Abstract
The reductive part of the catalytic cycle of cytochrome c oxidase from Paracoccus denitrificans was examined by using time-resolved potential measurements on black lipid membranes. Proteoliposomes were adsorbed to the black lipid membranes and RuII(2,2′-bipyridyl)32+ was used as photoreductant to measure flash-induced membrane potential generation. Single-electron reduction of the oxidized wild-type cytochrome c oxidase reveals two phases of membrane potential generation (τ1 ≈ 20 μs and τ2 ≈ 175 μs) at pH 7.4. The fast phase is not sensitive to cyanide and is assigned to electron transfer from CuA to heme a. The slower phase is inhibited completely by cyanide and shows a kinetic deuterium isotope effect by a factor of 2–3. Although two enzyme variants mutated in the so-called D pathway of proton transfer (D124N and E278Q) show the same time constants and relative amplitudes as the wild-type enzyme, in the K pathway variant K354M, τ2 is increased to 900 μs. This result suggests uptake of a proton through the K pathway during the transition from the oxidized to the one-electron reduced state. After the second laser flash under anaerobic conditions, a third electrogenic phase with a time constant of ≈1 ms appears. The amplitude of this phase grows with increasing flash number. We explain this growth by injection of a second electron into the single-electron reduced enzyme. On multiple flashes, both D pathway mutants behave differently compared with the wild type and two additional slow phases of τ3 ≈ 2 ms and τ4 ≈ 15 ms are observed. These results suggest that the D pathway is involved in proton transfer coupled to the uptake of the second electron.
Cytochrome c oxidase (COX) catalyzes electron transfer from ferrocytochrome c to molecular oxygen and reduces the latter to water (for recent reviews, see refs. 1–4). Electrons from cytochrome c are first accepted by a binuclear CuA center, which is located at the outer surface of the mitochondrial or bacterial membrane. From CuA, the electrons are transferred to heme a, then to heme a3 and CuB, which form the binuclear site. The hemes and CuB are buried within the transmembrane part of the enzyme. Dioxygen is bound to the heme a3 iron atom of the doubly reduced binuclear site.
During each turnover, four “chemical” protons are needed for water formation. Additionally, four protons are translocated (“pumped”) across the membrane (5). The determination of the crystal structure of COX from the soil bacterium Paracoccus denitrificans (6, 7) revealed two proton transfer pathways to the active site where oxygen is converted to water. The so-called K pathway leads directly into the binuclear center to a (putative) OH− bound to CuB involving the highly conserved residues K354, T351, Y280 (P. denitrificans numbering is used throughout the paper), and the hydroxyl group of the heme a3 hydroxyethylfarnesyl side chain. The D pathway proceeds from D124 to E278 via a number of conserved polar residues (N199, N113, N131, Y35, S134, and S193) and possibly bound water molecules.
It has been shown that COX functions as a proton pump (8). However, there is still a controversial discussion concerning the question as to which steps in the catalytic cycle are coupled to proton translocation and which pathway is used for the uptake of the different protons at the various stages of the catalytic cycle. Until recently, it was generally accepted that two protons are pumped both during the transition from the P (peroxy) to the F (oxoferryl) state and between the F and O (oxidized) intermediates (9). This view has been challenged recently (10). Three of the underlying assumptions (input parameters) for deriving this stoichiometry are most likely incorrect (11). A reanalysis of the data pertaining to the coupling of individual electron transfer steps to proton pumping suggests that the F → O transition is coupled to pumping of only one proton. Then it is likely that one proton is already pumped during the double reduction of O (10, 11).
A controversially discussed issue is the relative location of the heme groups with respect to the membrane dielectric. The x-ray crystallographic structure determination has shown that both heme groups are located at the same distance from the membrane surface but considerably closer to the outer than to the inner side. Analysis of the data for the mitochondrial COX suggests a relative dielectric location of the heme groups of 33–50% from the cytosolic surface (11).
