Abstract
The deuterostome phyla include Echinodermata, Hemichordata, and Chordata. Chordata is composed of three subphyla, Vertebrata, Cephalochordata (Branchiostoma), and Urochordata (Tunicata). Careful analysis of a new 18S rDNA data set indicates that deuterostomes are composed of two major clades: chordates and echinoderms + hemichordates. This analysis strongly supports the monophyly of each of the four major deuterostome taxa: Vertebrata + Cephalochordata, Urochordata, Hemichordata, and Echinodermata. Hemichordates include two distinct classes, the enteropneust worms and the colonial pterobranchs. Most previous hypotheses of deuterostome origins have assumed that the morphology of extant colonial pterobranchs resembles the ancestral deuterostome. We present a molecular phylogenetic analysis of hemichordates that challenges this long-held view. We used 18S rRNA to infer evolutionary relationships of the hemichordate classes Pterobranchia and Enteropneusta. Our data show that pterobranchs may be derived within enteropneust worms rather than being a sister clade to the enteropneusts. The nesting of the pterobranchs within the enteropneusts dramatically alters our view of the evolution of the chordate body plan and suggests that the ancestral deuterostome more closely resembled a mobile worm-like enteropneust than a sessile colonial pterobranch.
Recent molecular phylogenies suggest that deuterostomes include only echinoderms, hemichordates, and chordates (1–4), because chaetognaths (5) and lophophorates (6–8) are likely to be protostomes. The chordates are composed of three subphyla, Vertebrata, Cephalochordata, and Urochordata. Evolutionary relationships among the deuterostomes and debates over chordate ancestry have challenged zoologists for over a hundred years (9–15). Urochordate and hemichordate evolutionary relationships are central to understanding chordate evolution, but morphological disparities among taxa and a poor fossil record have hampered research efforts. Current phylogenetic analyses show that the urochordates are monophyletic (16) and suggest that Urochordata is a separate phylum of Deuterostomia, rather than a subphylum of Chordata.
Theories of chordate origins have been expertly reviewed (9–15) and are briefly mentioned here. One of the most cited theories of chordate origins is Garstang's hypothesis that the aboral ciliated band of an auricularian-like larva could evolutionarily form a dorsal tubular nerve cord, such as that found in an ascidian tadpole larva (13). Then, chordates were thought to have evolved from an ancestral chordate tadpole larva that underwent paedomorphosis and now retains adult characteristics with the larval tail (13). Chordates have also been thought to evolve from a pterobranch-like ancestor (reviewed in refs. 9 and 10) or from calcichordates (14).
The phylum Hemichordata consists of the classes Enteropneusta (acorn worms), Pterobranchia (tube dwelling), and Planctosphaeroidea (planktonic) (17, 18). Hemichordates resemble echinoderms in nervous system anatomy (19, 20) and larval morphology (21, 22), sharing a tricoelomate body plan and a coelomic excretory hydropore (23). The enteropneusts are solitary (Fig. 1 A and B), reproduce sexually, and either have a tornaria larva or are direct developers (17, 21). The three body parts are the proboscis (protosome), collar (mesosome), and trunk (metasome) (17, 18). Enteropneust adults also exhibit chordate characteristics, including pharyngeal gill pores, a partially neurulated dorsal cord, and a stomochord that has some similarities to the chordate notochord (17, 18, 24). On the other hand, hemichordates lack a dorsal postanal tail and segmentation of the muscular and nervous systems (9, 12, 17).
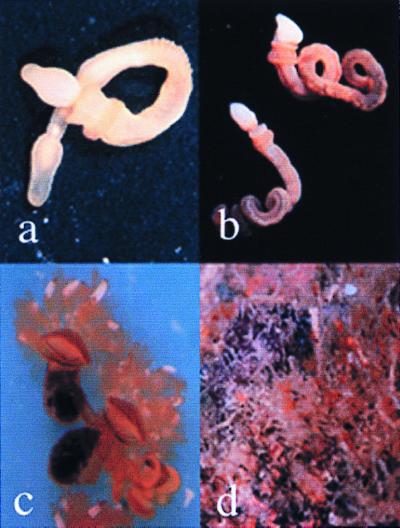
Figure 1.
Photographs of the adults of the hemichordate species represented in this study. (a) Ptychodera bahamensis; (b) Harrimania species; (c) Cephalodiscus gracilus individuals; (d) Cephalodiscus gracilus colony. Our results suggest that members of the family Ptychoderidae (a) form one clade of Enteropneusta, whereas the family Harrimanidae (b) plus Pterobranchia (c and d) form another.
Pterobranchs are colonial (Fig. 1 C and D), live in secreted tubular coenecia, and reproduce via a short-lived planula-shaped larvae or by asexual budding (17, 18). When first dredged from the deep sea, Rhabdopleura was classified as a bryozoan (in 1873; described in ref. 17). Later, Cephalodiscus was discovered, recognized as another pterobranch, and together they were considered similar to bryozoans and phoronids, with whom they shared lophophore-like ciliated feeding tentacles (8, 17). Morphological analysis of the lophophore structure (8, 9) combined with molecular data indicating that lophophorates are protostomes (6, 7) suggests that lophophore feeding structures in these disparate taxa may be due to convergence (8).
The monotypic Planctosphaeroidea (25) are transparent spherical larvae for which adults have not been described (17, 21). Planctosphaera pelagica is a large gelatinous larva with an extensive array of ciliated bands and may represent a tornaria distorted by hypertrophy because of a long planktonic life (25). P. pelagica is not represented in this study, because attempts to amplify by PCR the 18S rRNA genes failed (M. Hart, personal communication).
