Abstract
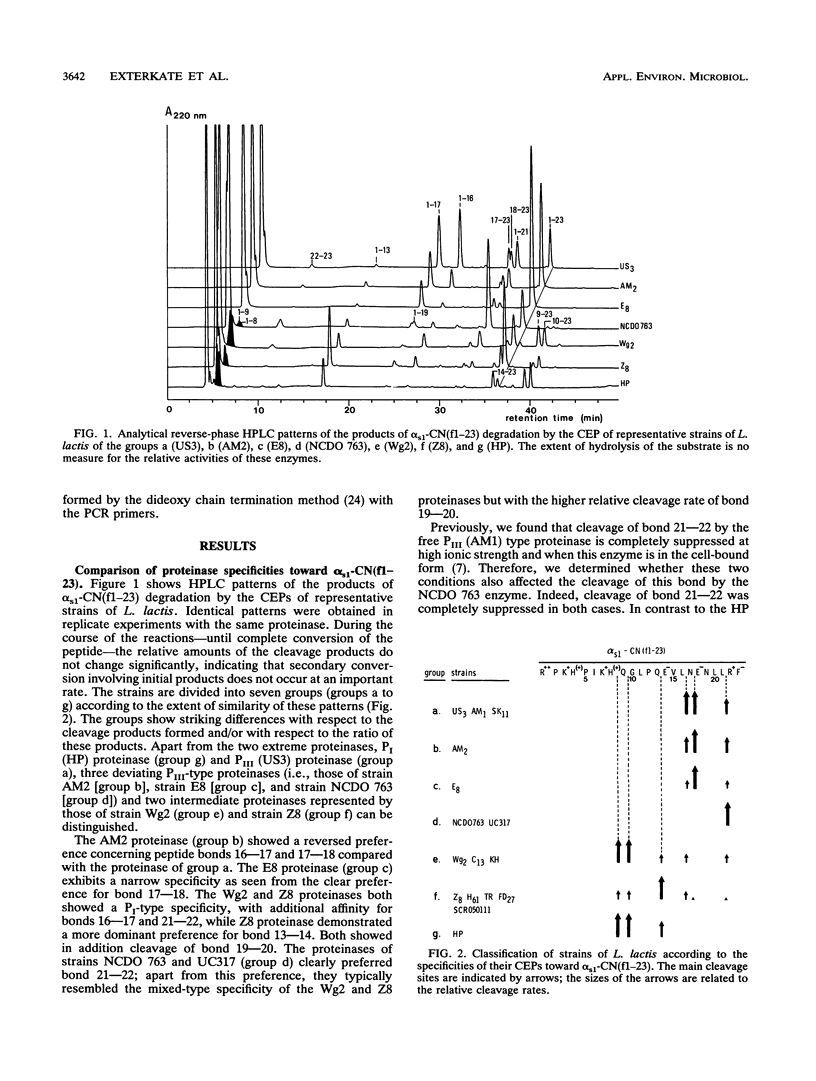
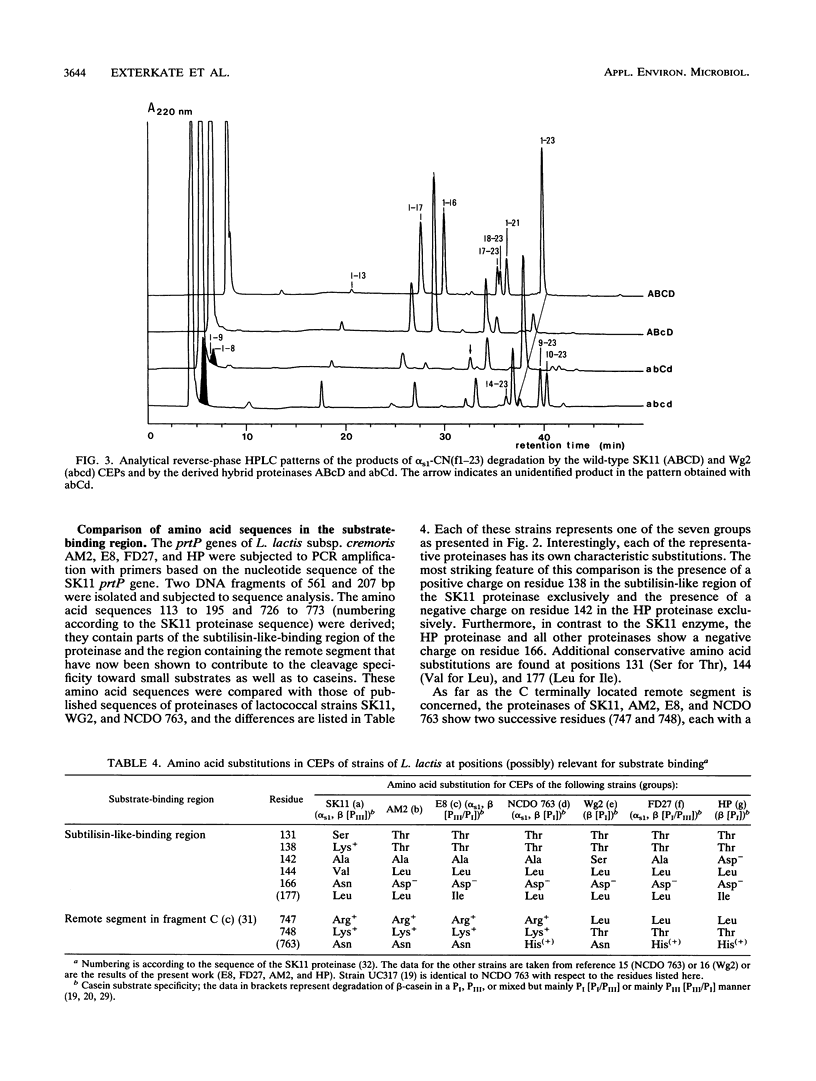
The biochemical and genetical diversity of the subtilisin-like cell envelope proteinase (CEP) among Lactococcus lactis strains was investigated. The specificities of the proteinases of 16 strains toward the important cheese peptide alpha s1-casein fragment 1 to 23 and toward two differently charged chromophoric peptides have been determined. On the basis of the results, these strains could be classified into seven groups. The contribution to the specificity of specific residues in the large C-terminal segment, which differentiates this proteinase from most other members of the subtilisin family, was established with hybrid proteinases, even in the case of the small substrates. These remote residues and the subtilisin-like substrate-binding region are therefore assumed to be spatially close to each other and together constitute most of the binding region of CEP. DNA sequence analysis of fragments of the gene (prtP) encoding segments of the proteinase which contain the relevant residues of the substrate-binding region shows that among the strains studied, this binding region is the most negatively charged in the CEP group represented by strain HP and the positively charged in the CEP group represented by strains AM1 and SK11. Consequently, these two proteinase groups show the most divergent specificities. Each of the proteinases of the other groups shows a different intermediate specificity which in part is the reflection of an intermediate charge in the binding region. However, the results suggest that amino acid residues outside the segments known to be part of the CEP-binding region also contribute to specificity.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chothia C. Principles that determine the structure of proteins. Annu Rev Biochem. 1984;53:537–572. doi: 10.1146/annurev.bi.53.070184.002541. [DOI] [PubMed] [Google Scholar]
- Coolbear T., Reid J. R., Pritchard G. G. Stability and Specificity of the Cell Wall-Associated Proteinase from Lactococcus lactis subsp. cremoris H2 Released by Treatment with Lysozyme in the Presence of Calcium Ions. Appl Environ Microbiol. 1992 Oct;58(10):3263–3270. doi: 10.1128/aem.58.10.3263-3270.