Abstract
In contrast to the highly mutagenic pyrimidine(6–4)pyrimidone photoproduct, its Dewar valence isomer (Dewar product) has low mutagenic potential and produces a broad range of mutations [LeClerc, J. E., Borden, A. & Lawrence, C. W. (1991) Proc. Natl. Acad. Sci. USA 88, 9685–9689]. To determine the origin of the mutagenic property of the Dewar product, we used experimental NMR restraints and molecular dynamics to determine the solution structure of a Dewar-lesion DNA decamer duplex. This DNA decamer duplex (DW/GA duplex) contains a mismatched base pair between the 3′ T residue of the Dewar lesion (T6) and an opposed G residue (G15). The 3′ T (T6) of the Dewar lesion formed stable hydrogen bonds with the opposing G15 residue. However, the helical bending and unwinding angles of the DW/GA duplex were much larger than those of a second duplex that contains the Dewar lesion and opposing A15 and A16 residues (DW/AA duplex). The DW/GA duplex showed poorer stacking interactions at the two bases of the Dewar product and at the adjacent A7⋅T14 base pair than did the DW/AA duplex. These structural features imply that no thermal stability or conformational benefit is obtained by incorporating a G instead of an A opposite the 3′ T of the Dewar lesion. These properties may thus facilitate the preferential incorporation of an A in accordance with the A rule during translesion replication and lead to the low frequency of 3′ T→C mutations observed at this site.
Irradiation of DNA with UV light produces a variety of photoproducts that block DNA replication (1–6). When an SOS response is induced by UV irradiation, these photoproducts are bypassed with modest efficiency by DNA polymerase and give rise to mutations (5, 6). This process has been termed translesion replication (TR) or bypass synthesis, and mutations are caused by the tendency of DNA polymerase to insert an incorrect nucleotide opposite the lesion during TR (7, 8).
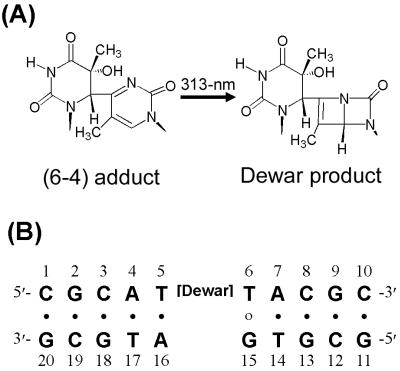
Although both the pyrimidine (6–4)pyrimidone photoproduct [(6–4) adduct], which is one of the major classes of UV-induced DNA photoproducts (9, 10), and its Dewar valence isomer (Dewar product) (Fig. 1A) cause mutations during TR, their mutagenic properties differ. The (6–4) adduct is highly mutagenic and yields a specific mutation (6, 11, 12). In SOS-induced Escherichia coli cells, the marked preference for the insertion of a guanine residue opposite the 3′ T of (6–4) adducts during TR leads to a predominant 3′ T→C transition with 85% replicating error frequency (6). In contrast, the Dewar product is less mutagenic and induces a broader range of mutations than does the (6–4) adduct (6, 12). In SOS-induced E. coli cells, adenine was found to be incorporated into the site opposite the 3′ T of the Dewar product with a frequency of 72%, whereas G was incorporated only 21% of the time (6). Thus, the 3′ T→C transition, which is the major class of mutations induced by the Dewar product, is produced with 13% replicating error frequency during TR (6).
Figure 1.
Chemical structures analyzed in this study. (A) Chemical structures of the (6–4) adduct and the Dewar product. (B) DNA sequence context of the DW/GA duplex.
Enzyme repair rates of 49-residue oligonucleotides that contain site-specific thymine photoproducts were reported to depend on the type of photoproduct present. The Dewar product and the (6–4) adduct were repaired at nearly identical rates (13). The recognition of the Dewar and (6–4) lesions by the E. coli uvrA subunit of the uvr(A)BC endonuclease was similar to that of human DNA damage-binding protein, with relative binding affinities of 4:9 (14). The lower binding affinity of the Dewar product compared with the (6–4) adduct was explained by a previous structural analysis study in which the Dewar product was shown to distort B-form helical DNA to a lesser extent than the (6–4) adduct (15). In addition, it has been reported that the Dewar product is more easily bypassed by a particular DNA polymerase and hence constitutes less of a block to DNA polymerase than does the (6–4) adduct (16).