Hemichordates have been considered a sister group to echinoderms (14, 17, 26) and to chordates (15). Recent cladistic analysis of morphological data sets has suggested that the pterobranchs are basal deuterostomes, whereas enteropneusts are an early offshoot of the chordate lineage, suggesting that the phylum Hemichordata is polyphyletic (15, 27). Molecular and morphological analyses of axis specification suggest that the enteropneusts may be inverted dorsoventrally with respect to protostomes (28–31). We embarked on a project to study the phylogenetic relationships among hemichordates, especially to examine the relationship of the pterobranchs to the enteropneusts. The data from this hemichordate analysis, combined with urochordate sequences published elsewhere (16), allow us to analyze further the evolutionary relationships among the deuterostomes.
Our results suggest that deuterostomes are comprised of two distinct clades: the chordates and the hemichordates + echinoderms. Each of the four deuterostome taxa are clearly monophyletic. However, examination of life history traits reveals that both urochordates (16, 32) and hemichordates (17, 18) can exhibit either a solitary sexual mode or a colonial life history that allows both asexual budding and sexual reproduction. These changes in life history have had profound effects on body plans and may have confounded previous morphological analyses. We discuss recent molecular and developmental comparisons of hemichordate and urochordate embryos and suggest further experiments that are likely to be informative about the evolution of the chordate body plan.
Materials and Methods
Animals.
Cephalodiscus gracilis and Ptychodera bahamensis were identified and collected from Bermuda by C.B.C. and stored in 70% ethanol. Saccoglossus species (C.B.C.) and Harrimania species (C.B.C.) were collected subtidally from Barkley Sound, British Columbia and have not yet been described (33). B.J.S.'s lab extracted DNA and entirely sequenced complete 18S rDNA from the hemichordates and the ascidians Ascidia ceratodes, Herdmania curvata, Phallusia mammillata, and Molgula oculata (ref. 16; Table 2).
Table 2.
Terminal branch lengths with four-cluster analysis
| Taxon | GenBank accession no. | Branch length |
|---|---|---|
| Cephalodiscus gracilus | AF236798 | 0.2063 |
| Oikopleura species 1 | D14360 | 0.1947 |
| Oikopleura species 2 | AB013015 | 0.1895 |
| Oikopleura dioica | AB013014 | 0.1881 |
| Stichopus japonicus | D14364 | 0.1735 |
| Herdmania curvata | AF165827 | 0.1555 |
| Molgula oculata | L12432 | 0.1449 |
| Saccoglossus species | AF236800 | 0.1442 |
| Doliolum nationalis | AB013012 | 0.1431 |
| ≤0.1400 | ||
| Saccoglossus kowalevskii | L28054 | 0.1314 |
| Harrimania species | AF236799 | 0.1313 |
| Thalia democratica | D14366 | 0.1267 |
| ≤0.1200 | ||
| Styela plicata | M97577 | 0.1200 |
| Halocynthia roretzi | AB013016 | 0.1199 |
| Homo sapiens | M10098 | 0.1176 |
| Rattus norvegicus | K01593 | 0.1119 |
| Petromyzon marinus | M97575 | 0.1052 |
| ≤0.1000 | ||
| Phallusia mammillata | AF236803 | 0.0964 |
| Branchiostoma floridae | M97571 | 0.0959 |
| Strongylocentrotus purpuratus | L28056 | 0.0934 |
| Ophioplocus japonicus | D14361 | 0.0887 |
| Asterias amurensis | D14358 | 0.0887 |
| Ciona intestinalis | AB013017 | 0.0874 |
| Ascidia ceratodes | L12378 | 0.0862 |
| Pyrosoma atlanticum | AB013011 | 0.0847 |
| Balanoglossus carnosus | D14359 | 0.0684 |
| Antedon serrata | D14357 | 0.0646 |
| Ptychodera bahamensis | AF236802 | 0.0518 |
The length of the terminal branch leading to each deuterostome sequence was calculated in turn with four-cluster analysis (phyltest 2.0; 40) by using Anemonia sulcata, Tenebrio molitor, and Nephtys hombergii as reference taxa.
Sequences.
The 18S ribosomal gene was chosen because of the large number of sequences available for representative deuterostomes (Table 2) and because it has been useful for resolving urochordate phylogenetic relationships (16, 32, 34). New sequences were obtained by PCR amplification and subsequent DNA sequencing with 18S BS 5′-CTGGTTGATCCTGCCAG-3′ and 18S PH 5′-TAATGATCCATCTGCAGGTTCACCT-3′ and other internal primers as previously described (16).
Alignments and Analysis.
Sequences were aligned according to a secondary structure model of the eukaryotic small ribosomal subunit (35) and are available at http://chuma.cas.usf.edu/∼garey/alignments/alignment.html. Maximum parsimony (MP), minimum evolution (ME), and neighbor joining analyses (NJ) were carried out by using paup (36) and mega (37) with 100 bootstrap replicates. Sites with gaps were excluded from the analyses. The α parameter for evolutionary distances using a γ distribution was calculated by using maximum likelihood (ML) analysis in paup. ML analyses were carried out by using a model of substitution that incorporated four rate categories by using the dnaml program in phylip (38) as previously described (16). NJ and ME analyses were carried out by using Jukes and Cantor, γ-corrected and uncorrected Kimura two-parameter distances, and Paralinear distances. Alternate topologies were tested with a combination of macclade (39) and paup (36). The length of the terminal branch leading to each deuterostome sequence was calculated in turn with four-cluster analysis (phyltest 2.0; 40) by using sequences from Anemonia sulcata, Tenebrio molitor, and Nephtys hombergii as reference taxa (Kimura two-parameter model with γ shape parameter = 0.26). Additional distance and MP analyses were carried out by using gambit (http://www.lifesci.ucla.edu/mcdbio/Faculty/Lake/Research/Programs/) (41) with a paralinear distance model of nucleotide substitution that also corrects for site-to-site variation in evolutionary rate (42). In the gambit analyses, 1,000 bootstrap replicates of cells 0–12 were used with the probability of obtaining the best tree set to 99.9%. Sites with gaps at more than 50% of the taxa in the alignment were omitted from the analyses.
