Abstract
Functional supramolecular chemistry at the single-molecule level. Single strands of DNA can be captured inside α-hemolysin transmembrane pore protein to form single-species α-HL·DNA pseudorotaxanes. This process can be used to identify a single adenine nucleotide at a specific location on a strand of DNA by the characteristic reductions in the α-HL ion conductance. This study suggests that α-HL-mediated single-molecule DNA sequencing might be fundamentally feasible.
Keywords: Single-molecule DNA seuqencing, transmembrane pore, pseudorotaxanes, supramolecular chemistry, channel conductance
There has been an intriguing suggestion that Staphylococcus aureus α-hemolysin (α-HL), a stable heptameric transmembrane protein pore[1], might be of use as the sensor element in a rapid pore-mediated single-molecule DNA sequencing process.[2] Although there remain several unmet requirements before such a process can be realized[2a,3], the most fundamental concern has been whether α-HL, or for that matter any other natural or man-made nanopore structure[4], is capable of recognizing DNA with single nucleobase resolution. In this study, we have probed the nucleobase resolution capacity of α-HL by threading and holding stably a given strand of ss-DNA inside the α-HL pore in the form of a single-species α-HL·DNA pseudorotaxane.[5] By using block copolymers of DNA and homopolymeric strands with position-specific single-nucleotide substitutions, we have found that a single adenine nucleotide at a specific location on a strand of poly d(C) can be distinguished by its characteristic effect in reducing the α-HL ion conductance. The discovery that α-HL can recognize ss-DNA with single nucleobase resolution strengthens the case for its utility in rapid single-molecule DNA sequencing.
It has been proposed that it might be possible to rapidly establish the sequence of an individual strand of ss-DNA as it traverses through an α-HL pore by recording the distinct ion channel conductance perturbations caused by the sequential passage of each nucleotide.[2,6] Toward this goal, in an important early study, it was shown that the transient reductions in ion channel conductance could be used to distinguish homopurine from homopyrimidine ss-nucleic acid strands.[2b,c] However, in part because of the experimental limitations imposed by the rapid rates of pore-mediated ss-nucleic acid transport, it has not yet been possible to use transient DNA transport measurements to establish the nucleobase resolution capacity of α-HL.[2,3] We envisioned that this issue might be resolved by analyzing ion channel conductance reductions caused by a given strand of ss-DNA while it is captured and held stably inside an α-HL pore in the form of a rotaxane or pseudorotaxane.[5]
The DNA sequences employed in our studies each have a long ss-DNA segment for threading the α-HL pore and a stable terminal hairpin structure for holding the thread in a pseudorotaxane configuration (Figure 1a).[5b] Because the hairpin duplex structure is wider than the α-HL internal lumen, a given DNA strand can only thread the pore with its free single-stranded terminus.[5] Therefore, depending on from which end an α-HL pore is threaded and whether the hairpin structure is placed on the 3’ or 5’ terminus of the DNA thread, four distinct topoisomeric forms of α-HL·DNA pseudorotaxanes can be formed.[5b] In this study we report on the characteristics of the topoisomers of α-HL·DNA pseudorotaxanes that are prepared by threading DNA from the cis side (Figure 1a). In a typical experiment, the DNA sample is introduced at the cis side of a lipid bilayer containing a single oriented α-HL pore and the threading process is initiated by applying positive transmembrane holding potentials (see supporting information for details). Formation of the α-HL·DNA pseudorotaxane is signified by the reduction in ion channel conductance caused by the presence of the ss-DNA inside the pore (Figure 1b).[2,5] The DNA strand can be held stably in the pseudorotaxane configuration as long as suitable positive transmembrane holding potential is maintained (see supporting information).[5b]
Figure 1.
