Abstract
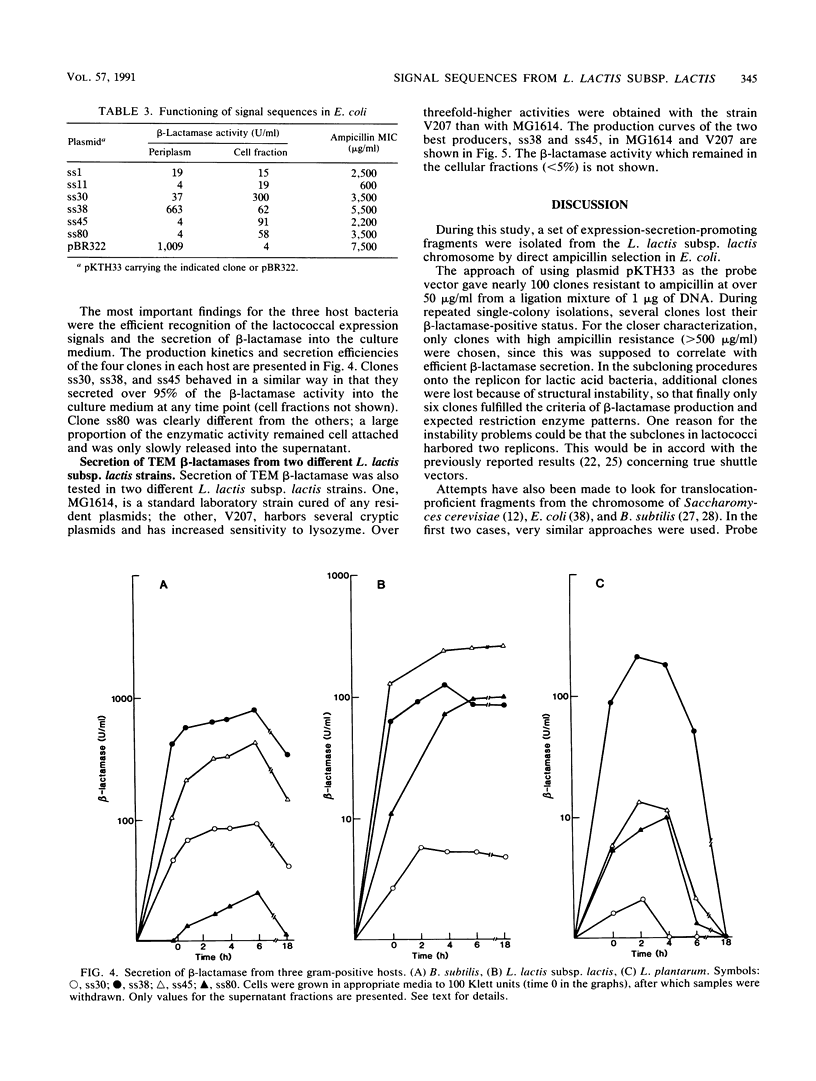
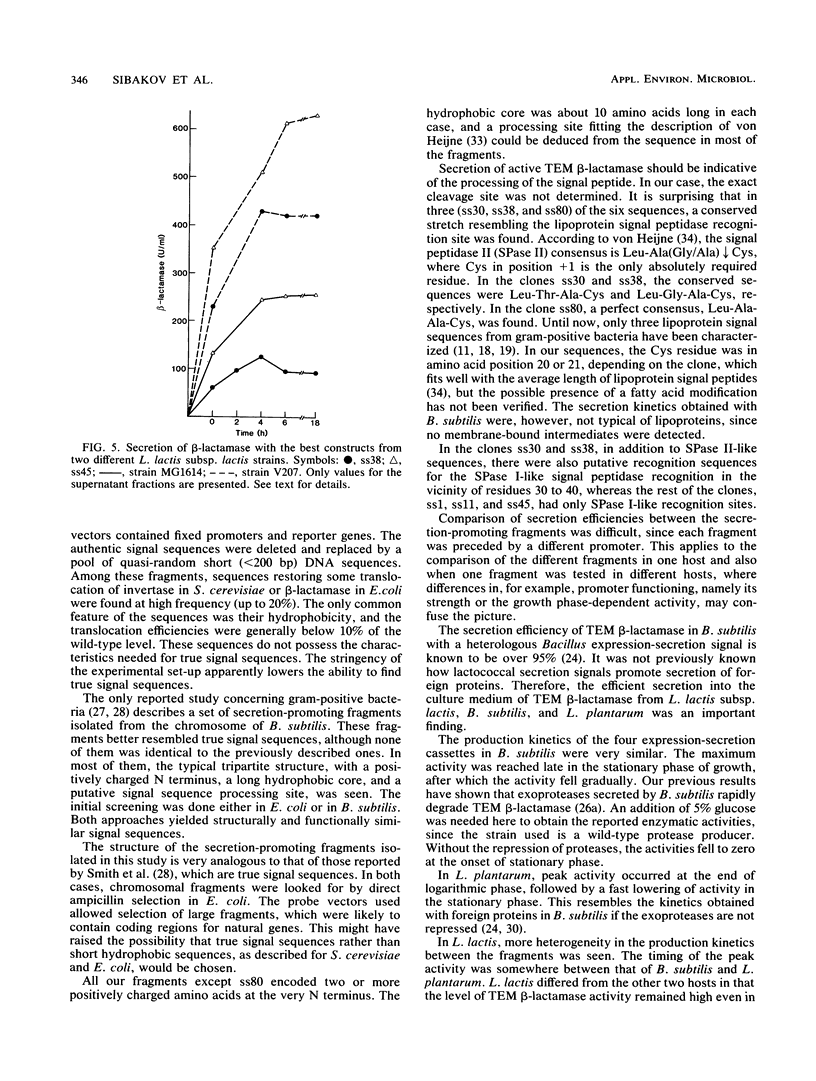
With TEM beta-lactamase as a reporter gene, a set of expression-secretion-promoting fragments were isolated from the chromosome of Lactococcus lactis subsp. lactis. The fact that only translocated beta-lactamase renders cells resistant to ampicillin allowed direct ampicillin selection with an Escherichia coli vector (pKTH33). The clones showing the greatest ampicillin resistance were subcloned onto a replicon capable of replication in lactic acid bacteria (pVS2), and the nucleotide sequences of the relevant fragments were determined. The structure of the secretion-promoting fragments in general resembled that of gram-positive true signal sequences, with a strongly positively charged N terminus, a long hydrophobic core, and a putative signal peptidase recognition site. The promoterlike sequences preceding the signal sequences matched well with those of previously published lactococcal promoters. In addition to E. coli, the functioning of these expression-secretion cassettes was studied in three gram-positive hosts: Bacillus subtilis, L. lactis, and Lactobacillus plantarum. Efficient expression and secretion of TEM beta-lactamase into the culture medium of each gram-positive host was obtained. Furthermore, when a strain of L. lactis subsp. lactis showing increased sensitivity to lysozyme was compared with a standard laboratory strain, threefold-higher secreted enzyme activities were detected.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. G., McKay L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates E. E., Gilbert H. J., Hazlewood G. P., Huckle J., Laurie J. I., Mann S. P. Expression of a Clostridium thermocellum endoglucanase gene in Lactobacillus plantarum. Appl Environ Microbiol. 1989 Aug;55(8):2095–2097. doi: 10.1128/aem.55.8.2095-2097.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Gasson M. J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983 Apr;154(1):1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Contente S., Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Hussain M., Pastor F. I., Lampen J. O. Cloning and sequencing of the blaZ gene encoding beta-lactamase III, a lipoprotein of Bacillus cereus 569/H. J Bacteriol. 