Results and Discussion
Relationships Within Deuterostomia.
The complete alignment of 28 deuterostome species and 5 outgroup taxa contained 2,161 sites including those with gaps or 1,472 sites after all sites with a gap at any taxon were deleted. Initial results with the entire data set of 28 deuterostome taxa by using MP, ME, and NJ analyses were mixed (Table 1). In nearly all analyses, each of the four deuterostome taxa (echinoderms, hemichordates, urochordates, vertebrates + cephalochordates) were well-supported monophyletic groups (Table 1). In some cases, hemichordates and echinoderms grouped together, but chordates and urochordates rarely grouped together (Table 1). Testing of alternate MP and ME topologies by using macclade revealed that there was virtually no difference in the length of trees with different topological arrangements of the four main taxa (data not shown).
Table 1.
Bootstrap support for deuterostome phylogenetic trees
| Tree algorithm | Uro | Chor | Hemi | Echin | Hemi + Echin | Chor + Uro | Basal Deut? |
|---|---|---|---|---|---|---|---|
| Full dataset (29 deuterostomes) | |||||||
| NJ, Kimura | 100 | 60 | 94 | 100 | 67 | — | Chor |
| NJ, Kimura with γ | 100 | 65 | 89 | 99 | — | — | Echin |
| MP | 96 | 83 | 93 | 89 | 80 | 59 | 2 + 2 |
| ME, Kimura | 100 | 68 | 92 | 99 | 56 | — | Chor |
| ME, Kimura with γ | 99 | 66 | 93 | 97 | — | — | Chor |
| ME, Paralinear | 99 | 56 | 95 | 97 | — | — | Chor |
| ≤0.1400 subs/site (19 deuterostomes) | |||||||
| NJ, Kimura | 100 | 70 | 100 | 100 | — | — | Chor |
| NJ, Kimura with γ | 100 | 72 | 100 | 100 | — | — | Echin |
| MP | 100 | 79 | 93 | 88 | 66 | 54 | 2 + 2 |
| ME, Kimura | 100 | 71 | 99 | 98 | — | — | poly |
| ME, Kimura with γ | 100 | 68 | 100 | 98 | — | — | poly |
| ME, Paralinear | 100 | 58 | 99 | 100 | — | — | poly |
| ≤0.1200 subs/site (16 deuterostomes) | |||||||
| NJ, Kimura | 100 | 71 | 100 | 100 | 80 | — | Chor |
| NJ, Kimura with γ | 100 | 67 | 100 | 99 | 64 | — | Chor |
| MP | 100 | 67 | 100 | 96 | 86 | 65 | 2 + 2 |
| ME, Kimura | 100 | 63 | 100 | 100 | 83 | — | Chor |
| ME, Kimura with γ | 100 | 74 | 100 | 100 | 70 | — | poly |
| ME, Paralinear | 100 | 63 | 100 | 100 | 89 | — | Chor |
| ML | 100 | 53 | 100 | 100 | 100 | — | Chor |
| gambit and paralinear outgroup | |||||||
| Nephtys (0.1518) | 100 | 71 | 98 | 90 | 86 | 83 | 2 + 2 |
| Glycera (0.1432) | 100 | 74 | 98 | 94 | 78 | 85 | 2 + 2 |
| Artemia (0.2478) | 100 | 68 | 86 | 99 | 86 | — | poly |
| Tenebrio (0.2327) | 100 | — | 100 | 87 | 89 | — | poly |
| Anemonia (0.3373) | 99 | 59 | 93 | 98 | 73 | — | Chor |
Summary of analyses of the full dataset (28 deuterostomes), a subset including those taxa with a branch length ≤0.1400 substitution per site, and a deuterostome subset with branch lengths of ≤0.1200 substitution per site. Bootstrap values are shown for the monophyly of urochordates (Uro), chordates (including cephalochordates, Chor), hemichordates (Hemi), and echinoderms (Echin). Bootstrap values for a monophyletic hemichordates + echinoderms and a monophyletic chordates + urochordates are shown when greater than 50%. The basal deuterostome in each analysis is shown unless there was a polytomy (poly). Instances where hemichordates + echinoderms was a sister group to chordates + urochordates are shown labeled “2 + 2.” Methods include NJ and ME trees with Kimura two-parameter, Kimura two-parameter with γ correction, or paralinear distances, and MP. ML and gambit analysis were carried out only for the smallest dataset (≤0.1200 subs per site). gambit was used with paralinear distances and a correction for site-to-site variation by using a variety of outgroups. The Kimura two-parameter γ corrected distances between each outgroup and Antedon are shown in parentheses. The most reliable analyses are shown in bold.
