Abstract
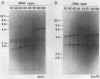
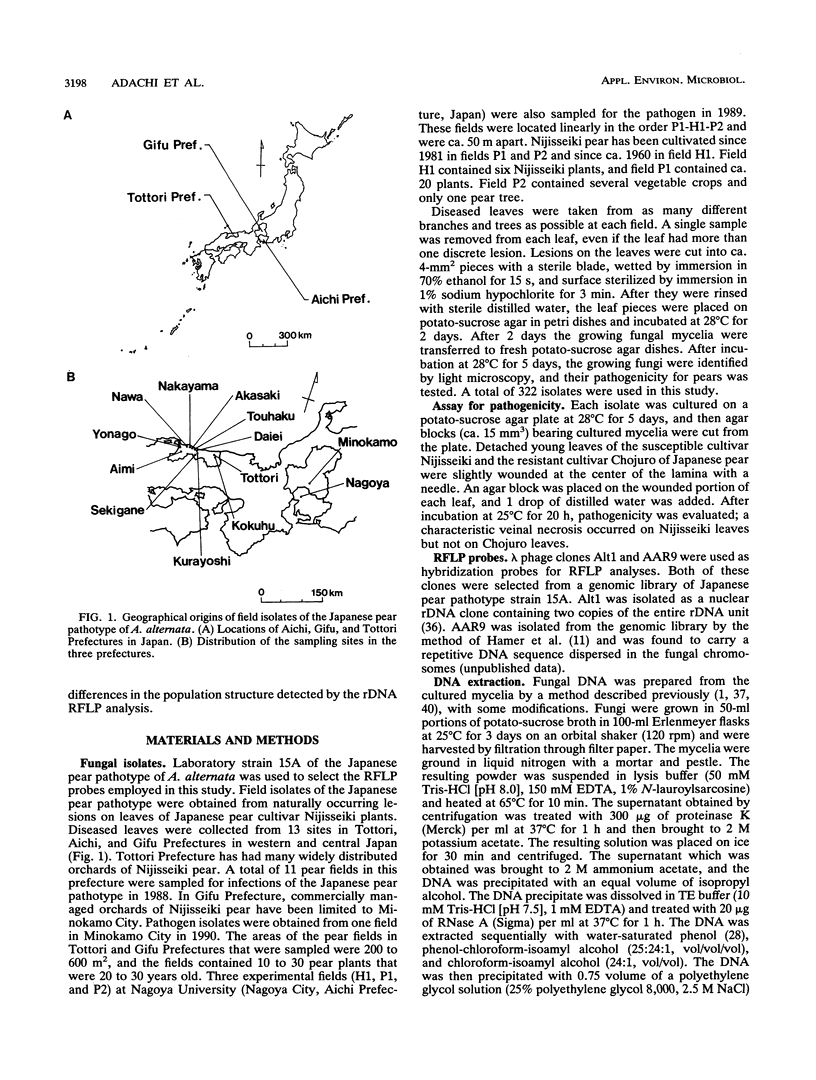
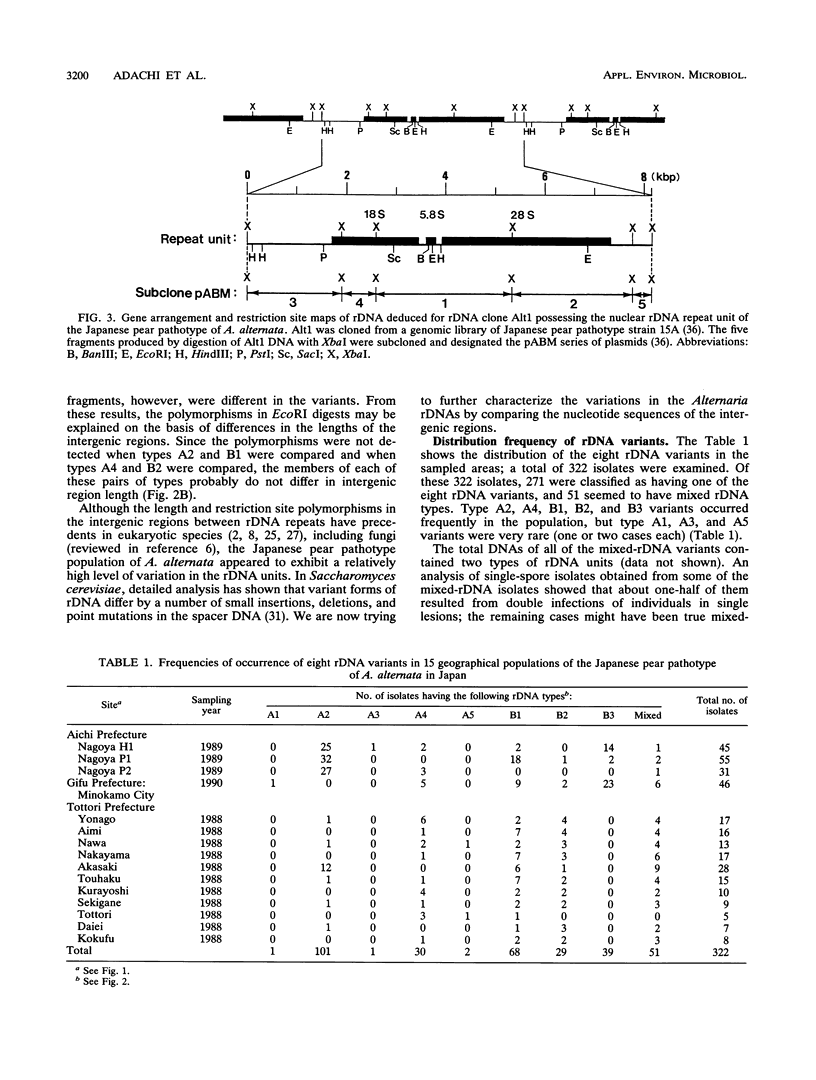
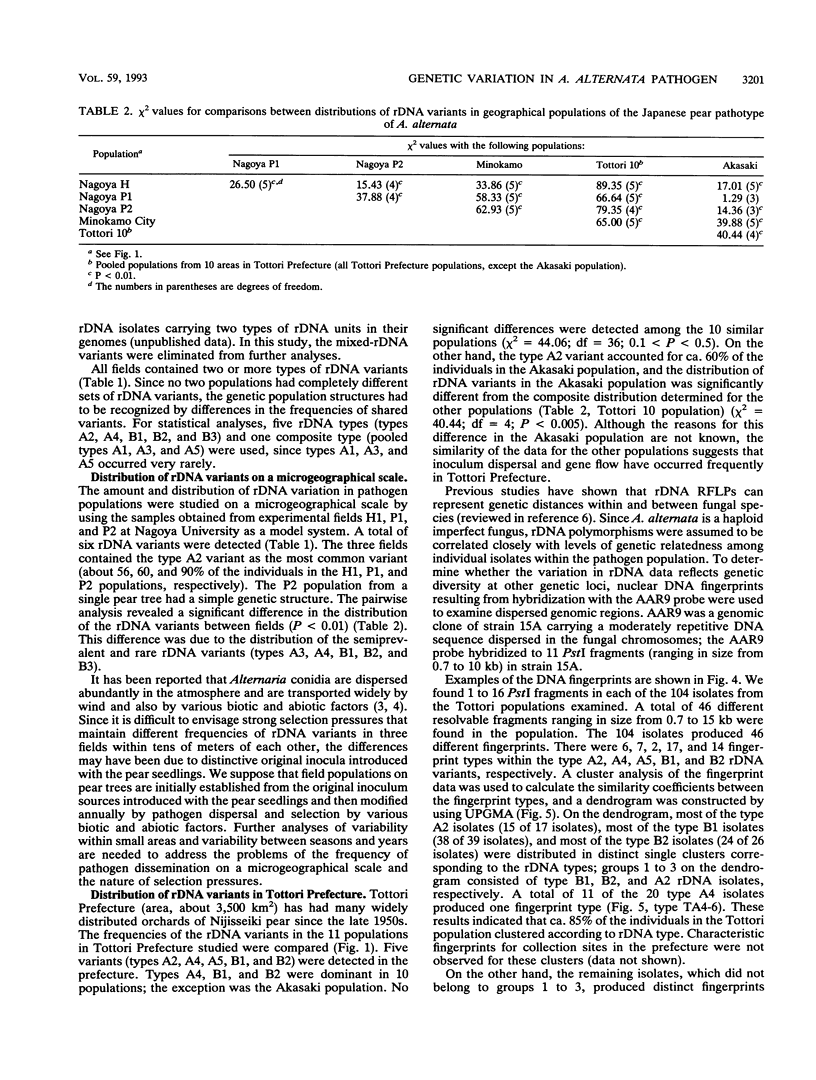
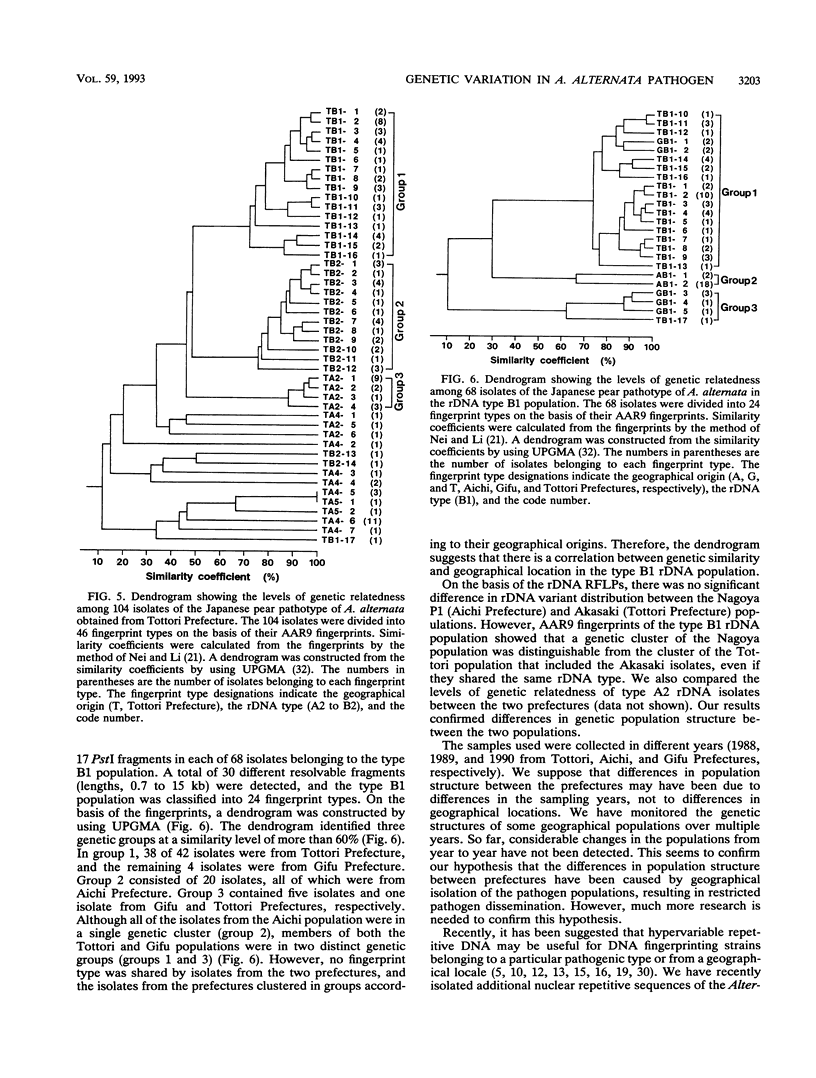
A restriction fragment length polymorphism analysis of nuclear ribosomal RNA genes (rDNA) was used to measure the amount and distribution of genetic variability in populations of the Japanese pear pathotype of Alternaria alternata on both micro- and macrogeographical scales. A total of 322 isolates were obtained from 13 areas in Aichi, Gifu, and Tottori Prefectures in central and western Japan. The restriction fragment length polymorphism analysis revealed that the pathogen populations contained at least eight rDNA variants. The eight variant types differed in the lengths and in the presence of the restriction sites in spacer DNA outside the coding regions for rRNAs. A total of 271 isolates were classified into the eight types. The remaining 51 isolates were determined to have mixed rDNA types. Single pear fields typically contained two to five types of rDNA variants. The frequencies of rDNA variants in 11 populations in Tottori Prefecture were compared; in this prefecture orchards