1987 Feb;169(2):579–586. doi: 10.1128/jb.169.2.579-586.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C. A., Preuss D., Grisafi P., Botstein D. Many random sequences functionally replace the secretion signal sequence of yeast invertase. Science. 1987 Jan 16;235(4786):312–317. doi: 10.1126/science.3541205. [DOI] [PubMed] [Google Scholar]
- Klessen C., Schmidt K. H., Ferretti J. J., Malke H. Tripartite streptokinase gene fusion vectors for gram-positive and gram-negative procaryotes. Mol Gen Genet. 1988 May;212(2):295–300. doi: 10.1007/BF00334699. [DOI] [PubMed] [Google Scholar]
- Koivula T., Sibakov M., Palva I. Isolation and characterization of Lactococcus lactis subsp. lactis promoters. Appl Environ Microbiol. 1991 Feb;57(2):333–340. doi: 10.1128/aem.57.2.333-340.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok J., Leenhouts K. J., Haandrikman A. J., Ledeboer A. M., Venema G. Nucleotide sequence of the cell wall proteinase gene of Streptococcus cremoris Wg2. Appl Environ Microbiol. 1988 Jan;54(1):231–238. doi: 10.1128/aem.54.1.231-238.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J Biol Chem. 1981 Nov 10;256(21):11283–11291. [PubMed] [Google Scholar]
- Nielsen J. B., Lampen J. O. Membrane-bound penicillinases in Gram-positive bacteria. J Biol Chem. 1982 Apr 25;257(8):4490–4495. [PubMed] [Google Scholar]
- O'Callaghan C., Morris A. Inhibition of beta-lactamases by beta-lactam antibiotics. Antimicrob Agents Chemother. 1972 Dec;2(6):442–448. doi: 10.1128/aac.2.6.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostroff G. R., Pène J. J. Molecular cloning with bifunctional plasmid vectors in Bacillus subtilis. I. Construction and analysis of B. subtilis clone banks in Escherichia coli. Mol Gen Genet. 1984;193(2):299–305. doi: 10.1007/BF00330684. [DOI] [PubMed] [Google Scholar]
- Palva I. Molecular cloning of alpha-amylase gene from Bacillus amyloliquefaciens and its expression in B. subtilis. Gene. 1982 Jul-Aug;19(1):81–87. doi: 10.1016/0378-1119(82)90191-3. [DOI] [PubMed] [Google Scholar]