Four-cluster analysis revealed the branch length from an internal node to each deuterostome taxon (Table 2). This shows that the number of substitutions per site varies considerably among the different deuterostome taxa used in this study. Results reported in Table 2 were used to produce two subsets of the alignment including only those sequences with branches ≤0.1400 substitution per site (19 taxa), or ≤0.1200 substitution per site (16 taxa). This approach of excluding taxa with the most rapidly evolving 18S rDNA sequences has been used previously to minimize unequal rate effects (43, 44). The bootstrap support for key nodes of phylogenetic trees generated with the different deuterostome subsets is shown in Table 1. As in the complete alignment, each of the four main deuterostome taxa (echinoderms, hemichordates, urochordates, vertebrates + cephalochordates) were generally well supported monophyletic groups, although the topology among the four groups varied (Table 1). With the subset of taxa with ≤0.12 substitution per site, there is moderate to strong support for a sister-group relationship between hemichordates and echinoderms (Table 1) with all methods (NJ, MP, ME, and ML). There was very little support for a sister-group relationship between chordates and urochordates.
The increasing support for a hemichordate + echinoderm clade from the alignment subset with the slowest evolving taxa (≤0.1200 substitution per site) suggested that unequal evolutionary rate was a significant source of error in the analyses. Further reduction of the data set (e.g., ≤0.1000 substitution per site) was not useful because it completely eliminated all of the vertebrate sequences from analyses. Therefore, bootstrapper's gambit (42), a method known to be insensitive to unequal evolutionary rates and site-to-site variation in evolutionary rate, was used for the final analysis on the ≤0.1200 substitution per site subset (16 deuterostome taxa). gambit revealed that hemichordates + echinoderms formed a sister group to chordates + urochordates with high bootstrap support (Table 1 and Fig. 2). This analysis may be sensitive to the length of the branch leading to the nondeuterostome outgroup, therefore, the branch length from the slow evolving deuterostome Antedon serrata to each outgroup was calculated and is shown at the bottom of Table 1. The two annelid sequences (Nephtys hombergii and Glycera americana) were the slowest evolving outgroups and, as expected, yielded the most highly supported topology. When longer-branched outgroups such as the arthropods Artemia salina and Tenebrio molitor or the diploblast Anemonia sulcata were used, the resolution of the tree degraded, and support for chordates + urochordates was lost (Table 1).
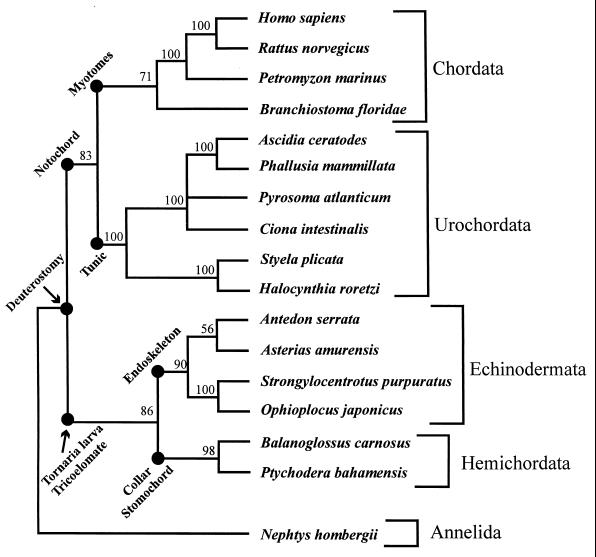
Figure 2.
Phylogenetic tree of the deuterostomes when sequences with similar evolutionary rates (16 taxa ≤0.12 substitutions per site) were analyzed with gambit. Key characters are mapped to the deeper nodes. The deuterostomes form two great clades, one containing the hemichordates and echinoderms and the other made up of urochordates and chordates (cephalochordates and vertebrates). Major differences in adult body plan between Cephalochordata + Vertebrata (myotomes) and Urochordata (tunic) are marked. These results, combined with morphological data, suggest that Chordata should be restricted to Cephalochordata + Vertebrata and that Urochordata is an independent phylum and the sister group to Chordata. Note that the tripartate coelom of hemichordates is considered homologous to the three pairs of echinoderm coeloms.
Our analysis of the slow evolving rDNA sequences of the 16 deuterostome taxa by using gambit as shown in Fig. 2 is the first molecular study to define clearly the evolutionary relationships among all four major groups of deuterostomes. We found Vertebrata to be monophyletic and a sister clade to Cephalochordata, as suggested by previous studies (45). Urochordata is monophyletic as shown previously (16) and forms a sister group to Vertebrata + Cephalochordata. The sister relationship between urochordates and vertebrates + cephalochordates is supported by morphological evidence. The presence of a notochord unites urochordates, cephalochordates, and vertebrates (9–13, 26, 45, 46), although some authors differentiate the notochord of vertebrates + cephalochordates from the urochord of urochordates (15). The characters that set vertebrates + cephalochordates apart from urochordates are the presence of myotomes in vertebrates + cephalochordates and the presence of the tunic in the urochordates (Fig. 2; 9–12, 26, 45–48). Urochordates are considered members of Chordata because the tadpole larva exhibits the chordate body plan. However, differences in adult body plan and life histories of urochordates compared with vertebrates + cephalochordates justify a reappraisal of the inclusion of urochordates within the phylum Chordata despite the presence of a notochord.
