Abstract
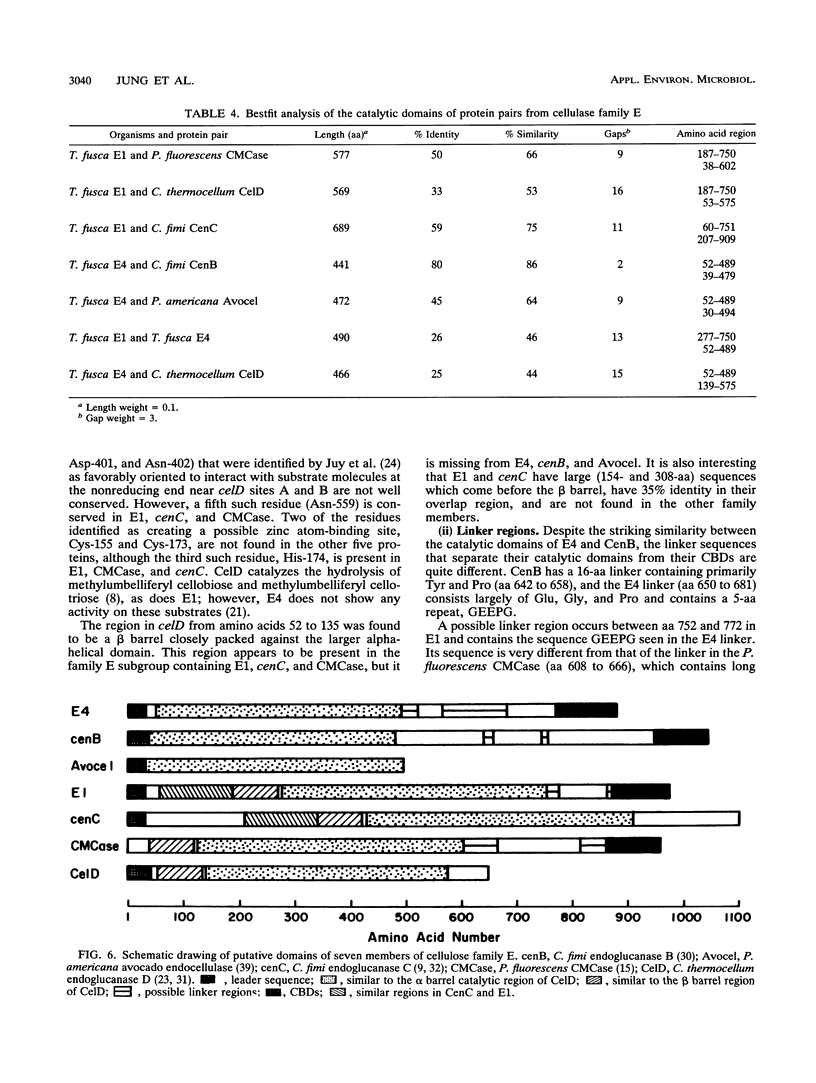
Two genes encoding cellulases E1 and E4 from Thermomonospora fusca have been cloned in Escherichia coli, and their DNA sequences have been determined. Both genes were introduced into Streptomyces lividans, and the enzymes were purified from the culture supernatants of transformants. E1 and E4 were expressed 18- and 4-fold higher, respectively, in S. lividans than in E. coli. Thin-layer chromatography of digestion products showed that E1 digests cellotriose, cellotetraose, and cellopentaose to cellobiose and a trace of glucose. E4 is poor at degrading cellotriose and cleaves cellopentaose to cellotetraose and glucose or cellotriose and cellobiose. It readily cleaves cellotetraose to cellobiose. E1 shows 59% identity to Cellulomonas fumi CenC in a 689-amino-acid overlap, and E4 shows 80% identity to the N terminus of C. fimi CenB in a 441-amino-acid overlap; all of these proteins are members of cellulase family E. Alignment of the amino acid sequences of Clostridium thermocellum celD, E1, E4, and four other members of family E demonstrates a clear relationship between their catalytic domains, although there is as little as 25% identity between some of them. Residues in celD that have been identified by site-directed mutagenesis and chemical modification to be important for catalytic activity are conserved in all seven proteins. The catalytic domains of E1 and E4 are not similar to those of T. fusca E2 or E5, but all four enzymes share similar cellulose-binding domains and have the same 14-bp inverted repeat upstream of their initiation codons. This sequence has been identified previously as the binding site for a protein that regulates induction.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bilofsky H. S., Burks C. The GenBank genetic sequence data bank. Nucleic Acids Res. 1988 Mar 11;16(5):1861–1863. doi: 10.1093/nar/16.5.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner F. R., Fiandt M., Hass K. K., Twose P. A., Szybalski W. Deletions and insertions in the immunity region of coliphage lambda: revised measurement of the promoter-startpoint distance. Virology. 1974 Dec;62(2):458–471. doi: 10.1016/0042-6822(74)90407-3. [DOI] [PubMed] [Google Scholar]
- Béguin P. Molecular biology of cellulose degradation. Annu Rev Microbiol. 1990;44:219–248. doi: 10.1146/annurev.mi.44.100190.001251. [DOI] [PubMed] [Google Scholar]
- Chauvaux S., Béguin P., Aubert J. P. Site-directed mutagenesis of essential carboxylic residues in Clostridium thermocellum endoglucanase CelD. J Biol Chem. 1992 Mar 5;267(7):4472–4478. [PubMed] [Google Scholar]
- Chirico W. J., Brown R. D., Jr Separation of [1-3H]cellooligosaccharides by thin-layer chromatography: assay for cellulolytic enzymes. Anal Biochem. 1985 Nov 1;150(2):264–272. doi: 10.1016/0003-2697(85)90509-3. [DOI] [PubMed] [Google Scholar]
- Claeyssens M., Henrissat B. Specificity mapping of cellulolytic enzymes: classification into families of structurally related proteins confirmed by biochemical analysis. Protein Sci. 1992 Oct;1(10):1293–1297. doi: 10.1002/pro.5560011008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho J. B., Moser B., Kilburn D. G., Warren R. A., Miller R. C., Jr Nucleotide sequence of the endoglucanase C gene (cenC) of Cellulomonas fimi, its high-level expression in Escherichia coli, and characterization of its products. Mol Microbiol. 1991 May;5(5):1221–1233. doi: 10.1111/j.1365-2958.1991.tb01896.x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Ghangas G. S., Wilson D. B. Cloning of the Thermomonospora fusca Endoglucanase E2 Gene in Streptomyces lividans: Affinity Purification and Functional Domains of the Cloned Gene Product. Appl Environ Microbiol. 