Abstract
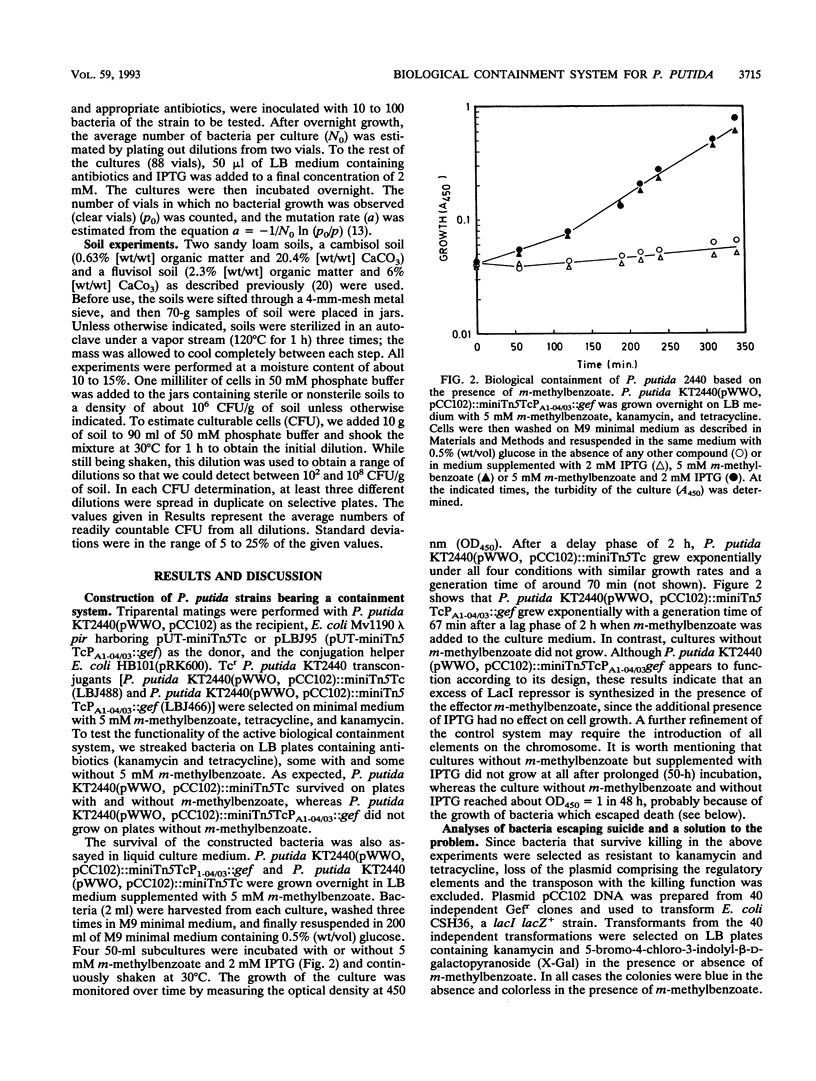
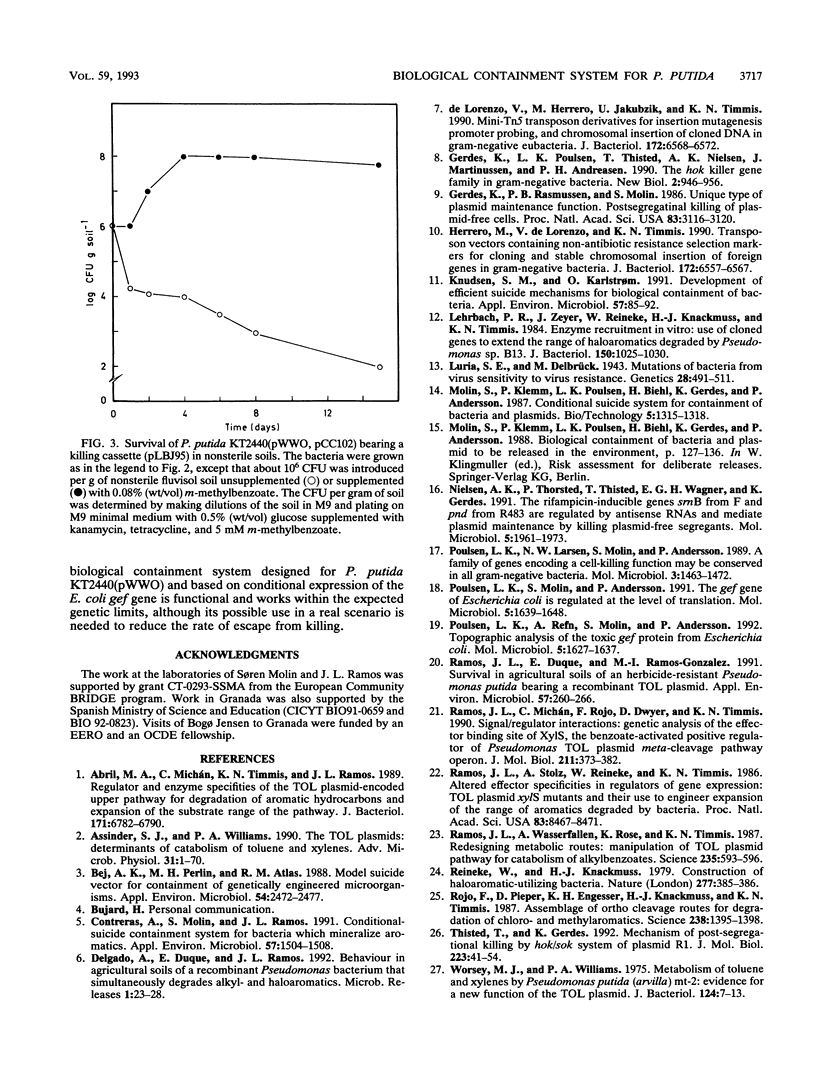
A model substrate-dependent suicide system to biologically contain Pseudomonas putida KT2440 is reported. The system consists of two elements. One element carries a fusion between a synthetic lac promoter (PA1-04/03) and the gef gene, which encodes a killing function. This element is contained within a transposaseless mini-Tn5 transposon so that it can be integrated at random locations on the Pseudomonas chromosome. The second element, harbored by plasmid pCC102, is designed to control the first and bears a fusion between the promoter of the P. putida TOL plasmid-encoded meta-cleavage pathway operon (Pm) and the lacI gene, encoding the Lac repressor, plus xylS2, coding for a positive regulator of Pm. In liquid culture under optimal growth conditions and in sterile and nonsterile soil microcosms, P. putida KT2440 (pWWO) bearing the containment system behaves as designed. In the presence of a XylS effector, such as m-methylbenzoate, the LacI protein is synthesized, preventing the expression of the killing function. In the absence of effectors, expression of the PA1-04/03::gef cassette is no longer prevented and a high rate of cell killing is observed. Fluctuation test analyses revealed that mutants resistant to cell killing arise at a frequency of around 10(-5) to 10(-6) per cell per generation. Mutations are linked to the killing element rather than to the regulatory one. In bacteria bearing two copies of the killing cassette, the rate of appearance of mutants resistant to killing decreased to as low as 10(-8) per cell per generation.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abril M. A., Michan C., Timmis K. N., Ramos J. L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol. 1989 Dec;171(12):6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assinder S. J., Williams P. A. The TOL plasmids: determinants of the catabolism of toluene and the xylenes. Adv Microb Physiol. 1990;31:1–69. doi: 10.1016/s0065-2911(08)60119-8. [DOI] [PubMed] [Google Scholar]