- Primrose S. B., Ehrlich S. D. Isolation of plasmid deletion Mutants and study of their instability. Plasmid. 1981 Sep;6(2):193–201. doi: 10.1016/0147-619x(81)90066-4. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H., Bron S., Van Ee J., Venema G. Construction and use of signal sequence selection vectors in Escherichia coli and Bacillus subtilis. J Bacteriol. 1987 Jul;169(7):3321–3328. doi: 10.1128/jb.169.7.3321-3328.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H., de Jong A., Bron S., Venema G. Characterization of signal-sequence-coding regions selected from the Bacillus subtilis chromosome. Gene. 1988 Oct 30;70(2):351–361. doi: 10.1016/0378-1119(88)90207-7. [DOI] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmanen I., Lundström K., Lehtovaara P., Sarvas M., Ruohonen M., Palva I. Transcription and translation of foreign genes in Bacillus subtilis by the aid of a secretion vector. J Bacteriol. 1985 Apr;162(1):176–182. doi: 10.1128/jb.162.1.176-182.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Broome-Smith J. K. Identification of amino acid sequences that can function as translocators of beta-lactamase in Escherichia coli. Mol Microbiol. 1989 Oct;3(10):1361–1369. doi: 10.1111/j.1365-2958.1989.tb00117.x. [DOI] [PubMed] [Google Scholar]
- van de Guchte M., van der Vossen J. M., Kok J., Venema G. Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1989 Jan;55(1):224–228. doi: 10.1128/aem.55.1.224-228.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vossen J. M., van der Lelie D., Venema G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl Environ Microbiol. 1987 Oct;53(10):2452–2457. doi: 10.1128/aem.53.10.2452-2457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 1989 May;2(7):531–534. doi: 10.1093/protein/2.7.531. [DOI] [PubMed] [Google Scholar]
- von Wright A., Taimisto A. M., Sivelä S. Effect of Ca2+ ions on plasmid transformation of Streptococcus lactis protoplasts. Appl Environ Microbiol. 1985 Oct;50(4):1100–1102. doi: 10.1128/aem.50.4.1100-1102.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wright A., Tynkkynen S., Suominen M. Cloning of a Streptococcus lactis subsp. lactis Chromosomal Fragment Associated with the Ability To Grow in Milk. Appl Environ Microbiol. 1987 Jul;53(7):1584–1588. doi: 10.1128/aem.53.7.1584-1588.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]