The chordate features of ascidian tadpole larvae develop during embryogenesis from morphological and genetic pathways similar to chordates (49, 50). Recent phylogenetic evidence from 18S rDNA data shows that the appendicularians, or larvaceans, are likely to be a sister group to the ascidians and thaliacians (16, 51). In light of these results, the ancestral urochordate may have been either pelagic or sessile. Urochordate adults are morphologically distinct from vertebrates and cephalochordates (46–48). Both sessile ascidians (46, 47) and pelagic tunicates (48) contain a characteristic extracellular coat, or tunic, that protects the adult. There is also a fundamental difference in life history traits between urochordates and other chordates. Both vertebrates and cephalochordates are solitary and sexual organisms, whereas urochordates have evolved a colonial lifestyle several times independently (16, 32, 52). All colonial urochordates can also reproduce asexually by budding or may reproduce sexually (46, 47). Although this is common in urochordates, it is virtually absent from other chordates. This morphological and ecological evidence, coupled with our molecular results (16), suggests that Urochordata should be considered an independent phylum that is the sister group to Vertebrata + Cephalochordata. The phylum name Chordata should be restricted to Cephalochordata + Vertebrata, and Urochordata should be raised to phylum status.
This study found strong support for hemichordate monophyly and for an echinoderm + hemichordate clade. The echinoderm + hemichordate clade, obtained by molecular data here and elsewhere (2–4, 53), is also strongly supported by larval morphological evidence. For many years, the enteropneust tornaria was considered the larva of an echinoderm, in particular an auricularia of a holothoroid or bipinnaria of an asteroid (11, 17). These large gelatinous larvae share a preoral feeding band that creates an upstream feeding current by using monociliated cells (21, 54) and a perioral ciliated band that manipulates food into the esophagus. The three coelomic sacs in hemichordates and echinoderms are both organized anterior to posterior as protocoel (echinoderm axocoel), mesocoels (echinoderm hydrocoels), and metacoels (echinoderm somatocoels) (9, 21). For an extensive comparison of an echinoderm auricularia to hemichordate tornaria see (9, 21, 22).
Relationships Within Hemichordata.
All of the phylogenetic trees showed strong support for the monophyly of the hemichordates within the deuterostomes (Fig. 2; Table 1). The pterobranch Rhabdopleura normani 18S rDNA sequence is shorter (572 bp; ref. 4) than the near complete sequences used above (Table 1). Therefore, we used a truncated alignment of all of the hemichordate sequences, including Rhabdopleura normani (645 sites, with gaps). The short-branched echinoderm Antedon serrata was used as an outgroup. Trees generated with this data set (Fig. 3) were consistent with the hemichordate topology resulting from the analysis of full-length sequences without Rhabdopleura normani (not shown). Relative branch lengths of the hemichordates varied from 0.2063 to 0.0518 substitutions per site (Table 2). Excluding taxa with long branches was impractical for these analyses because of the limited number of taxa sampled, and the tree shown in Fig. 3 shows the branches drawn to scale to emphasize the variation in evolutionary rate between taxa.
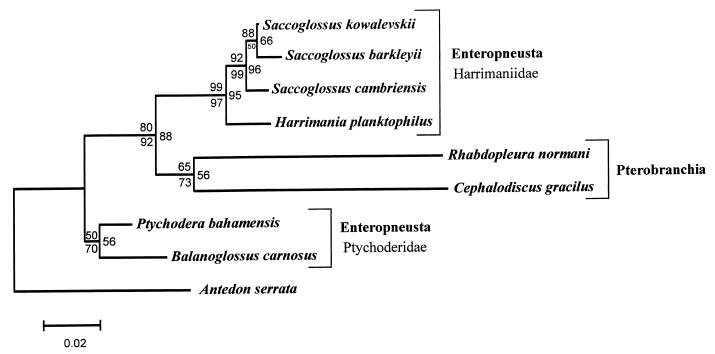
Figure 3.
Analyses of hemichordate phylogeny. Branches are drawn to scale (Kimura two-parameter distances) to emphasize the potential for artifacts because of unequal rate effects. The same topology was obtained from NJ with Kimura two-parameter distances (bootstrap values above each branch), gambit paralinear distances with correction for site-to-site variation (bootstrap values below each branch), and gambit MP (bootstrap values to the right of each branch). See text for details. Hemichordate classes (bold) and families are indicated (Right). S. barkleyii and H. planktophilus are undescribed species (33).
The enteropneust worms consistently formed two clades, also with high bootstrap support (Fig. 3). These two clades correspond to two hemichordate families, the large and complex worms in the Ptychoderidae and the relatively small and simple Harrimaniidae. We obtained several different taxa from each family in an effort to improve the phylogenetic signal. Surprisingly, the colonial class Pterobranchia was a sister group to the harrimaniid worms (Fig. 3).
Colonial Pterobranchs May Have Evolved from an Enteropneust-Like Ancestor.
The most startling finding in this study was inclusion of the hemichordate class Pterobranchia within the class Enteropneusta (Fig. 3). Prevailing hypotheses suggested that pterobranchs are either basal deuterostomes (9, 10, 15, 27) or are plesiomorphic hemichordates (17, 18). This 18S rDNA study suggests that pterobranchs may be derived from within the enteropneust clade. This is the only phylogenetic tree topology of echinoderm, pterobranchs, enteropneusts, and chordates that has NOT been suggested by other authors but is well supported by bootstrap analyses. Because of the long branches leading to the pterobranchs (Fig. 3), we cannot rule out the possibility that the position of the pterobranchs within the enteropneusts is an artifact of unequal evolutionary rates. However, if long branch attraction were occurring, one would expect the long branches leading to the pterobranchs to have been attracted to the relatively long branch leading to the outgroup rather than the shorter branches among the enteropneusts. If this topology is correct, the pterobranchs may have evolved from an enteropneust-like ancestor. This solitary to colonial switch in lifestyle, also seen in urochordates (16, 32, 52), involves a dramatic decrease in body size, the interaction of individuals, and the ability to reproduce asexually as well as sexually (17, 18). The enteropneust to pterobranch evolutionary transition may become apparent with examination of the range of morphologies within the enteropneusts.