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exterkate F. A., Alting A. C., Slangen C. J. Specificity of two genetically related cell-envelope proteinases of Lactococcus lactis subsp. cremoris towards alpha s1-casein-(1-23)-fragment. Biochem J. 1991 Jan 1;273(Pt 1):135–139. doi: 10.1042/bj2730135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exterkate F. A. Differences in short peptide-substrate cleavage by two cell-envelope-located serine proteinases of Lactococcus lactis subsp. cremoris are related to secondary binding specificity. Appl Microbiol Biotechnol. 1990 Jul;33(4):401–406. doi: 10.1007/BF00176654. [DOI] [PubMed] [Google Scholar]
- Exterkate F. A., de Veer G. J. Partial Isolation and Degradation of Caseins by Cell Wall Proteinase(s) of Streptococcus cremoris HP. Appl Environ Microbiol. 1985 Feb;49(2):328–332. doi: 10.1128/aem.49.2.328-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holck A., Naes H. Cloning, sequencing and expression of the gene encoding the cell-envelope-associated proteinase from Lactobacillus paracasei subsp. paracasei NCDO 151. J Gen Microbiol. 1992 Jul;138(7):1353–1364. doi: 10.1099/00221287-138-7-1353. [DOI] [PubMed] [Google Scholar]
- Hugenholtz J., Exterkate F., Konings W. N. The Proteolytic Systems of Streptococcus cremoris: an Immunological Analysis. Appl Environ Microbiol. 1984 Dec;48(6):1105–1110. doi: 10.1128/aem.48.6.1105-1110.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiwaki M., Ikemura H., Shimizu-Kadota M., Hirashima A. Molecular characterization of a cell wall-associated proteinase gene from Streptococcus lactis NCDO763. Mol Microbiol. 1989 Mar;3(3):359–369. doi: 10.1111/j.1365-2958.1989.tb00181.x. [DOI] [PubMed] [Google Scholar]
- Kok J., Leenhouts K. J., Haandrikman A. J., Ledeboer A. M., Venema G. Nucleotide sequence of the cell wall proteinase gene of Streptococcus cremoris Wg2. Appl Environ Microbiol. 1988 Jan;54(1):231–238. doi: 10.1128/aem.54.1.231-238.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers O. P., Boot H. J., de Vos W. M. Improved site-directed mutagenesis method using PCR. Nucleic Acids Res. 1991 Aug 25;19(16):4558–4558. doi: 10.1093/nar/19.16.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J., Vos P., Hayes F., Daly C., de Vos W. M., Fitzgerald G. Cloning and partial sequencing of the proteinase gene complex from Lactococcus lactis subsp. lactis UC317. J Gen Microbiol. 1992 Apr;138(4):709–718. doi: 10.1099/00221287-138-4-709. [DOI] [PubMed] [Google Scholar]