- Bej A. K., Perlin M. H., Atlas R. M. Model suicide vector for containment of genetically engineered microorganisms. Appl Environ Microbiol. 1988 Oct;54(10):2472–2477. doi: 10.1128/aem.54.10.2472-2477.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras A., Molin S., Ramos J. L. Conditional-suicide containment system for bacteria which mineralize aromatics. Appl Environ Microbiol. 1991 May;57(5):1504–1508. doi: 10.1128/aem.57.5.1504-1508.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado A., Duque E., Ramos J. L. Behavior in agricultural soils of a recombinant Pseudomonas bacterium that simultaneously degrades alkyl- and haloaromatics. Microb Releases. 1992 Jun;1(1):23–28. [PubMed] [Google Scholar]
- Gerdes K., Poulsen L. K., Thisted T., Nielsen A. K., Martinussen J., Andreasen P. H. The hok killer gene family in gram-negative bacteria. New Biol. 1990 Nov;2(11):946–956. [PubMed] [Google Scholar]
- Gerdes K., Rasmussen P. B., Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci U S A. 1986 May;83(10):3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero M., de Lorenzo V., Timmis K. N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990 Nov;172(11):6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen S. M., Karlström O. H. Development of efficient suicide mechanisms for biological containment of bacteria. Appl Environ Microbiol. 1991 Jan;57(1):85–92. doi: 10.1128/aem.57.1.85-92.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrbach P. R., Zeyer J., Reineke W., Knackmuss H. J., Timmis K. N. Enzyme recruitment in vitro: use of cloned genes to extend the range of haloaromatics degraded by Pseudomonas sp. strain B13. J Bacteriol. 1984 Jun;158(3):1025–1032. doi: 10.1128/jb.158.3.1025-1032.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A. K., Thorsted P., Thisted T., Wagner E. G., Gerdes K. The rifampicin-inducible genes srnB from F and pnd from R483 are regulated by antisense RNAs and mediate plasmid maintenance by killing of plasmid-free segregants. Mol Microbiol. 1991 Aug;5(8):1961–1973. doi: 10.1111/j.1365-2958.1991.tb00818.x. [DOI] [PubMed] [Google Scholar]
- Poulsen L. K., Larsen N. W., Molin S., Andersson P. A family of genes encoding a cell-killing function may be conserved in all gram-negative bacteria. Mol Microbiol. 1989 Nov;3(11):1463–1472. doi: 10.1111/j.1365-2958.1989.tb00131.x. [DOI] [PubMed] [Google Scholar]
- Poulsen L. K., Refn A., Molin S., Andersson P. The gef gene from Escherichia coli is regulated at the level of translation. Mol Microbiol. 1991 Jul;5(7):1639–1648. doi: 10.1111/j.1365-2958.1991.tb01911.x. [DOI] [PubMed] [Google Scholar]