Ptychoderidae (class Enteropneusta) is monophyletic and forms a sister group to the family Harrimaniidae + class Pterobranchia in our analyses (Fig. 3). Ptychoderids have paired dorsolateral ridges of the anterior trunk that house the gonads, hepatic sacs and well developed gill slit skeletal bars with synapticles, or supporting cross bars, which run horizontal between primary bars and secondary bars. Ptychoderids are large worms, up to several feet long, and typically develop via a tornaria larva (21, 22).
In contrast, family Harrimaniidae contains small worms, usually less than 6 inches long, which do not possess the morphological complexities of Ptychoderids and have no hepatic sacs and no genital ridges or synapticles (17, 33). Within harrimaniid worms, comparison of different genera shows a general evolutionary trend in reduction of body size and other similarities to the pterobranchs. For instance, harrimaniid juveniles have a postanal ventral tail, and in pterobranchs this tail makes up the stalk of the colonial individuals (17, 55). Furthermore, pterobranchs are filter feeders (17) and Harrimania species can also feed by filtering seawater (33). Both pterobranchs and Stereobalanus (Harrimaniidae) have two rather than one protoceol duct and pore (17, 33, 55). A reduction in the gill slits is also obvious, as Stereobalanus and Cephalodiscus have two slits, whereas Rhabdopleura has none (17, 33). Harrimaniids and pterobranchs also show reduction and disappearance of atria and coelomic diverticula and reduction in the number and size of gonads (17, 33, 56). In each case, the pterobranchs show the most extreme reduction in size and complexity. For example, Cephalodiscus and Rhabdopleura brood two and one embryos, respectively (17). These results suggest that comparative studies between harrimaniid genera (Saccoglossus, Harrimania, Stereobalanus, Protoglossus) may allow further insight into the simplification of the enteropneust body plan that may have accompanied the evolution of the pterobranchs.
Ascidian Tadpole Larvae Develop like Chordate Embryos.
It has long been recognized that ascidian tadpole larvae develop in a similar manner to chordate embryos (49, 50). The notochord is a mesodermal tissue that forms by the process of convergence and extension. Furthermore, the presumptive ascidian notochord cells develop and differentiate from a gene cascade that is initiated in all chordate embryos by the t-box transcription factor, brachyury T (57). This gene is known to be important in notochord development in both vertebrates and cephalochordates (58).
The notochord was a key tissue in the evolution of the chordates because it serves as a structural tissue in the tadpole larva and also signals to the ectoderm overlying it to develop into the dorsal neural tube. The gene cascades that are initiated during neural development in all chordate embryos have been remarkably conserved (29, 31). The ability to clone homologous genes and examine their embryonic expression in a temporal and spatial manner in different phyla has led to clues about the gene cascades that may be coopted several times during development. The chordate molecular markers for notochord tissue, neural tissue, and pharyngeal slits allow examination of hemichordate embryos for the expression of these genes during development.
Do Enteropneusts Share Developmental and Genetic Pathways with Chordates?
Adult enteropneusts exhibit some chordate characteristics, namely pharyngeal gill pores, a stomochord (17, 24), and an endostyle-like structure in the pharynx (30). In contrast, echinoderms do not contain these structures. Therefore, it is parsimonious to consider an enteropneust-like ancestor as the prototype from which Echinodermata (along one lineage) and Chordata (along another) evolved. Pax1- and Pax9-related genes in urochordates (Ciona and Halocynthia) and an enteropneust (Ptychodera flava) are expressed in the pharyngeal epithelium of developing gill pores in both phyla (59), suggesting that these structures may be homologous.
However, the expression of brachyury T in enteropneust larvae turned out to be a more complicated story. Although brachyury T is expressed exclusively in the notochord lineage in ascidian embryos (57), when examined in starfish (60) and enteropneust (61) larvae, expression was seen in the coelomic pouches and posterior gut. In sea urchins, there is an even more derived expression pattern (62) in secondary mesenchyme. These results do not rule out the possibility that the enteropneust stomochord is homologous to the urochordate notochord, because the downstream genes may be activated by a different transcription factor than brachyury T in the hemichordate larvae. This suggests that further studies of embryonic development in hemichordates are necessary to distinguish homologous and nonhomologous structures in larvae and adults.
Hemichordates lack a dorsal postanal tail and segmentation of the major functional systems, such as the muscular and nervous systems, characteristic of chordates. A detailed analysis of the development of the stomochord, dorsal hollow nerve, and pharynx of enteropneust worms may allow insight into the evolutionary origin of these structures. If chordate-like features of enteropneusts come from similar developmental and/or molecular pathways, then similar structures are the result of common origins rather than convergence. Functional experiments will then be necessary to prove that hemichordate developmental genes were coopted for different structures in adult echinoderms compared with chordates. Our phylogenetic results show that pterobranch hemichordates may have been derived within enteropneusts, suggesting that enteropneusts are basal hemichordates. Further developmental studies will be important in revealing how the evolutionarily successful chordate body plan may have evolved from a worm-like deuterostome ancestor.