The nucleotide substitution mutations induced by DNA lesions during TR result from the misinstructive or noninstructive properties of the distorted templates (17). Opposite the abasic site (the prototypical noninstructional DNA lesion), an A residue is incorporated with a frequency of 77% during TR in E. coli (18). This preferential incorporation of an A residue opposite an abasic lesion is referred to as the “A rule” (19–21). NMR studies suggest that the A residue opposite an abasic site stacks better in an intrahelical configuration than do other bases and thus causes no helical distortion (22–24).
The term “misinstructive” is used to indicate the existence of physical interactions between a distorted DNA template and an incoming dNTP that lead to the incorporation of a mismatched nucleotide opposite the lesion (6). It has been reported that the (6–4) adduct is a misinstructional DNA lesion, and the 3′ T→C transition induced by the (6–4) adduct is caused by physical interactions and features such as hydrogen bonding, high thermal stability, and no backbone distortion (25). Thermodynamic studies have shown that the ΔG0 value of a DNA helix containing the 3′ T⋅G base pair of the (6–4) adduct is much lower than that of the 3′ T⋅A base pair, making the 3′ T⋅G base pair more favorable. In contrast, this difference in ΔG0 values for the Dewar product is very small (26). These observations could explain at least in part the differences in the mutagenic properties of the two types of photoproducts; however, these thermodynamic findings still do not explain why the Dewar product is less lethal than the (6–4) adduct. To gain a complete understanding of why an A residue is inserted preferentially at the 3′ T site of the Dewar lesion, structural studies of a DNA duplex containing a Dewar product in a mutated sequence context are required.
In this report, we determined the conformational influence of the 3′ T⋅G base pair in a DNA decamer duplex that contains a mismatched base pair between the 3′ T of the Dewar product and an opposed G residue (represented by the DW/GA duplex; Fig. 1B). These results were then compared with those of the 3′ T⋅A base pair in a DW/AA duplex, which were described in our previous study (15). In addition, the conformational differences between two Dewar lesion-containing duplexes were compared with those of the (6–4) adduct in (6–4)/AA and (6–4)/GA duplexes, which were established previously (25, 27). This structural comparison provides insight into the mechanisms that determine base selection during TR, and these observed structural differences can account for the low mutagenicity and specificity of the Dewar product.
Materials and Methods
Sample Preparation.
The (6–4) adduct-containing DNA decamer duplex [(6–4)/GA duplex] was prepared as described (25). The (6–4)/GA duplex was dissolved in a D2O solution containing 10 mM sodium phosphate (pH 6.6) and 200 mM NaCl. Photoisomerization of the (6–4) adduct to its Dewar valence isomer in the duplex was performed by direct UV irradiation at 313 nm in an NMR tube. Complete conversion was confirmed by observing the disappearance of the T6-H6 resonance and the upfield shift of the T6–methyl resonance in an 1H-NMR experiment.
NMR Experiments.
All NMR data sets generated with the DW/GA duplex were collected with a Bruker DMX-600 spectrometer (Korea Basic Science Institute, Taejon, Korea). Details of NMR experiments and data processing can be found in our earlier published studies of a photoproduct-containing DNA duplex (15, 27). Nuclear Overhauser effect (NOE) distance restraints from nonexchangeable protons were obtained from two-dimensional NOE spectroscopy (NOESY) experiments with mixing times of 50, 80, and 300 ms in a D2O buffer solution. Exchangeable proton NOEs were determined by using NOESY spectra in H2O buffer with 120- and 400-ms mixing times. Watson–Crick-type hydrogen-bonding restraints were imposed in each base pair except the T5⋅A16 and T6⋅G15 base pairs.
Structure Calculation.
The structure of the DW/GA duplex was calculated by using the program x-plor 3.1 (28) with restrained molecular dynamics (RMD). We initially generated the normal A- and B-form starting structures with modification of the Dewar product at the T5-T6 position. These structures were subjected to the RMD and simulated annealing protocols. The first stage of computation began with energy minimization, followed by 10 ps of molecular dynamics (MD) at 1,000 K. The force constants for the distance restraints were gradually increased over 10 cycles of 1-ps dynamics. The final values of the force constants were 100 kcal⋅mol−1 Å−2. The system was subsequently cooled to 300 K over 10 cycles of 0.5-ps dynamics followed by energy minimization. The final stage involved 20 ps of RMD at 300 K, and the structures were subsequently energy minimized. Fourteen structures (seven of the B-form and seven of the A-form initial structures) were chosen on the basis of the lowest NOE violations and total energies.