- Monnet V., Ley J. P., Gonzàlez S. Substrate specificity of the cell envelope-located proteinase of Lactococcus lactis subsp. lactis NCDO 763. Int J Biochem. 1992 May;24(5):707–718. doi: 10.1016/0020-711x(92)90004-k. [DOI] [PubMed] [Google Scholar]
- Reid J. R., Ng K. H., Moore C. H., Coolbear T., Pritchard G. G. Comparison of bovine beta-casein hydrolysis by PI and PIII-type proteinases from Lactococcus lactis subsp. cremoris [corrected]. Appl Microbiol Biotechnol. 1991 Dec;36(3):344–351. doi: 10.1007/BF00208154. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter I., Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967 Apr 20;27(2):157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Siezen R. J., de Vos W. M., Leunissen J. A., Dijkstra B. W. Homology modelling and protein engineering strategy of subtilases, the family of subtilisin-like serine proteinases. Protein Eng. 1991 Oct;4(7):719–737. doi: 10.1093/protein/4.7.719. [DOI] [PubMed] [Google Scholar]
- Smid E. J., Poolman B., Konings W. N. Casein utilization by lactococci. Appl Environ Microbiol. 1991 Sep;57(9):2447–2452. doi: 10.1128/aem.57.9.2447-2452.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser S., Exterkate F. A., Slangen C. J., de Veer G. J. Comparative Study of Action of Cell Wall Proteinases from Various Strains of Streptococcus cremoris on Bovine alpha(s1)-, beta-, and kappa-Casein. Appl Environ Microbiol. 1986 Nov;52(5):1162–1166. doi: 10.1128/aem.52.5.1162-1166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P., Boerrigter I. J., Buist G., Haandrikman A. J., Nijhuis M., de Reuver M. B., Siezen R. J., Venema G., de Vos W. M., Kok J. Engineering of the Lactococcus lactis serine proteinase by construction of hybrid enzymes. Protein Eng. 1991 Apr;4(4):479–484. doi: 10.1093/protein/4.4.479. [DOI] [PubMed] [Google Scholar]
- Vos P., Simons G., Siezen R. J., de Vos W. M. Primary structure and organization of the gene for a procaryotic, cell envelope-located serine proteinase. J Biol Chem. 1989 Aug 15;264(23):13579–13585. [PubMed] [Google Scholar]
- de Vos W. M., Vos P., de Haard H., Boerrigter I. Cloning and expression of the Lactococcus lactis subsp. cremoris SK11 gene encoding an extracellular serine proteinase. Gene. 1989 Dec 21;85(1):169–176. doi: 10.1016/0378-1119(89)90477-0. [DOI] [PubMed] [Google Scholar]