1988 Oct;54(10):2521–2526. doi: 10.1128/aem.54.10.2521-2526.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes N. R., Henrissat B., Kilburn D. G., Miller R. C., Jr, Warren R. A. Domains in microbial beta-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991 Jun;55(2):303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Gilbert H. J. The nucleotide sequence of a carboxymethylcellulase gene from Pseudomonas fluorescens subsp. cellulosa. Mol Gen Genet. 1988 Jul;213(1):112–117. doi: 10.1007/BF00333406. [DOI] [PubMed] [Google Scholar]
- Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991 Dec 1;280(Pt 2):309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B., Claeyssens M., Tomme P., Lemesle L., Mornon J. P. Cellulase families revealed by hydrophobic cluster analysis. Gene. 1989 Sep 1;81(1):83–95. doi: 10.1016/0378-1119(89)90339-9. [DOI] [PubMed] [Google Scholar]
- Hu Y. J., Wilson D. B. Cloning of Thermomonospora fusca genes coding for beta 1-4 endoglucanases E1, E2 and E5. Gene. 1988 Nov 30;71(2):331–337. doi: 10.1016/0378-1119(88)90050-9. [DOI] [PubMed] [Google Scholar]
- Joliff G., Béguin P., Aubert J. P. Nucleotide sequence of the cellulase gene celD encoding endoglucanase D of Clostridium thermocellum. Nucleic Acids Res. 1986 Nov 11;14(21):8605–8613. doi: 10.1093/nar/14.21.8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao G., Ghangas G. S., Jung E. D., Wilson D. B. DNA sequences of three beta-1,4-endoglucanase genes from Thermomonospora fusca. J Bacteriol. 1991 Jun;173(11):3397–3407. doi: 10.1128/jb.173.11.3397-3407.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin E. S., Wilson D. B. Identification of a celE-binding protein and its potential role in induction of the celE gene in Thermomonospora fusca. J Bacteriol. 1988 Sep;170(9):3843–3846. doi: 10.1128/jb.170.9.3843-3846.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke A., Braun C., Gilkes N. R., Kilburn D. G., Miller R. C., Jr, Warren R. A. Unusual sequence organization in CenB, an inverting endoglucanase from Cellulomonas fimi. J Bacteriol. 1991 Jan;173(1):308–314. doi: 10.1128/jb.173.1.308-314.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S., Béguin P., Aubert J. P. Transcription of Clostridium thermocellum endoglucanase genes celF and celD. J Bacteriol. 1991 Jan;173(1):80–85. doi: 10.1128/jb.173.1.80-85.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser B., Gilkes N. R., Kilburn D. G., Warren R. A., Miller R. C., Jr Purification and characterization of endoglucanase C of Cellulomonas fimi, cloning of the gene, and analysis of in vivo transcripts of the gene. Appl Environ Microbiol. 1989 Oct;55(10):2480–2487. doi: 10.1128/aem.55.10.2480-2487.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai R., Horinouchi S., Beppu T. Cloning and nucleotide sequence of a cellulase gene, casA, from an alkalophilic Streptomyces strain. Gene. 1988 May 30;65(2):229–238. doi: 10.1016/0378-1119(88)90459-3. [DOI] [PubMed] [Google Scholar]
- Shen S. H., Chrétien P., Bastien L., Slilaty S. N. Primary sequence of the glucanase gene from Oerskovia xanthineolytica. Expression and purification of the enzyme from Escherichia coli. J Biol Chem. 1991 Jan 15;266(2):1058–1063. [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomme P., Chauvaux S., Béguin P., Millet J., Aubert J. P., Claeyssens M. Identification of a histidyl residue in the active center of endoglucanase D from Clostridium thermocellum. J Biol Chem. 1991 Jun 5;266(16):10313–10318. [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. B. Biochemistry and genetics of actinomycete cellulases. Crit Rev Biotechnol. 1992;12(1-2):45–63. doi: 10.3109/07388559209069187. [DOI] [PubMed] [Google Scholar]