Recent experiments (12) have been used to reinterpret data from a previous publication (9). Starting from a fully reduced, four-electron-containing enzyme, it has been found that the electric field generated during oxidation is lower than that generated during a subsequent rereduction. From this finding, it has been concluded that only two protons are pumped during the oxidative phase, and another two during rereduction. It is claimed that the latter proton pumping during the reductive phase is energetically linked to the oxidative phase, and pumping of two protons is triggered only by the first rereducing electron. For the interpretation of the data, a relative location of the heme groups with respect to the membrane dielectric of 32% has been used. This interpretation of the experiment from ref. 12 has been challenged recently (13).
Another point still under debate is the role of the proton transfer pathways. The “histidine cycle” model (14), proposed to explain pumping of two protons per P → F and F → O transition each (9), emphasizes strictly separate pathways for delivery of chemical and pumped protons from the inner side. In agreement with mutagenesis data (15, 16), which have shown that mutation of the D pathway residue D124 to Asn leads to a slower turnover and to a complete loss of proton pumping, whereas mutations of the K pathway residues K354 and T351 result in an inactive enzyme, Iwata et al. (6) associated the K pathway with uptake of substrate protons and the D pathway with pumped protons. Later, it was proposed that the K pathway is required for the initial double reduction of COX in the catalytic cycle (eu-oxidase phase) whereas the peroxidase half-reaction is serviced by the D pathway (17).
In this paper, we describe the analysis of electrogenic steps associated with the transfer of the first two electrons and examine the effects of three mutations (K354 M, D124N, and E278Q) in COX of Paracoccus denitrificans. The results suggest that the K and D pathways are both required during the first part of the catalytic cycle, i.e., the O → R transition.
Materials and Methods
Materials.
Diphytanoyl lecithin and octadecylamine for black lipid membrane (BLM) preparation were purchased from Avanti Biochemicals (Alabaster, AL) and Riedel-de-Haen (Seelze, Germany). Liposomes were formed from Escherichia coli lipids (acetone/ether preparation; Avanti Biochemicals). [RuII(2,2′-bipyridyl)3]2+ (Rubpy) was obtained from Aldrich. Other chemicals were from Sigma and Fluka.
Site-Directed Mutants.
Mutagenesis was performed as described (18). Bacterial membranes were isolated (19), and COX was purified by streptavidin affinity chromatography (20).
Electrometric Measurements.
COX was reconstituted into E. coli lipid liposomes at a lipid/protein ratio of 20 (wt/wt) as described by Gropp et al. (21) by using 2% (wt/vol) sodium cholate as detergent in 50 mM Hepes-KOH (pH 7.4). BLM experiments were made as described (22) with a time resolution of about 1 μs. Rubpy (80 μM) was used as light-inducible electron donor (23, 24). A short laser pulse (wavelength 450 nm; duration ≈ 10 ns) converts the ruthenium complex to the excited state, which is able to transfer an electron to COX. To prevent electron backflow to the generated Ru(III)bpy, 300 μM EDTA acts as a sacrificial electron donor. All measurements were performed under anaerobic conditions. The Faraday cage containing the BLM cell and all solutions were degassed and flushed with argon. Both compartments of the thermostated (25°C) Teflon cell were filled with 1.3 ml 50 mM Hepes-KOH (pH 7.4), and 150 mM β-d-glucose, 40 μg/ml glucose oxidase (Fluka), and 25 μg/ml catalase (Sigma) were added to remove residual oxygen.
For the D2O experiment, the liposomes and the buffers were prepared in D2O. The pD of the buffer was estimated by adding 0.4 to the pH meter reading (25).
Results and Discussion
Single-Electron Reduction (O → E Transition).
Wild-type (WT) oxidase.