containing the susceptible pear are common. Except for one collection site, there were no significant differences in the composition of the rDNA variants among the populations. This suggests that dispersal of inocula has occurred frequently in Tottori Prefecture. In contrast, significantly different distributions were observed in the three prefectures, indicating that gene flow between prefectures might be limited by geographical isolation. DNA fingerprints resulting from hybridization with a moderately repetitive DNA sequence of the fungus revealed greater genetic variability and geographical differences in genetic population structure even within the same rDNA type.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akins R. A., Lambowitz A. M. General method for cloning Neurospora crassa nuclear genes by complementation of mutants. Mol Cell Biol. 1985 Sep;5(9):2272–2278. doi: 10.1128/mcb.5.9.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluster P. D., Marinković D., Allard R. W., Ayala F. J. Correlations between development rates, enzyme activities, ribosomal DNA spacer-length phenotypes, and adaptation in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1987 Jan;84(2):610–614. doi: 10.1073/pnas.84.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Hamer J. E., Farrall L., Orbach M. J., Valent B., Chumley F. G. Host species-specific conservation of a family of repeated DNA sequences in the genome of a fungal plant pathogen. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9981–9985. doi: 10.1073/pnas.86.24.9981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M., Romao J., Marchetti M. A., Hamer J. E. DNA Fingerprinting with a Dispersed Repeated Sequence Resolves Pathotype Diversity in the Rice Blast Fungus. Plant Cell. 1991 Jan;3(1):95–102. doi: 10.1105/tpc.3.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milgroom M. G., Lipari S. E., Powell W. A. DNA fingerprinting and analysis of population structure in the chestnut blight fungus, Cryphonectria parasitica. Genetics. 1992 Jun;131(2):297–306. doi: 10.1093/genetics/131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Li W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T. D., Botstein D. Simple Mendelian inheritance of the reiterated ribosomal DNA of yeast. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5091–5095. doi: 10.1073/pnas.74.11.