The mean structure of the RMD-refined structures was next optimized by using full relaxation matrix refinement (based on NOE intensity) with x-plor. NOE volumes from 738 cross peaks of three mixing times of 50, 80, and 160 ms were used as restraints. The first stage of computation began with 10 ps of MD at 500 K. The force constants for the distance restraints were gradually decreased to zero, and those of the intensity restraints were increased to 100 kcal⋅mol−1 over 10 cycles of 0.5-ps dynamics. The system was subsequently cooled to 300 K over 5 cycles of 0.5-ps dynamics, which was followed by energy minimization. The final stage involved 10 ps of MD at 300 K, and the structures were subsequently energy minimized. Eight structures were chosen on the basis of the lowest total energies. The helical parameters of the refined structures were calculated by using the program curves (29).
Results
NMR Resonance Assignment.
The nonexchangeable base and sugar protons in the DW/GA duplex were assigned according to their intraresidue and sequential NOE connectivities. Saturation of the 5–6 double bond of the T5 base, which resulted from the formation of a covalent bond between the T5-C6 and T6-C4 carbons, led to the upfield shift of the T5-H6 resonance (4.34 ppm). Photoconversion of the pyrimidone ring (T6 base) to the Dewar valence form led to significant upfield shifts of its H6 proton (7.96→4.71 ppm) and methyl (2.33→2.06 ppm) resonances.
The exchangeable protons in the DW/GA duplex were assigned by analyzing NOESY data in an H2O buffer solution. One striking feature of the temperature-dependent imino proton spectra in H2O buffer (unpublished data) was the persistence of the G15 imino proton resonance for experiments performed at 1–20°C. These spectra were similar to those of the DW/AA duplex (30). These results indicate that the hydrogen-bonding interaction between the 3′ T of the Dewar product and the imino proton of the opposite G15 residue was obviously present, but the stability of the overall helix was not improved by substituting a G for an A opposite the 3′ T site of the Dewar product.
Structure Determination.
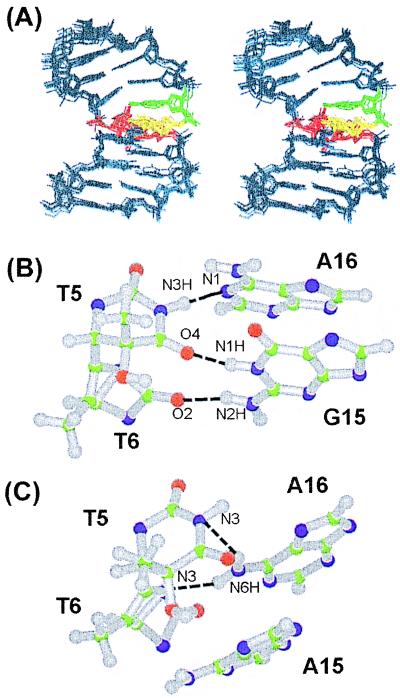
The solution structure of the DW/GA duplex was calculated according to the protocols outlined in Materials and Methods. A total of 374 distances was restrained in the RMD calculation; these distance restraints consisted of 285 interproton distances derived from NOESY data in D2O buffer, 15 interproton distances obtained from H2O-NOESY cross peaks, and 74 distances for the Watson–Crick base pairs of the flanking residues. A converged subset of 14 structures refined by RMD was identified on the basis of low NOE violations and total energies. These structures exhibited pairwise rms deviation values of 1.09 Å ± 0.25 for all heavy atoms. Full relaxation matrix refinements of the mean structure yielded a well-converged subset of eight refined structures. The eight superimposed refined structures of the DW/GA duplex are plotted in Fig. 2A and exhibited pairwise rms deviation values of 0.68 Å ± 0.19 for all heavy atoms.
Figure 2.
(A) Superimposed stereo view of eight intensity-refined structures of the DW/GA duplex. The two thymine residues (T5, T6) of the Dewar product are colored red, and their opposite G15 and A16 residues are colored yellow and green, respectively. The other flanking residues are colored dark blue. The hydrogen atoms are excluded in this figure. (B) Ball-and-stick view of the two base pairs of the Dewar product in the DW/GA and (C) DW/AA duplexes. Balls are colored by using the accepted atomic color representation: gray, hydrogen; green, carbon; blue, nitrogen; red, oxygen. The dotted lines indicate the hydrogen bonds, as determined with the program insight ii.
Overall Helical Conformation.