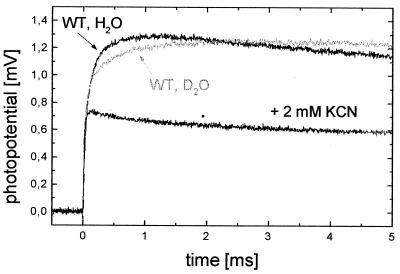
Fig. 1 shows the electrogenic response linked to single-electron photochemical reduction of the WT enzyme by Rubpy. After the flash, a rapid negative transient is observed, which is also present in the absence of COX and is an artifact of Rubpy photolysis in the vicinity of the BLM. Then, an electric potential is generated across the BLM corresponding to translocation of positive charges from the intracellular to the extracellular side of COX and/or negative charges in the opposite direction. Subsequently, the potential decays with a time constant of ≈250 ms caused by the passive discharge of the liposomes.
Figure 1.
Membrane potential generation coupled to single-electron photoreduction of liposome-reconstituted WT COX starting from the oxidized state. The anaerobic buffer contains 50 mM Hepes-KOH (pH 7.4), 150 mM β-d-glucose, 40 μg/ml glucose oxidase, 25 μg/ml catalase, 80 μM Rubpy, 300 μM EDTA, and 200 μM ferricyanide (T = 25°C). After recording a trace (WT, H2O), potassium cyanide, was added, and a second trace (+2 mM KCN) was recorded. The gray curve (WT, D2O) shows the effect of solvent exchange from H2O to D2O on the slow phase monitored at pH-meter reading 7.4, which means that the pD value is 0.4 units higher.
In the rising part of the signal, two phases (τ1 ≈ 20 μs and τ2 ≈ 175 μs) can be observed. The fast phase (τ1) cannot be inhibited by KCN, and in agreement with Konstantinov et al. (17), who found a 15-μs phase for the reduction of heme a by CuA during the F → O transition in Rhodobacter sphaeroides, this phase is assigned to electron transfer from CuA to heme a. The time constant of the slow phase (τ2; gray curve in Fig. 1) becomes larger in D2O (τ2 ≈ 350 μs). Slowing down by a factor of 2 to 3 is in the typical range of a kinetic deuterium isotope effect (25). It suggests that this phase is linked to proton transfer. Addition of 2 mM KCN completely inhibits the slower reaction in H2O as well as in D2O. Therefore, this phase is probably linked to processes at the binuclear center without necessarily involving its reduction (see below). Isotope effect, amplitudes (see Table 1), sign, and KCN sensitivity of the slow phase (τ2) of the signal support the concept that a proton is taken up from the intracellular side of COX during this phase. This notion is supported by the results obtained with the mutant enzymes (see below).
Table 1.
Time constants and relative amplitudes of membrane potential generation on photoreduction of P. denitrificans COX (wild type and variants)
| Reaction | WT, H2O | WT, D2O | D124N | E278Q | K354M |
|---|---|---|---|---|---|
| O → E | τ1 = 21.7 ± 5.5 μs | τ1 = 20.6 ± 4.6 μs | τ1 = 19.7 ± 2.9 μs | τ1 = 22.7 ± 6.2 μs | τ1 = 18.0 ± 3.8 μs |
| α1 = 0.57 ± 0.10 | α1 = 0.67 ± 0.15 | α1 = 0.58 ± 0.08 | α1 = 0.62 ± 0.11 | α1 = 0.79 ± 0.08 | |
| τ2 = 176 ± 50 μs | τ2 = 352 ± 71 μs | τ2 = 163 ± 32 μs | τ2 = 139 ± 29 μs | τ2 = 859 ± 170 μs | |
| α2 = 0.43 ± 0.10 | α2 = 0.33 ± 0.15 | α2 = 0.42 ± 0.08 | α2 = 0.38 ± 0.11 | α2 = 0.21 ± 0.08 | |
| O → R | τ1 = 19.6 ± 3.3 μs | τ1 = 21.8 ± 5.1 μs | τ1 = 28.7 ± 6.5 μs | τ1 = 28.8 ± 5.3 μs | τ1 = 31.9 ± 5.6 μs |