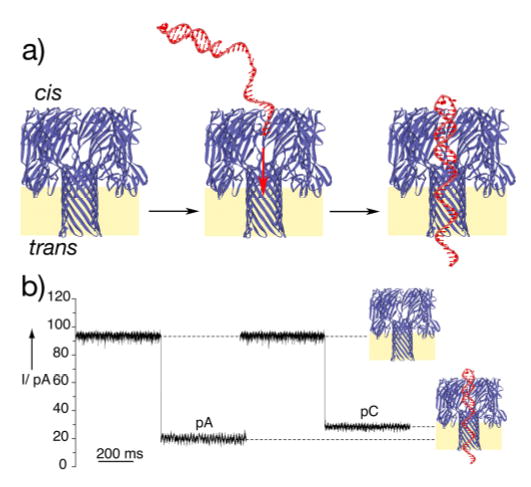
a) Schematic representation of the single-species α– HL·DNA pseudorotaxane formation. By employing sequences with a long ss-DNA segment having a hairpin structure at one end, the SS-DNA can thread the pore under positive applied potentials and held stably in a pseudorotaxane configuration by the interactions of the DNA duplex segment at the pore entrance. The heptameric transmembrane pore structure is depicted in a cutaway side view. b) Ion conductance blockades caused by capture of poly d(A) and poly d(C) single strands (sequences 1 and 2, respectively). The typical events shown are from ion conductance traces recorded at +170 mV when the thread molecules (1 μM) were added to the cis chamber. Traces were recorded under symmetrical conditions (500 mM KCl, 5 mM MOPS pH 7.5), Bessel filtered at 5 kHz, and sampled at 200 μs.
Initial experiments were performed with α-HL·DNA pseudorotaxanes of Poly d(A) and Poly d(C) (sequences 1-4) to ascertain whether homopurine and homopyrimidine DNA strands held inside the α-HL pore can give rise to distinct ion conductance blockades. Current versus voltage curves, measured from +10 to +170 mV, indicated that the homopurine and homopyrimidine pseudorotaxanes could be readily distinguished at applied potentials of greater than 100 mV (Figure S1). The residual current IR at +170 mV (calculated as a percentage of the DNA free α-HL current measured during the same experiment) is 22±3% and 31±3% for poly d(A) (sequence 1) and poly d(C) (sequence 2) pseudorotaxanes, respectively (Figure 2). Interestingly, topoisomeric α-HL·DNA pseudorotaxanes of poly d(A) and poly d(C) that differ only in the orientation of the captured DNA strand (threading from the 3’-terminus using 1 or 3 versus threading from the 5’-terminus using 2 or 4) gave similar characteristic “A-type” or “C-type” ion channel blockades, respectively (Figure 2).
Figure 2.
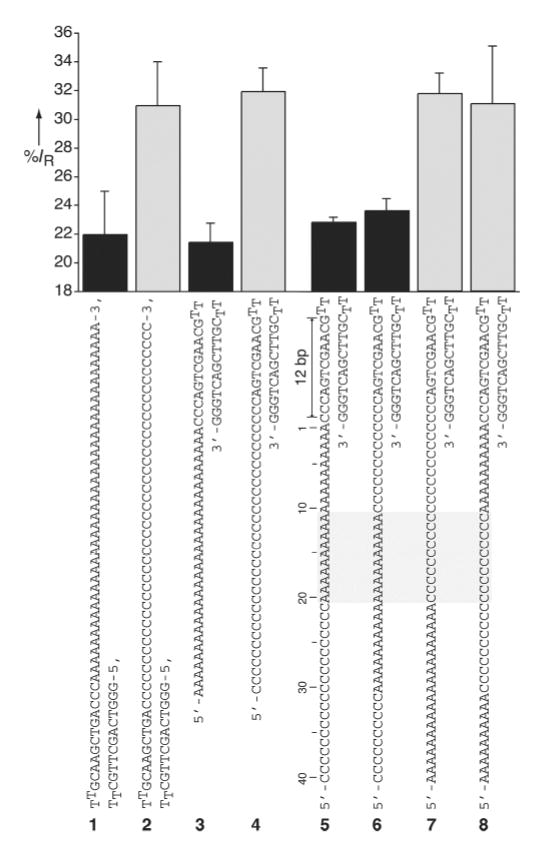
The averaged residual currents for various α-HL·DNA pseudorotaxanes measured at +170 mV. The left panel depicts the percent residual currents measured for poly d(A) and poly d(C) threads in two topoisomeric α-HL·DNA pseudorotaxane configurations. Note that when the α-HL pore is threaded with a homopolymeric DNA segment either from the free 3’-terminus (1 or 2) or from 5’-terminus (3 or 4), similar characteristic A-type or C-type ion channel blockades are observed. The right panel depicts the observed residual currents for α-HL·DNA pseudorotaxanes prepared using four different ss-DNA block copolymer threads (5–6). The region of the ss-DNA segment recognized by the α-HL channel (10 to 20 nucleotides away from the edge of the hairpin segment) is marked by the gray rectangle. For clarity A-type and C-type residual currents are shown as black and gray bars, respectively.