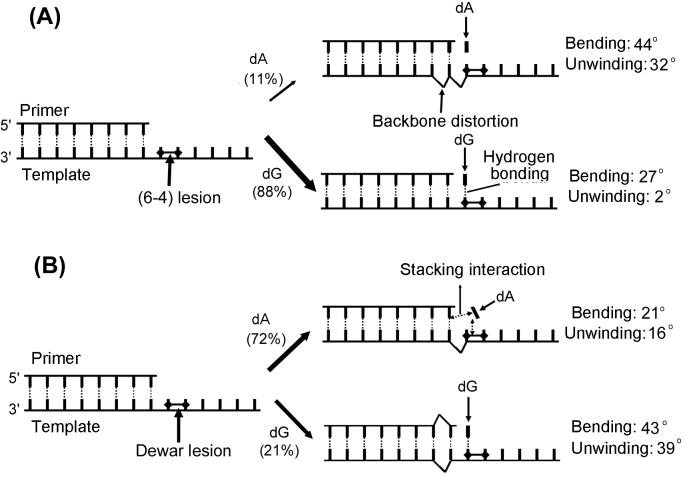
Structural calculation showed that the formation of the Dewar product caused a significant bending (overall bending of 43° ± 5) and unwinding (39° ± 5) of the DNA helix of the DW/GA duplex. These values are much higher than those of the DW/AA duplex (see Fig. 4). It is noteworthy that the helical distortions caused by the (6–4) adduct in the (6–4)/GA duplex are much less significant than those of the (6–4)/AA duplex (see Fig. 4). This finding indicates that in contrast to the (6–4) adduct, the insertion of a G residue opposite the 3′ T of the Dewar product distorted the DNA helix more severely than did the 3′ T⋅A base pairing.
Figure 4.
Summary of conformational features after base insertion opposite the 3′ T site of (A) the (6–4) adduct and (B) the Dewar product during TR. The conformational features represented in this figure are indicated. The values of the helical bending and unwinding of the model duplexes studied herein and previously are shown (Right). The incorporating frequencies of the A and G residues opposite the 3′ T site of the photoproducts during TR (6) are shown in parentheses at the center of the figure.
Hydrogen Bonding Feature at the Dewar Lesion Site.
The T5⋅A16 base pair of the Dewar lesion in the DW/GA duplex was shown to deviate from the standard Watson–Crick base pairing geometry, supported by the absence of the NOE cross peak between the A16-H2 and T5-imino proton resonances (unpublished data). Our structural model showed that the imino proton of the T5 formed a hydrogen bond (albeit a weak one) to the N1 nitrogen of the opposite A16 residue (Fig. 2B) (heteroatom separation of 3.2 Å ± 0.2). However, the O4 carbonyl oxygen of the T5 residue formed a hydrogen bond, not to the amino proton of the opposite A16, but to the imino proton of the G15 residue (heteroatom separation of 2.9 Å ± 0.1) (Fig. 2B). This caused (i) the T5⋅A16 base pair to have a high propeller twist (−58° ± 7), and (ii) unwinding in the G15/A16 base step (base twist of 10° ± 3).
The hydrogen bonding feature of the 3′ T (T6) residue of the Dewar product in the DW/GA duplex contrasts with that in the DW/AA duplex. For the DW/GA duplex, the O2 carbonyl oxygen of T6 formed a hydrogen bond with the amino proton of the opposite G15 residue (heteroatom separation of 2.9 Å ± 0.1) (Fig. 2B). However, in the DW/AA duplex, the N3 nitrogen of the T6 residue formed a hydrogen bond with the amino proton of the A16 residue, rather than with the opposing A15 residue (Fig. 2C).
We reported recently that the strong hydrogen-bonding interaction between the 3′ T of the (6–4) adduct and the opposite G15 residue stabilizes the overall helix much more than that does a 3′ T⋅A base pair (25). However, although the 3′ T of the Dewar product can form a hydrogen bond to the opposite G15 residue, this interaction improved only slightly the thermal stability of the overall helix relative to that of the 3′ T⋅A base pair (see results of temperature-dependent NMR experiment, described above). These results are in agreement with thermodynamics studies in which the ΔG0 value of a DNA helix containing a 3′ T⋅G base pair of the (6–4) adduct is much lower than that of a DNA helix containing a 3′ T⋅A base pair, whereas the corresponding difference in ΔG0 values for the Dewar product is very small (26).
Backbone Conformation.
The distortions of the backbone conformation caused by the formation of the Dewar product were significantly different in the two duplexes. The glycosidic bond torsion angles of T5 in the DW/GA duplex remained in the anti conformation (χ = −166° ± 1), whereas the high-anti conformation is predominant for the corresponding residue in the DW/AA duplex (15). The deoxyribose ring conformations of both T residues of the Dewar lesions also differ for the two duplexes. C3′-endo sugar puckers were observed at T5 in the DW/GA duplex (pseudorotation P = 33° ± 1) and at T6 in the DW/AA duplex, whereas S-type sugar puckers were adopted by the T6 residue of the DW/GA duplex and the T5 residue of the DW/AA duplex.