- Poulsen L. K., Refn A., Molin S., Andersson P. Topographic analysis of the toxic Gef protein from Escherichia coli. Mol Microbiol. 1991 Jul;5(7):1627–1637. doi: 10.1111/j.1365-2958.1991.tb01910.x. [DOI] [PubMed] [Google Scholar]
- Ramos J. L., Duque E., Ramos-Gonzalez M. I. Survival in soils of an herbicide-resistant Pseudomonas putida strain bearing a recombinant TOL plasmid. Appl Environ Microbiol. 1991 Jan;57(1):260–266. doi: 10.1128/aem.57.1.260-266.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J. L., Michan C., Rojo F., Dwyer D., Timmis K. Signal-regulator interactions. Genetic analysis of the effector binding site of xylS, the benzoate-activated positive regulator of Pseudomonas TOL plasmid meta-cleavage pathway operon. J Mol Biol. 1990 Jan 20;211(2):373–382. doi: 10.1016/0022-2836(90)90358-S. [DOI] [PubMed] [Google Scholar]
- Ramos J. L., Stolz A., Reineke W., Timmis K. N. Altered effector specificities in regulators of gene expression: TOL plasmid xylS mutants and their use to engineer expansion of the range of aromatics degraded by bacteria. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8467–8471. doi: 10.1073/pnas.83.22.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J. L., Wasserfallen A., Rose K., Timmis K. N. Redesigning metabolic routes: manipulation of TOL plasmid pathway for catabolism of alkylbenzoates. Science. 1987 Jan 30;235(4788):593–596. doi: 10.1126/science.3468623. [DOI] [PubMed] [Google Scholar]
- Reineke W., Knackmuss H. J. Construction of haloaromatics utilising bacteria. Nature. 1979 Feb 1;277(5695):385–386. doi: 10.1038/277385a0. [DOI] [PubMed] [Google Scholar]
- Rojo F., Pieper D. H., Engesser K. H., Knackmuss H. J., Timmis K. N. Assemblage of ortho cleavage route for simultaneous degradation of chloro- and methylaromatics. Science. 1987 Dec 4;238(4832):1395–1398. doi: 10.1126/science.3479842. [DOI] [PubMed] [Google Scholar]
- Thisted T., Gerdes K. Mechanism of post-segregational killing by the hok/sok system of plasmid R1. Sok antisense RNA regulates hok gene expression indirectly through the overlapping mok gene. J Mol Biol. 1992 Jan 5;223(1):41–54. doi: 10.1016/0022-2836(92)90714-u. [DOI] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975 Oct;124(1):7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V., Herrero M., Jakubzik U., Timmis K. N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990 Nov;172(11):6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]