Given that in an α-HL·DNA pseudorotaxane a significant portion of the ss-DNA is held stably inside the protein’s pore,[5] we sought to determine what part of the encased DNA thread could factor in the production of the observed ion conductance differences between poly d(A) and poly d(C) oligonucleotides. Accordingly, we prepared single species α-HL·DNA pseudorotaxanes using four different DNA block copolymers (sequences 5-8) each designed to place stretches of poly d(A) and poly d(C) at discrete locations along the DNA thread (Figure 2). The measured residual currents at +170 mV for each of the DNA block copolymer/α-HL pseudorotaxanes fall clearly into either “A-type” or “C-type” signal categories (Figure 2). Therefore, by a simple comparative sequence analysis, it can be concluded that the nucleobase(s) recognized by α-HL must be located in the region 10 to 20 nucleotides away from the edge of the hairpin structure (Figure 2). Additional studies with similar DNA block copolymer threads, but having shorter poly d(C) and poly d(A) segments, further refined the recognition site to around nucleotide 20 (data not shown).
To address whether the α-HL pore structure is capable of recognizing DNA with single nucleobase resolution, we designed five poly d(C) threads (sequences 9-13, Figure 3), each having a single deoxyadenosine at a unique position within the putative recognition site located 18 to 22 nucleotides away from the edge of the 5’-stem-loop structure. Each DNA strand was used to prepare the corresponding single species α-HL·DNA pseudorotaxanes. Furthermore, each of the pseudorotaxanes was analyzed several times to obtain statistically significant numbers of events for calculating the residual current (IR at +170 mV, Figure S2). The measured IR distribution was roughly bimodal and could be classified into two groups corresponding to the A-type (IR < 27%) and the C-type (IR > 27%) events (Figure 3). A given event had current characteristics similar to either poly d(A) or poly d(C) pseudorotaxanes (Figure 1b) and did not interconvert from one type to the other during the analysis period. The data clearly show that when deoxyadenosine is at positions 18, 19, 21, or 22 (sequences 9, 10, 12, 13) the majority of the ion conductance blockades correspond to the C-type signal. However, when a single deoxyadenosine is positioned 20 nucleotides away from the edge of the stem-loop segment (sequence 11) in spite of being flanked on either side by long stretches of poly d(C), most observed events corresponded to the A-type signal (Figure 3). Therefore, these studies demonstrate that the α-HL pore structure can recognize ss-DNA site-specifically with a single nucleobase resolution.
Figure 3.
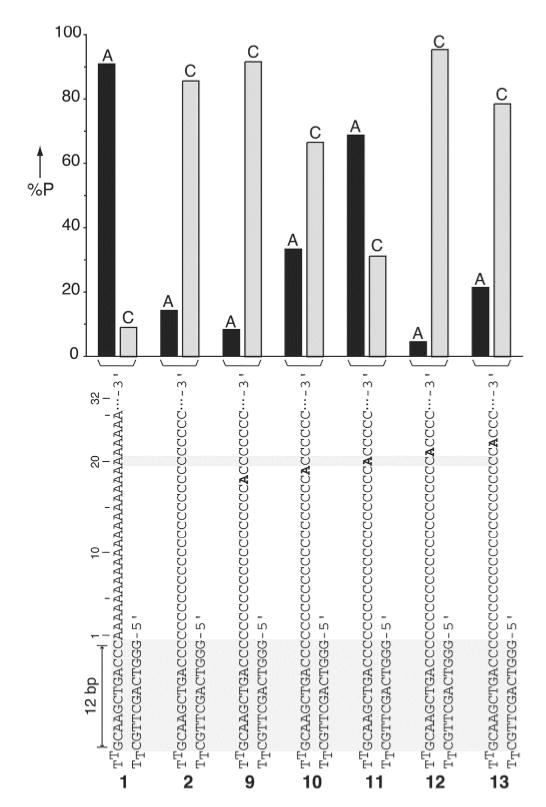
Percent probability of the A-type (IR < 27%) and the C-type (IR > 27%) residual currents for single deoxyadenosine-substituted poly d(C) DNA strands captured inside the α-HL pore as a pseudorotaxane. Percent probability was calculated by the number of A-type or C-type events measured at +170 mV divided by the total number of events (n) recorded for a given strand (n = 11, 21, 22, 32, 22, and 14 for strands 1, 2, 9, 10, 11, 12, 13, respectively). The percent probability of A-type and C-type events for each strand are given as black and gray bars respectively.