The most striking aspect of these data is that the Dewar products in the two duplexes caused deviations of the backbone torsion angles from the values of canonical B-form DNA at different sites in the duplexes. In the DW/AA duplex, the backbone conformation involving the phosphorous atom between the T6 and A7 residues, which is related to the backbone torsion angles of T6, is significantly distorted (Table 1). The cis orientation of the ɛ and gauche+ orientation of the ζ torsion angles reduce the intersugar spacing between the T6 and A7 residues but enlarge the interbase spacing of the same site, which can be confirmed by comparing some distances. The T6-C4′ to A7-C4′ and T6-N1 to A7-N9 distances are 4.9 Å and 5.7 Å, whereas they are 6.1 Å and 4.4 Å in normal B-DNA. This spacing feature introduces a helical bend, which allows the unusual hydrogen-bonding interaction observed between the T6 and A16 residues. In the DW/GA duplex, the hydrogen-bonding interaction between T6 of the Dewar product and the opposite G15 residue does not require this spacing feature or the backbone distortion in the T6-p-A7 step, so that all of the backbone torsion angles of T6 are in the range of those of canonical B-DNA (Table 1). Surprisingly, the torsion angles of the A7 and G13 residues of the DW/GA duplex, which involved the two phosphorous atoms between the A7⋅T14 and C8⋅G13 base pairs, showed deviation from the values of B-DNA (Table 1). In the DW/GA duplex, the β torsion angles underwent transitions from trans to gauche+ conformations, and the ζ angle values changed from the gauche− conformation to the more typical trans conformations. The gauche− to trans transition of the ζ angles of the A7 and G13 residue in the DW/GA duplex is consistent with the downfield shifts of their 31P resonances (A7 and G13: −2.84 ppm). These backbone distortions between the A7⋅T14 and C8⋅G13 base pairs caused significant widening of the major groove at this site. This major groove widening can be confirmed by the two interphosphorous distances of the A7 to T14 and C8 to G13 residues, which are 19.4 Å ± 0.3 and 20.4 Å ± 0.3, respectively; the corresponding values for B-DNA and the DW/AA duplex are 18.2 Å and 18.4 Å, respectively.
Table 1.
Backbone torsion angles of photoproduct-containing DNA duplexes
| Torsion angles† | DNA duplexes*
|
||||
|---|---|---|---|---|---|
| DW/AA | DW/GA | (6–4)/AA | (6–4)/GA | ||
| α, ° (−61) | T6 | −76 | −80 ± 5 | 149 | −78 |
| A7 | −57 | −37 ± 22 | −74 | −63 | |
| T14 | 69 | −66 ± 1 | −76 | −76 | |
| G13 | −66 | −39 ± 3 | −57 | −57 | |
| β, ° (180) | T6 | 134 | −174 ± 8 | −146 | −168 |
| A7 | −176 | 134 ± 24 | 139 | 175 | |
| T14 | −144 | 174 ± 4 | 159 | −116 | |
| G13 | −141 | 128 ± 17 | 180 | 174 | |
| ɛ, ° (173) | T6 | −13 | 179 ± 6 | −170 | −173 |
| A7 | 155 | −160 ± 23 | −73 | 158 | |
| T14 | −118 | −171 ± 4 | −160 | 170 | |
| G13 | −171 | −160 ± 14 | −172 | 173 | |
| ζ, ° (−91) | T6 | 113 | −89 ± 5 | −83 | −96 |
| A7 | −85 | −165 ± 28 | 157 | −78 | |
| T14 | −93 | −110 ± 16 | −87 | −96 | |
| G13 | −91 | −172 ± 13 | −111 | −93 | |
Base-Stacking Interaction.