| τ2 = 154 ± 45 μs | τ2 = 304 ± 75 μs | τ2 = 170 ± 53 μs | τ2 = 168 ± 36 μs | τ2 = 897 ± 181 μs | |
| τ3 = 1.1 ± 0.3 ms | τ3 = 2.0 ± 0.6 ms | τ3 = 2.1 ± 0.5 ms | τ3 = 1.5 ± 0.6 ms | τ3 = 4.3 ± 0.7 ms | |
| τ4 = 17.6 ± 5.4 ms | τ4 = 14.9 ± 5.9 ms | ||||
| (+KCN) | τ1 = 20.9 ± 5.0 μs | τ1 = 20.2 ± 4.7 μs | τ1 = 21.0 ± 4.9 μs | τ1 = 19.6 ± 5.2 μs | τ1 = 18.7 ± 5.4 μs |
A different interpretation has been put forward recently by Siletsky et al. (26). On the basis of similar experiments that used bovine heart COX, they reported that “single electron reduction of the O form is not associated with the KCN-sensitive electrogenic proton transfer steps but only with the KCN-insensitive microsecond phase (τ1 in our notation) of heme a reduction by CuA”. The additionally observed KCN-sensitive slow phase in the millisecond time range was interpreted in terms of a proton translocation in a small fraction of COX present initially in the F or P state. According to this interpretation, the slow phase (τ2 in our notation) is only the result of reducing contaminants present in the phospholipids (27), which generate a variable steady-state fraction of F and P states. However, this interpretation can be clearly ruled out for our experiments, which were performed under anaerobic conditions (checked with an oxygen electrode), and the enzyme was converted completely to the oxidized state O by ferricyanide addition (confirmed by absorption spectroscopy; data not shown). We, therefore, believe that our results represent substantial evidence for proton uptake during the O → E transition. In a similar way, uptake of protons monitored by potential measurements has been demonstrated previously for the F → O transition (17, 24).
A remaining question is whether electron transfer from CuA to heme a or the subsequent reduction of the binuclear center leads to proton uptake. It has been suggested previously that, on single-electron reduction of COX, the electron proceeds only to heme a (23, 28). In this case, the experiments presented in our study provide evidence for proton uptake during electron transfer to heme a in agreement with the results of electrostatic calculations (A.K., C. R. D. Lancaster, and H.M., unpublished work). The inhibition of proton uptake by KCN residing in the binuclear center is not easily understood and has to involve an as-yet unknown mechanism possibly at the terminal end of the K pathway, e.g., absence of the proton-accepting group.
D pathway mutants.
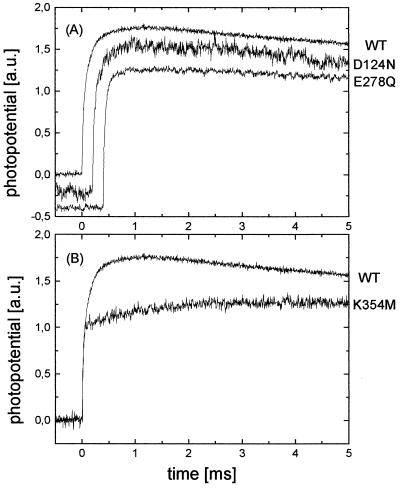
In Fig. 2A, the electrogenic processes associated with flash-induced reduction of the mutant enzymes D124N and E278Q are compared with those of the WT enzyme. Both mutants show the same time constants τ and relative amplitudes α as the WT enzyme (see Table 1). Thus, these mutations seem to have no influence on processes during the O → E transition, and the D pathway is not needed for proton transfer linked to the uptake of the first electron.
Figure 2.
Effect of mutations on the membrane potential generation during O → E transition by P. denitrificans COX. