Abstract
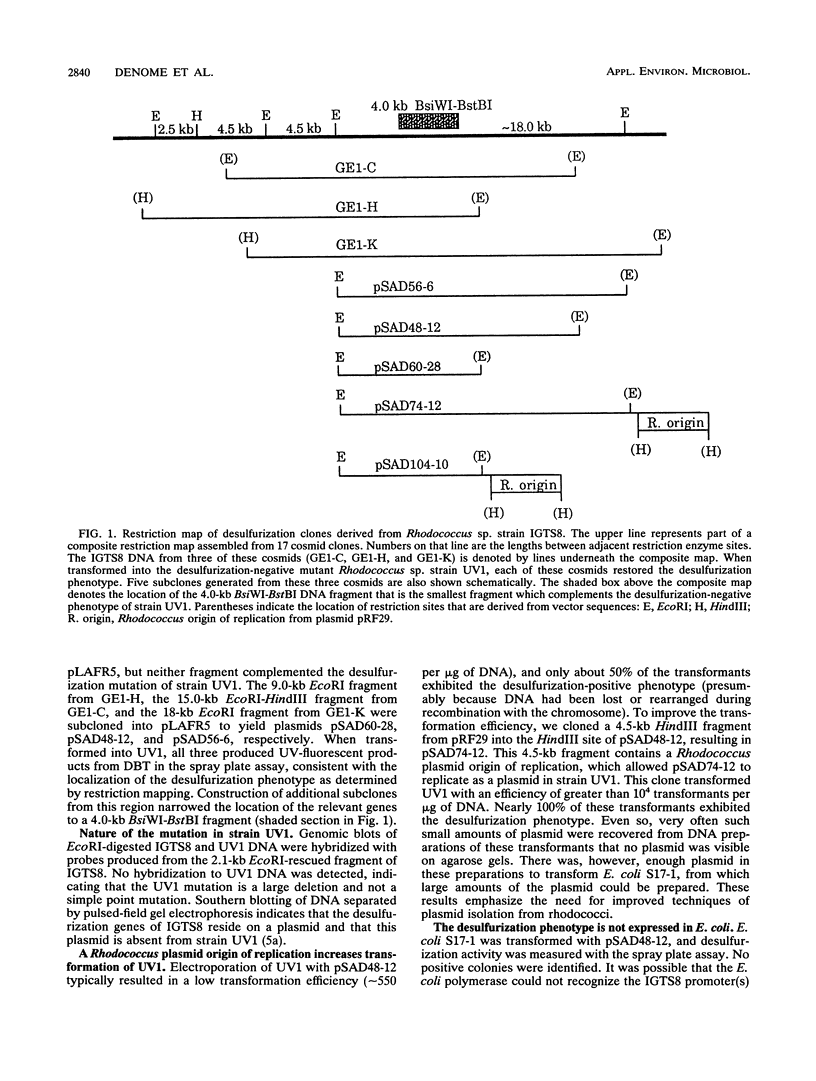
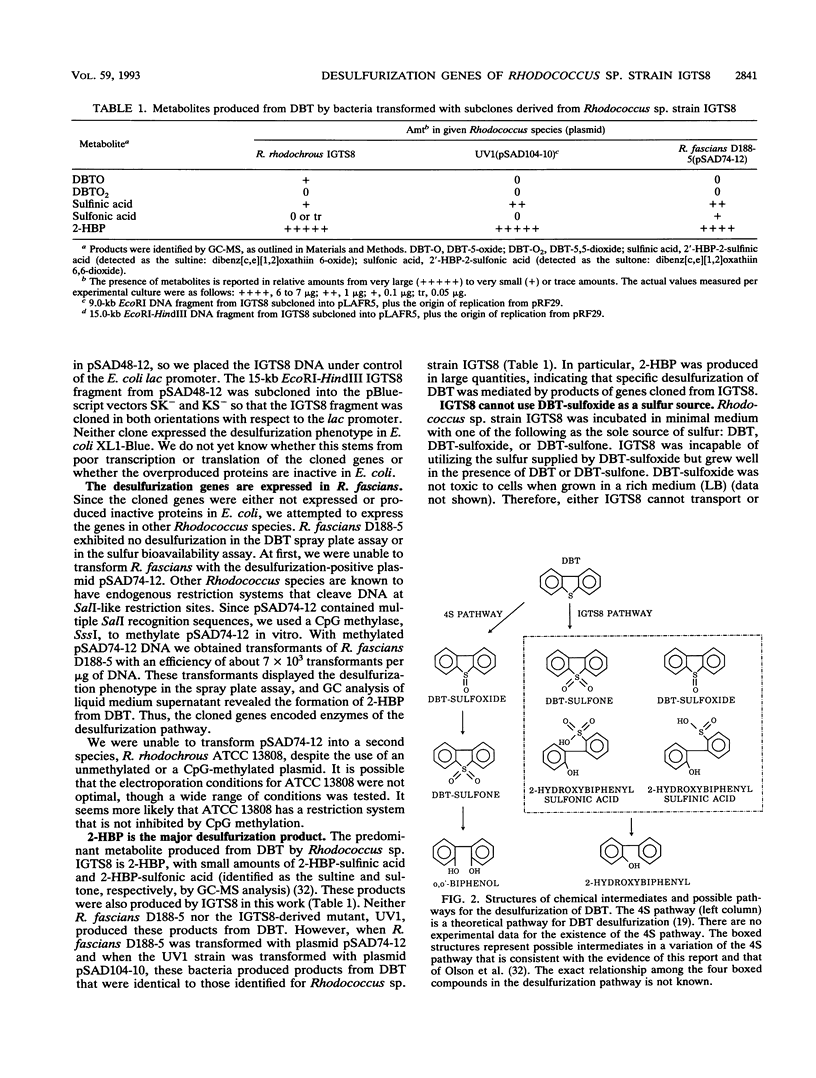
The gram-positive bacterium Rhodococcus sp. strain IGTS8 is able to remove sulfur from certain aromatic compounds without breaking carbon-carbon bonds. In particular, sulfur is removed from dibenzothiophene (DBT) to give the final product, 2-hydroxybiphenyl. A genomic library of IGTS8 was constructed in the cosmid vector pLAFR5, but no desulfurization phenotype was imparted to Escherichia coli. Therefore, IGTS8 was mutagenized, and a new strain (UV1) was selected that had lost the ability to desulfurize DBT. The genomic library was transferred into UV1, and several colonies that had regained the desulfurization phenotype were isolated, though free plasmid could not be isolated. Instead, vector DNA had integrated into either the chromosome or a large resident plasmid. DNA on either side of the inserted vector sequences was cloned and used to probe the original genomic library in E. coli. This procedure identified individual cosmid clones that, when electroporated into strain UV1, restored desulfurization. When the origin of replication from a Rhodococcus plasmid was inserted, the efficiency with which these clones transformed UV1 increased 20- to 50-fold and they could be retrieved as free plasmids. Restriction mapping and subcloning indicated that the desulfurization genes reside on a 4.0-kb DNA fragment. Finally, the phenotype was transferred to Rhodococcus fascians D188-5, a species normally incapable of desulfurizing DBT. The mutant strain, UV1, and R. fascians produced 2-hydroxybiphenyl from DBT when they contained appropriate clones, indicating that the genes for the entire pathway have been isolated.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Consevage M. W., Porter R. D., Phillips A. T. Cloning and expression in Escherichia coli of histidine utilization genes from Pseudomonas putida. J Bacteriol. 1985 Apr;162(1):138–146. doi: 10.1128/jb.162.1.138-146.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desomer J., Crespi M., Van Montagu M. Illegitimate integration of non-replicative vectors in the genome of Rhodococcus fascians upon electrotransformation as an insertional mutagenesis system. Mol Microbiol. 1991 Sep;5(9):2115–2124. doi: 10.1111/j.1365-2958.1991.tb02141.x. [DOI] [PubMed] [Google Scholar]