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghai-Maroof M. A., Soliman K. M., Jorgensen R. A., Allard R. W. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci U S A. 1984 Dec;81(24):8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolik-Utlaut S., Petes T. D. Recombination of plasmids into the Saccharomyces cerevisiae chromosome is reduced by small amounts of sequence heterogeneity. Mol Cell Biol. 1983 Jul;3(7):1204–1211. doi: 10.1128/mcb.3.7.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge T., Kobayashi H., Nishimura S. Organization of ribosomal RNA genes in Alternaria alternata Japanese pear pathotype, a host-selective AK-toxin-producing fungus. Curr Genet. 1989 Oct;16(4):267–272. doi: 10.1007/BF00422113. [DOI] [PubMed] [Google Scholar]
- Tsuge T., Nishimura S., Kobayashi H. Efficient integrative transformation of the phytopathogenic fungus Alternaria alternata mediated by the repetitive rDNA sequences. Gene. 1990 Jun 15;90(2):207–214. doi: 10.1016/0378-1119(90)90181-p. [DOI] [PubMed] [Google Scholar]
- Workman P. L., Niswander J. D. Population studies on southwestern Indian tribes. II. Local genetic differentiation in the Papago. Am J Hum Genet. 1970 Jan;22(1):24–49. [PMC free article] [PubMed] [Google Scholar]