The above observations imply that in the α-HL·DNA pseudorotaxanes the ss-DNA thread must be held in a specific or closely related conformations inside the pore to allow correct positioning of the recognized nucleotide at the putative α-HL sensing element. Therefore, by estimating how the threaded DNA hairpin is pinned to the protein and the conformation of the ss-DNA inside the pore barrel it should be possible to propose where on the α-HL pore the nucleobase sensing site might be located. Considering that DNA hairpins with stems longer than 8-bp can enter the vestibule and bump up against the internal pore constriction,[8a] the 12-bp stem hairpin strands used in our studies are most likely pinned to the α-HL pore constriction by the edge of their stem duplex structure (Figure 1a). This view is also consistent with a study in which covalently tethered DNA molecules were used to probe the internal α-HL pore structure and dimensions.[9] Therefore, in such a configuration the position of the recognized nucleotide (counting from the edge of the stem structure) should be independent of the number of stem base pairs (if > 8-bp). Consistent with the above hypothesis, α-HL·DNA pseudorotaxanes prepared using hairpin DNA strands having 13-bp (strands 14 and 15) or 10-bp (strands 16 to 18) stem segments retained their ability to report on a single deoxyadenosine placed 20 nucleotides away from the edge of the hairpin (Figures 4, S3). Furthermore, we speculate that the estimated length of the single DNA strand within the barrel is somewhat shorter than a DNA strand with the same number of bases within B-form dsDNA. ssDNA, by comparison with dsDNA, is highly contractile and at forces below ~8 pN would be shortened compared to B-DNA.[10] Such small forces seem reasonable taking into account that the effective charge per nucleotide within the α-HL pore is only ~0.1e as suggested by recent studies.[11] Therefore, the above rationale and observations suggest that the α-HL recognition site is most likely located near its trans opening (Figure 1a).[1]
Figure 4.
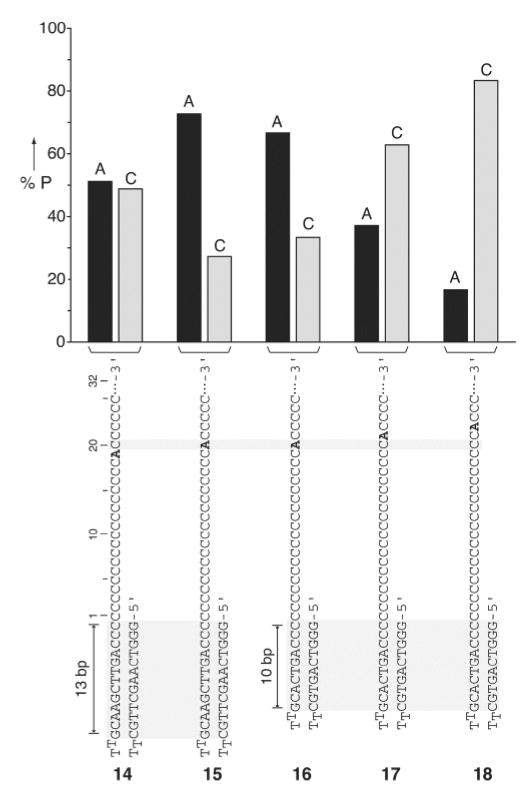
Percent probability of the A-type and the C-type events for single deoxyadenosine-substituted poly d(C) DNA hairpins sequences having either 13- or 10-basepair stem duplexes (strands 14–15, and 16–18, respectively) measured at +170 mV (n = 43, 44, 69, 35, 66 for 14–18, respectively). For clarity the A-type (IR < 27%) and C-type (IR > 27%) events are shown as black and grey bars, respectively.