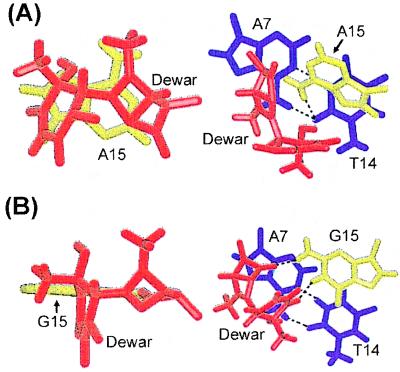
We also studied the base-stacking interactions at the Dewar lesion sites and 3′ flanking regions of the DW/GA and DW/AA duplexes. Base-stacking interactions correlate with thermal stability of a DNA helix. In the DW/AA duplex, the A15 base opposite the 3′ T of the Dewar product is nicely stacked with the covalently linked bases of the Dewar lesion and the A7⋅T14 base pair (Fig. 3A). However, in the DW/GA duplex, the hydrogen-bonding interaction of the G15 residue with both T5 and T6 of the Dewar product makes stacking between these bases impossible and also disrupts the stacking interaction between G15 and the adjacent A7⋅T14 base pair (Fig. 3B) by causing overwinding of the T14-G15 step (base twist of 46° ± 2). The disruption of these base-stacking interactions at the Dewar lesion site and 3′ flanking region destabilizes the DNA helix. Thus, the thermal stability of a DNA helix containing a 3′ T⋅G base pair of the Dewar product is not greater than that of a DNA duplex that contains a 3′ T⋅A base pair, even though the former shows more stable hydrogen-bonding interactions than does the latter.
Figure 3.
Comparative stick view of the stacking interactions of (A) the A15 residue (DW/AA duplex) and (B) the corresponding G15 residue (DW/GA duplex) opposite the 3′ T of the Dewar product with two bases of the Dewar product (Left) and the adjacent A7⋅T14 base pair (Right). The two thymine bases (T5, T6) of the Dewar product are colored red, and the opposite G15 and A15 residues are colored yellow. The A7⋅T14 base pair is colored dark blue. The dotted lines indicate the hydrogen bonds, as determined with the program insight ii.
Discussion
The replicating error frequency of the 3′ T→C transition, which is the most frequent mutation induced by the Dewar lesion, is only 13% in E. coli (6). It has been suggested that the nucleotide substitution mutations induced by the (6–4) adduct and Dewar lesion during TR result from physical interactions between a distorted DNA template and an incoming dNTP (6). In the case of the (6–4) adduct, our previous study revealed that these physical interactions play a crucial role in determining the nature of the dNTP incorporated opposite the 3′ T of the (6–4) lesion (25).
The nature of hydrogen-bonding interactions between the 3′ T of the photoproduct and the opposite residue is crucial in determining which dNTP is incorporated opposite this 3′ T site. It has been proposed that the potential hydrogen bonding of the 3′ T⋅G base pair of the (6–4) adduct, which does not occur in a 3′ T⋅A base pair, explains in part the high frequency and specificity of the 3′ T→C transition induced by the (6–4) adduct (25). The solution structure of the DW/GA duplex in this study clearly shows that the 3′ T of the Dewar lesion forms a hydrogen bond with the opposite G residue. However, we find that the conformational change of the DNA duplex achieved by substituting a G for an A opposite the 3′ T site of the Dewar product contrasts with that of the (6–4) adduct. Comparison of this conformational change for the Dewar lesion with that for the (6–4) lesion can explain the low frequency of mutations that occur at the 3′ T site of the Dewar lesion (summarized in Fig. 4). First, an A residue opposite the 3′ T of the Dewar lesion shows better stacking interactions with the two bases of the Dewar product and the adjacent A⋅T base pair than does a G residue at this position (Fig. 3). This significant difference in base-stacking interactions of the G and A residues opposite the 3′ T was not found for the (6–4) adduct (25). The base-stacking interaction of the A residue opposite the 3′ T of the Dewar lesion in the DW/AA duplex contributed to the overall helical stability; thus the total difference in thermal stability between the DW/AA and DW/GA duplexes becomes very small, even though the hydrogen bonding of the 3′ T residue occurs only in the DW/GA duplex.
Second, both A and G residues opposite the 3′ T of the Dewar lesion caused backbone distortions in the 3′ flanking duplex region, although these distortions occurred at different sites (Table 1). This implies that backbone distortion of the duplex region (formed between the lesion-containing template and primer) is produced when the A or G residues are incorporated opposite the 3′ T of the Dewar lesion during TR. In contrast, only an A residue opposite the 3′ T of the (6–4) adduct produces backbone distortions (25). This result implies that during TR, the backbone of the duplex region is not distorted when a G residue is incorporated at the 3′ T of the (6–4) adduct, but it is severely distorted when an A is incorporated.
Third, in contrast to the (6–4) adduct, the magnitudes of the helical bending and unwinding caused by forming the Dewar lesion in the DW/GA duplex were much higher than those observed for the DW/AA duplex. As shown in Fig. 4, the severe distortion of the DNA duplex caused by the (6–4) adduct is reduced dramatically by substituting a G for an A opposite the 3′ T site of the (6–4) lesion. However, in the case of the Dewar lesion, the helical distortion is increased by this base substitution.