Acknowledgments
C.B.C. thanks his Ph.D. advisor, A. R. Palmer, and E. E. Ruppert for encouragement and advice. J.R.G. thanks Ken Hayes for help with the initial alignment. We thank Jim Lake for helpful discussions on the analyses. Anujit Bhai, Karen Burke da Silva, Jenn Huber, and Jonathan York all contributed to this project in the laboratory of B.J.S., generating the full-length urochordate and hemichordate 18S rRNA sequences reported here. Chris Winchell at Washington State University completed the Cephalodiscus gracilis sequence from DNA isolated in the Swalla lab. We thank the Molecular Evolution Seminar Group at Vanderbilt and the Institute of Molecular Evolutionary Genetics at Pennsylvania State University for their advice and discussions at various stages of this study and the National Science Foundation, National Science and Engineering Research Council of Canada, and the American Museum of Natural History Lerner Gray Fund for financial support.
Abbreviations
- MP
maximum parsimony
- ME
minimum evolution
- NJ
neighbor joining
- ML
maximum likelihood
Footnotes
References
- 1.Holland P W H, Hacker A M, Williams N A. Philos Trans R Soc London B. 1991;332:185–189. doi: 10.1098/rstb.1991.0048. [DOI] [PubMed] [Google Scholar]
- 2.Turbeville J M, Schulz J R, Raff R A. Mol Biol Evol. 1994;11:648–655. doi: 10.1093/oxfordjournals.molbev.a040143. [DOI] [PubMed] [Google Scholar]
- 3.Wada H, Satoh N. Proc Natl Acad Sci USA. 1994;91:1801–1804. doi: 10.1073/pnas.91.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halanych K. Mol Phylogenet Evol. 1995;4:72–76. doi: 10.1006/mpev.1995.1007. [DOI] [PubMed] [Google Scholar]
- 5.Halanych K. Syst Biol. 1996;45:223–246. [Google Scholar]
- 6.Halanych K M, Bacheller J D, Aguinaldo A M, Liva S M, Hillis D M, Lake J A. Science. 1995;267:1641–1643. doi: 10.1126/science.7886451. [DOI] [PubMed] [Google Scholar]
- 7.Mackey L Y, Winnepenninckx B, Backeljau I, De Wachter R, Emschermann P, Garey J R. J Mol Evol. 1996;42:552–559. doi: 10.1007/BF02352285. [DOI] [PubMed] [Google Scholar]
- 8.Halanych K. Biol Bull. 1996;190:1–5. doi: 10.2307/1542669. [DOI] [PubMed] [Google Scholar]
- 9.Willmer P. Invertebrate Relationships: Patterns in Animal Evolution. New York: Cambridge Univ. Press; 1990. [Google Scholar]
- 10.Gee H. Before the Backbone. Views on the Origin of the Vertebrates. London: Chapman & Hall; 1996. [Google Scholar]
- 11.Raff R A. The Shape of Life: Genes, Development, and the Evolution of Animal Form. Chicago: Univ. of Chicago Press; 1996. [Google Scholar]
- 12.Gerhart J, Kirschner M. Cells, Embryos and Evolution. Malden, MA: Blackwell Science; 1997. [Google Scholar]
- 13.Garstang W. Q J Micr Sci. 1928;72:51–187. [Google Scholar]
- 14.Jeffries R P S. The Ancestry of Vertebrates. London: British Museum of Natural History; 1986. [Google Scholar]
- 15.Nielsen C. Biol J Linn Soc. 1996;57:385–410. [Google Scholar]
- 16.Swalla B J, Cameron C B, Corley L S, Garey J R. Syst Biol. 2000;49:122–134. doi: 10.1080/10635150050207384. [DOI] [PubMed] [Google Scholar]
- 17.Hyman L H. The Invertebrates: Smaller Coelomate Groups. New York: McGraw–Hill; 1959. pp. 72–207. [Google Scholar]
- 18.Barrington E J W. The Biology of Hemichordata and Protochordata. London: Oliver & Boyd; 1965. [Google Scholar]
- 19.Bullock T H, Horridge G A. Structure and Function in the Nervous Systems of Invertebrates. San Francisco: Freeman; 1965. [Google Scholar]
- 20.Cameron C B, Mackie G O. Canada J Zool. 1996;74:15–19. [Google Scholar]
- 21.Hadfield M G. Reproduction of Marine Invertebrates. New York: Academic; 1975. [Google Scholar]
- 22.Peterson K J, Cameron R A, Davidson E H. Dev Biol. 2000;219:1–17. doi: 10.1006/dbio.1999.9475. [DOI] [PubMed] [Google Scholar]
- 23.Dautov S Sh, Nezlin L P. Biol Bull. 1992;183:463–475. doi: 10.2307/1542023. [DOI] [PubMed] [Google Scholar]
- 24.Balser E J, Ruppert E E. Acta Zoologica (Stockholm) 1990;71:235–249. [Google Scholar]
- 25.Hart M W, Miller R L, Madin L P. Mar Biol (Berlin) 1994;120:521–533. [Google Scholar]
- 26.Berrill N J. The Origin of Vertebrates. London: Oxford Univ. Press; 1955. [Google Scholar]
- 27.Eernisse D J, Albert J S, Anderson F E. Syst Biol. 1992;41(3):305–330. [Google Scholar]