The precision of the single nucleobase recognition is an important factor in single-molecule DNA sequencing. Careful analysis of the data describe above indicate that the precision of nucleotide sensing is not sufficiently high in the “native” α-HL pore for use in DNA sequencing. For example, while the observed residual currents indicate that an adenine substitution at either positions 18, 21, and 22 (sequences 9, 12, 13) is not recognized (Figure 3), the discriminating power between A and C at position 19 (using sequence 10) is noticeably less pronounced (Figures 3, S2). There are a number of factors that might influence the precision of the single nucleobase recognition. These include the conformational states and structural dynamics of a single strand of DNA held inside the pore structure under the influence of an electric force[9,12], and potential sequence-dependant DNA structural features that could effect the precise alignment of the recognized nucleobase with respect to the α-HL recognition. For instance, our data indicate that variations in the hairpin structure seem to somewhat shift the alignment of the captured DNA strand with respect to the α-HL sensing site either slightly forward (strands 16 and 17) or backward (strand 14) as compared to strand 11 (Figures 2, 3). These apparently small changes in relative strand positioning could be responsible for decreased discrimination between A and C in the case of strand 14 and diminished selectivity for the C-type events in strand 17. We note that issues concerning the precision of recognition arising from the base-dependent length of a ssDNA in an electric field would be less serious for DNA sequenced during transit through the pore, where the time-evolution of the conductance would be monitored, or for stepwise approaches where the DNA is fed into the pore by an enzyme or a nanoscopic device.[3,6] In these cases, the computer analyzing the signal would also be trained in base recognition within the context of neighboring sequences. Approaches based on current amplitude could also have advantages over a method based on the mean dwell time of each base at a recognition site.[3,12] Even if the mean dwell time for each of the four bases differed by an order of magnitude from the others, there would be a problematical overlap between the four dwell-time distributions.
In conclusion, our studies seem to support the notion that α-HL-mediated DNA sequencing might be fundamentally feasible. We believe that progress toward that goal would greatly benefit from targeted engineering of pores[7] which could provide the means for improved positional selectivity and signal discrimination between the four DNA nucleobases. Furthermore, the single-species α-HL·DNA rotaxane strategies described recently[5b,c] might also be of use in enhancing the fidelity of single nucleobase recognition via multiple-pass reading.
Supplementary Material
Footnotes
This work was supported by a grant from the Office of Naval Research (Grant no. MURI-99, N000149910717). We thank W. S. Horne for his generous assistance in molecular modeling.
References
- 1.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Science. 1996;274:1859. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 2.a) Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Proc Natl Acad Sci USA. 1996;93:13770. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Biophys J. 1999;77:3227. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Proc Natl Acad Sci USA. 2000;97:1079. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Howorka S, Cheley S, Bayley H. Nat Biotechnol. 2001;19:636. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 3.a) Bayley H, Martin CR. Chem Rev. 2000;100:2575. doi: 10.1021/cr980099g. [DOI] [PubMed] [Google Scholar]; b) Deamer DW, Akeson M. TIBTECH. 2000;18:147. doi: 10.1016/s0167-7799(00)01426-8. [DOI] [PubMed] [Google Scholar]; c) Deamer DW, Branton D. Acc Chem Res. 2002;35:817–825. doi: 10.1021/ar000138m. [DOI] [PubMed] [Google Scholar]; (d) Nakane JJ, Akeson M, Marziali A. J Phys: Condens Matter. 2003;15:R1365. [Google Scholar]