Our previous study concluded that the 3′ T→C transition induced by the (6–4) adduct is a misinstructive mutation caused by important physical features such as hydrogen bonding, high thermal stability, and no backbone distortion (25). However, this structural study of the DW/GA duplex revealed that the 3′ T of the Dewar product does not display more favorable physical interactions when an opposing A residue is replaced by a G residue. This finding indicates that the Dewar product does not exhibit a preference for insertion of a G opposite its 3′ T site during TR. It has been reported that during TR in E. coli, an A residue is incorporated opposite abasic sites with a frequency of 77% (18). This is in accordance with the A rule, where an A residue is incorporated opposite an abasic lesion with a 10-fold higher efficiency than a G residue (18). A physical basis for the A rule has been provided by NMR studies showing that A residues opposite abasic sites stack better in the intrahelical configuration than do other bases and cause no helical distortion (22–24). The G residue opposite the 3′ T of the Dewar product distorts more severely the overall helical configuration and shows poorer stacking interactions than does an A residue. Therefore, we conclude that the incorporation of an A residue opposite the 3′ T of the Dewar lesion is facilitated not by potential physical interaction such as hydrogen bonding but by the incorporation preference of the A residue in a noninstructional manner.
In summary, by using experimental NMR restraints and MD, we have demonstrated the solution structural features unique to the 3′ T⋅G base pair of the DW/GA duplex. The structural comparison between the 3′ T⋅G and 3′ T⋅A base pairs of both the (6–4) adduct and Dewar lesions offers an explanation of their differing mutagenic properties (Fig. 4). The incorporation of a G residue, which forms stable hydrogen bonds with the opposite 3′ T of the (6–4) adduct, restores the distorted backbone conformation and increases the thermal stability of the overall helix. Although the 3′ T of the Dewar lesion forms stable hydrogen bonds with the opposite G residue (DW/GA duplex), this G showed poorer stacking interactions with the two bases of the Dewar product and with the adjacent A⋅T base pair than does the corresponding A residue (DW/AA duplex). The stable hydrogen bonding of the G residue did not increase the thermal stability of the overall helix and also did not restore the distorted backbone conformation of the DNA helix caused by forming the Dewar lesion. In addition, the helical bending and unwinding angles of the DW/GA duplex were much higher than those of the DW/AA duplex. This demonstrates that, in contrast with the (6–4) adduct, no thermal stability or conformational benefits are achieved when a G is incorporated instead of an A opposite the 3′ T of the Dewar lesion. These structural properties may thus facilitate the noninstructive incorporation of an A in accordance with the A rule during TR and consequently may lead to the low frequency of 3′ T→C mutations at Dewar lesions.
Acknowledgments
This work was supported by the Korea Science and Engineering Foundation through the Center for Molecular Catalysis at Seoul National University and by the Molecular Medicine Research Group Program (MOST. 98-J03–01-01-A-01).
Abbreviations
- (6–4) adduct
pyrimidine (6–4) pyrimidone photoproduct
- Dewar product
Dewar valence isomer of the (6–4) adduct
- TR
translesion replication
- MD
molecular dynamics
- RMD
restrained MD
- NOE
nuclear Overhauser effect
- NOESY
NOE spectroscopy
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1qkg).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080057097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080057097
References
- 1.Brash D E. Photochem Photobiol. 1988;48:59–66. doi: 10.1111/j.1751-1097.1988.tb02786.x. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell D L. Photochem Photobiol. 1988;48:51–57. doi: 10.1111/j.1751-1097.1988.tb02785.x. [DOI] [PubMed] [Google Scholar]