- 28.Bergstrom J. Paleontol Res. 1997;1:1–14. [Google Scholar]
- 29.Nübler-Jung K, Arendt D. Curr Biol. 1996;6(4):352–353. doi: 10.1016/s0960-9822(02)00491-8. [DOI] [PubMed] [Google Scholar]
- 30.Ruppert E E, Cameron C B, Frick J E. Invert Biol. 1999;118:202–212. [Google Scholar]
- 31.Gerhart J. Proc Natl Acad Sci USA. 2000;97:4445–4448. doi: 10.1073/pnas.97.9.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wada H, Makabe K W, Nakauchi M, Satoh N. Biol Bull. 1992;183:448–455. doi: 10.2307/1542021. [DOI] [PubMed] [Google Scholar]
- 33.Cameron C B. Ph.D. thesis. Edmonton, Alberta, Canada: University of Alberta; 2000. [Google Scholar]
- 34.Hadfield K A, Swalla B J, Jeffery W R. J Mol Evol. 1995;40:413–427. doi: 10.1007/BF00164028. [DOI] [PubMed] [Google Scholar]
- 35.De Rijk P, De Wachter R. Comput Appl Biosci. 1993;9:735–740. doi: 10.1093/bioinformatics/9.6.735. [DOI] [PubMed] [Google Scholar]
- 36.Swofford D L. paup. Phylogenetic Analyses Using Parsimony (and Other Methods), Ver. 4. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 37.Kumar S, Tamara K, Nei M. Comput Appl Biosci. 1994;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- 38.Felsenstein J. phylip: Phylogeny Inference Package, Ver. 3.5. Seattle: Univ. of Washington; 1993. [Google Scholar]
- 39.Maddison W P, Maddison D R. macclade, Analysis of Phylogeny and Character Evolution, Ver. 3. Sunderland, MA: Sinauer; 1992. [Google Scholar]
- 40.Rzhetsky A, Kumar S, Nei M. Mol Biol Evol. 1995;12:163–167. doi: 10.1093/oxfordjournals.molbev.a040185. [DOI] [PubMed] [Google Scholar]
- 41.Lake J A. Proc Natl Acad Sci USA. 1995;92:9662–9666. doi: 10.1073/pnas.92.21.9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lake J A. Mol Biol Evol. 1998;15:1224–1231. doi: 10.1093/oxfordjournals.molbev.a026030. [DOI] [PubMed] [Google Scholar]
- 43.Aguinaldo A M A, Turbeville J M, Linford L S, Rivera M C, Garey J R, Raff R A, Lake J A. Nature (London) 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- 44.Blaxter M L, DeLey P, Garey J R, Liu L X, Scheldeman P, Vierstraete A, Vanfleteren J R, Mackey L Y, Dorris M, Frisse L M, et al. Nature (London) 1998;392:71–74. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- 45.Holland P W H. Israel J Zool. 1996;42:S247–S272. [Google Scholar]
- 46.Berrill N J. The Tunicata. London: Ray Society; 1950. [Google Scholar]
- 47.Van Name W G. Bull. Am. Mus. Nat. His. 1945. [Google Scholar]
- 48.Bone Q. The Biology of Pelagic Tunicates. Oxford: Oxford Univ. Press; 1998. [Google Scholar]
- 49.Jeffery W R, Swalla B J. In: Embryology: Constructing the Organism. Gilbert S, editor. Sunderland, MA: Sinauer; 1997. pp. 331–364. [Google Scholar]
- 50.Satoh N. Developmental Biology of Ascidians. New York: Cambridge Univ. Press; 1994. [Google Scholar]
- 51.Wada H. Mol Biol Evol. 1998;15:1189–1194. doi: 10.1093/oxfordjournals.molbev.a026026. [DOI] [PubMed] [Google Scholar]
- 52.Cohen C S, Saito Y, Weissman I L. Evolution. 1998;52:746–756. doi: 10.1111/j.1558-5646.1998.tb03699.x. [DOI] [PubMed] [Google Scholar]
- 53.Bromham L D, Degnan B M. Evol Dev. 1999;1:166–171. doi: 10.1046/j.1525-142x.1999.99026.x. [DOI] [PubMed] [Google Scholar]
- 54.Strathmann R R, Bonar D. Mar Biol (Berlin) 1976;34:317–324. [Google Scholar]
- 55.Dilly P N, Welsch U, Rehkamper G. Acta Zool. 1986;67(3):173–179. [Google Scholar]
- 56.Balser E J, Ruppert E E. Acta Zool. 1990;71:235–249. [Google Scholar]
- 57.Yasuo H, Satoh N. Dev Growth Differ. 1994;36:9–18. doi: 10.1111/j.1440-169X.1994.00009.x. [DOI] [PubMed] [Google Scholar]
- 58.Smith J. Trends Genet. 1999;15:154–158. doi: 10.1016/s0168-9525(99)01693-5. [DOI] [PubMed] [Google Scholar]
- 59.Ogasawara M, Hiroshi W, Heiko P, Satoh N. Development (Cambridge, UK) 1999;126:2539–2550. doi: 10.1242/dev.126.11.2539. [DOI] [PubMed] [Google Scholar]
- 60.Shoguchi E, Satoh N, Maruyama Y K. Mech Dev. 1999;82:185–189. doi: 10.1016/s0925-4773(99)00008-8. [DOI] [PubMed] [Google Scholar]
- 61.Peterson K, Cameron R A, Tagawa K, Satoh N, Davidson E H. Development (Cambridge, UK) 1999;126:85–95. doi: 10.1242/dev.126.1.85. [DOI] [PubMed] [Google Scholar]
- 62.Harada Y, Yasuo H, Satoh N. Development (Cambridge, UK) 1995;121:2747–2754. doi: 10.1242/dev.121.9.2747. [DOI] [PubMed] [Google Scholar]