- Desomer J., Dhaese P., Montagu M. V. Transformation of Rhodococcus fascians by High-Voltage Electroporation and Development of R. fascians Cloning Vectors. Appl Environ Microbiol. 1990 Sep;56(9):2818–2825. doi: 10.1128/aem.56.9.2818-2825.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desomer J., Dhaese P., Van Montagu M. Conjugative transfer of cadmium resistance plasmids in Rhodococcus fascians strains. J Bacteriol. 1988 May;170(5):2401–2405. doi: 10.1128/jb.170.5.2401-2405.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Foght J. M., Westlake D. W. Expression of dibenzothiophene-degradative genes in two Pseudomonas species. Can J Microbiol. 1990 Oct;36(10):718–724. doi: 10.1139/m90-121. [DOI] [PubMed] [Google Scholar]
- Frischauf A. M. Digestion of DNA: size fractionation. Methods Enzymol. 1987;152:183–189. doi: 10.1016/0076-6879(87)52019-5. [DOI] [PubMed] [Google Scholar]
- Holland H. L., Khan S. H., Richards D., Riemland E. Biotransformation of polycyclic aromatic compounds by fungi. Xenobiotica. 1986 Aug;16(8):733–741. doi: 10.3109/00498258609043564. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen N. T., Tamaki S., Kobayashi D., Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988 Oct 15;70(1):191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- Kiyohara H., Nagao K., Yana K. Rapid screen for bacteria degrading water-insoluble, solid hydrocarbons on agar plates. Appl Environ Microbiol. 1982 Feb;43(2):454–457. doi: 10.1128/aem.43.2.454-457.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R., O'Connell M., Labes M., Pühler A. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 1986;118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- Singer M. E., Finnerty W. R. Construction of an Escherichia coli-Rhodococcus shuttle vector and plasmid transformation in Rhodococcus spp. J Bacteriol. 1988 Feb;170(2):638–645. doi: 10.1128/jb.170.2.638-645.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]