- 4.a) Jirage KB, Hulteen JC, Martin CR. Science. 1997;278:655. [Google Scholar]; b) Schmidt C, Mayer M, Vogel H. Angew Chem. 2000;112:3267. doi: 10.1002/1521-3773(20000901)39:17<3137::aid-anie3137>3.0.co;2-d. Angew. Chem. Int. Ed. 2000, 39, 3137. [DOI] [PubMed] [Google Scholar]; c) Li J, Stein D, McMullan C, Branton D, Aziz MJ, Golovchenko JA. Nature. 2001;412:166. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]; d) Bayley H, Cremer PS. Nature. 2001;413:226. doi: 10.1038/35093038. [DOI] [PubMed] [Google Scholar]; e) Chen P, Mitsui T, Farmer DB, Golovchenko J, Gordon RG, Branton D. Nano Lett. 2004;4:1333. doi: 10.1021/nl0494001. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Chang H, Kosari F, Andreadakis G, Alam MA, Vasmatzis G, Bashir R. Nano Lett. 2004;4:1551. [Google Scholar]
- 5.a) Sauer-Budge AF, Nyamwanda JA, Lubensky DK, Branton D. Phys Rev Lett. 2003;90:238101. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]; b) Sanchez-Quesada J, Saghatelian A, Cheley S, Bayley H, Ghadiri MR. Angew Chem Int Ed. 2004;43:3063. doi: 10.1002/anie.200453907. Angew. Chem. 2004, 116, 3125. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Nakane J, Wiggin M, Marziali A. Biophys J. 2004;87:615. doi: 10.1529/biophysj.104.040212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Church G, Deamer DW, Branton D, Baldarelli R, Kasianowicz J. 5,795,782. U.S. Patent. 1995 1998
- 7.a) Cheley S, Braha O, Lu X, Conlan S, Bayley H. Protein Sci. 1999;8:1257. doi: 10.1110/ps.8.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kasianowicz JJ, Burden DL, Han LC, Cheley S, Bayley H. Biophys J. 1999;76:837. doi: 10.1016/S0006-3495(99)77247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Gu L-Q, Dalla Serra M, Vincent JB, Vigh G, Cheley S, Braha O, Bayley H. Proc Natl Acad Sci U S A. 2000;97:3959. doi: 10.1073/pnas.97.8.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Gu LQ, Cheley S, Bayley H. Science. 2001;291:636. doi: 10.1126/science.291.5504.636. [DOI] [PubMed] [Google Scholar]; e) Gu LQ, Cheley S, Bayley H. J Gen Physiol. 2001;118:481. doi: 10.1085/jgp.118.5.481. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Cheley S, Gu LQ, Bayley H. Chem Biol. 2002;9:829. doi: 10.1016/s1074-5521(02)00172-2. [DOI] [PubMed] [Google Scholar]
- 8.a) Vercoutere W, Winters-Hilt S, Olsen H, Deamer D, Haussler D, Akeson M. Nat Biotechnol. 2001;19:248. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]; b) Winters-Hilt S, Vercoutere W, DeGuzman VS, Deamer D, Akeson M, Haussler D. Biophys J. 2003;84:967. doi: 10.1016/S0006-3495(03)74913-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Vercoutere WA, Winters-Hilt S, DeGuzman VS, Deamer D, Ridino SE, Rodgers JT, Olsen HE, Marziali A, Akeson M. Nucleic Acids Res. 2003;31:1311. doi: 10.1093/nar/gkg218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howorka S, Bayley H. Biophys J. 2002;83:3202. doi: 10.1016/S0006-3495(02)75322-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Smith SB, Cui Y, Bustamante C. Science. 1996;271:795. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]; b) Bustamante C, Smith SB, Liphardt J, Smith D. Curr Op Struct Biol. 2000;10:279–285. doi: 10.1016/s0959-440x(00)00085-3. [DOI] [PubMed] [Google Scholar]
- 11.a) Sauer-Budge AF, Nyamwanda JA, Lubensky DK, Branton D. Phys Rev Lett. 2003;90:238101-1–238101-4. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]; b) Mathé J, Visram H, Viasnoff V, Rabin Y, Meller A. Biophys J. doi: 10.1529/biophysj.104.047274. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Lubensky DK, Nelson DR. Biophys J. 1999;77:1824. doi: 10.1016/S0006-3495(99)77027-X. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bates M, Burns M, Meller A. Biophys J. 2003;84:2366. doi: 10.1016/S0006-3495(03)75042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