- 3.Ananthaswamy H N, Pierceall W E. Photochem Photobiol. 1990;52:1119–1136. doi: 10.1111/j.1751-1097.1990.tb08452.x. [DOI] [PubMed] [Google Scholar]
- 4.Brash D E, Rudolph J A, Simon J A, Lin A, McKenna G J, Baden H P, Halperin A J, Potén J. Proc Natl Acad Sci USA. 1991;88:10124–10128. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee S K, Christensen R B, Lawrence C W, LeClerc J E. Proc Natl Acad Sci USA. 1988;85:8141–8145. doi: 10.1073/pnas.85.21.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LeClerc J E, Borden A, Lawrence C W. Proc Natl Acad Sci USA. 1991;88:9685–9689. doi: 10.1073/pnas.88.21.9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuchs R P P, Napolitano R L. Proc Natl Acad Sci USA. 1998;95:13114–13119. doi: 10.1073/pnas.95.22.13114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomer G, Reuven N B, Livneh Z. Proc Natl Acad Sci USA. 1998;95:14106–14111. doi: 10.1073/pnas.95.24.14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell D L, Nairn R S. Photochem Photobiol. 1989;49:805–819. doi: 10.1111/j.1751-1097.1989.tb05578.x. [DOI] [PubMed] [Google Scholar]
- 10.Pfeifer G P. Photochem Photobiol. 1997;65:270–283. doi: 10.1111/j.1751-1097.1997.tb08560.x. [DOI] [PubMed] [Google Scholar]
- 11.Gibbs P E M, Borden A, Lawrence C W. Nucleic Acids Res. 1995;23:1919–1922. doi: 10.1093/nar/23.11.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith C A, Wang M, Jiang N, Che L, Zhao X, Taylor J-S. Biochemistry. 1996;35:4146–4154. doi: 10.1021/bi951975c. [DOI] [PubMed] [Google Scholar]
- 13.Svoboda D L, Smith C A, Taylor J-S, Sancar A. J Biol Chem. 1993;268:10694–10700. [PubMed] [Google Scholar]
- 14.Reardon J T, Nichols A F, Keeney S, Smith C A, Taylor J-S, Linn S, Sancar A. J Biol Chem. 1993;268:21301–21308. [PubMed] [Google Scholar]
- 15.Lee J-H, Hwang G-S, Kim J-K, Choi B-S. FEBS Lett. 1998;428:269–274. doi: 10.1016/s0014-5793(98)00508-0. [DOI] [PubMed] [Google Scholar]
- 16.Taylor J-S. Pure Appl Chem. 1995;67:183–190. [Google Scholar]
- 17.Gibbs P E M, Kilbey B J, Banerjee S K, Lawrence C W. J Bacteriol. 1993;175:2607–2612. doi: 10.1128/jb.175.9.2607-2612.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang N, Taylor J-S. Biochemistry. 1993;32:472–481. doi: 10.1021/bi00053a011. [DOI] [PubMed] [Google Scholar]
- 19.Schaaper R M, Kunkel T A, Loeb L A. Proc Natl Acad Sci USA. 1983;80:487–491. doi: 10.1073/pnas.80.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boiteux S, Laval J. Biochemistry. 1982;21:6746–6751. doi: 10.1021/bi00269a020. [DOI] [PubMed] [Google Scholar]
- 21.Sagher D, Strauss B S. Biochemistry. 1983;22:4518–4526. doi: 10.1021/bi00288a026. [DOI] [PubMed] [Google Scholar]
- 22.Cuniasse P, Sowers L C, Eritja R, Kaplan B E, Goodman M F, Cognet J A, LeBret M, Guschlbauer W, Fazakerley G V. Nucleic Acids Res. 1987;15:8003–8022. doi: 10.1093/nar/15.19.8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalnik M W, Chang C N, Grollman A P, Patel D J. Biochemistry. 1988;27:924–931. doi: 10.1021/bi00403a013. [DOI] [PubMed] [Google Scholar]
- 24.Cuniasse P, Fazakerley G V, Guschlbauer W, Kaplan B E, Sowers L C. J Mol Biol. 1990;213:303–314. doi: 10.1016/S0022-2836(05)80192-5. [DOI] [PubMed] [Google Scholar]
- 25.Lee J-H, Hwang G-S, Choi B-S. Proc Natl Acad Sci USA. 1999;96:6632–6636. doi: 10.1073/pnas.96.12.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujiwara Y, Iwai S. Biochemistry. 1997;36:1544–1550. doi: 10.1021/bi9619942. [DOI] [PubMed] [Google Scholar]
- 27.Kim J-K, Choi B-S. Eur J Biochem. 1995;228:849–854. doi: 10.1111/j.1432-1033.1995.tb20331.x. [DOI] [PubMed] [Google Scholar]
- 28.Brünger A T. x-plor Ver. 3.1. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 29.Lavery R, Sklenar H. J Biomol Struct Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 30.Hwang G-S, Kim J-K, Choi B-S. Eur J Biochem. 1996;235:359–365. doi: 10.1111/j.1432-1033.1996.00359.x. [DOI] [PubMed] [Google Scholar]
- 31.Saenger W. Principles of Nucleic Acid Structure. New York: Springer; 1984. pp. 51–104. [Google Scholar]