucose Utilization by Pseudomonas fluorescens That Are Free-Living and That Are Attached to Surfaces. Appl Environ Microbiol. 1986 Oct;52(4):672–676. doi: 10.1128/aem.52.4.672-676.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni S. J., DeLong E. F., Olsen G. J., Pace N. R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988 Feb;170(2):720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks R. E., Amann R. I., Stahl D. A. Dual staining of natural bacterioplankton with 4',6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl Environ Microbiol. 1992 Jul;58(7):2158–2163. doi: 10.1128/aem.58.7.2158-2163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howgrave-Graham Alan R., Steyn Pieter L. Application of the Fluorescent-Antibody Technique for the Detection of Sphaerotilus natans in Activated Sludge. Appl Environ Microbiol. 1988 Mar;54(3):799–802. doi: 10.1128/aem.54.3.799-802.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell C. R., Hui Y. Design and testing of a functional group-specific DNA probe for the study of natural populations of acetogenic bacteria. Appl Environ Microbiol. 1991 Sep;57(9):2602–2609. doi: 10.1128/aem.57.9.2602-2609.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy W. F., Bryers J. D., Robbins J., Costerton J. W. Observations of fouling biofilm formation. Can J Microbiol. 1981 Sep;27(9):910–917. doi: 10.1139/m81-143. [DOI] [PubMed] [Google Scholar]
- McCoy W. F., Olson B. H. Analysis of the microbiological particulates in municipal drinking-water by scanning electron microscopy/X-ray energy spectroscopy. Zentralbl Bakteriol Mikrobiol Hyg B. 1987 Apr;183(5-6):511–529. [PubMed] [Google Scholar]
- McFeters G. A., Singh A., Byun S., Callis P. R., Williams S. Acridine orange staining reaction as an index of physiological activity in Escherichia coli. J Microbiol Methods. 1991;13:87–97. doi: 10.1016/0167-7012(91)90009-f. [DOI] [PubMed] [Google Scholar]
- Nickel J. C., Ruseska I., Wright J. B., Costerton J. W. Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob Agents Chemother. 1985 Apr;27(4):619–624. doi: 10.1128/aac.27.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway H. F., Olson B. H. Scanning electron microscope evidence for bacterial colonization of a drinking-water distribution system. Appl Environ Microbiol. 1981 Jan;41(1):274–287. doi: 10.1128/aem.41.1.274-287.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J., Keevil C. W. Immunogold and fluorescein immunolabelling of Legionella pneumophila within an aquatic biofilm visualized by using episcopic differential interference contrast microscopy. Appl Environ Microbiol. 1992 Jul;58(7):2326–2330. doi: 10.1128/aem.58.7.2326-2330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Röbbecke R., Fischeder R. Mycobacteria in biofilms. Zentralbl Hyg Umweltmed. 1989 Jun;188(3-4):385–390. [PubMed] [Google Scholar]
- Stahl D. A., Flesher B., Mansfield H. R., Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988 May;54(5):1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. Polymerase chain reaction: applications in environmental microbiology. Annu Rev Microbiol. 1991;45:137–161. doi: 10.1146/annurev.mi.45.100191.001033. [DOI] [PubMed] [Google Scholar]
- Vestal J. R., White D. C. Lipid analysis in microbial ecology: quantitative approaches to the study of microbial communities. Bioscience. 1989 Sep;39(8):535–541. [PubMed] [Google Scholar]
- Zarda B., Amann R., Wallner G., Schleifer K. H. Identification of single bacterial cells using digoxigenin-labelled, rRNA-targeted oligonucleotides. J Gen Microbiol. 1991 Dec;137(12):2823–2830. doi: 10.1099/00221287-137-12-2823. [DOI] [PubMed] [Google Scholar]
- van Loosdrecht M. C., Lyklema J., Norde W., Zehnder A. J. Influence of interfaces on microbial activity. Microbiol Rev. 1990 Mar;54(1):75–87. doi: 10.1128/mr.54.1.75-87.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]




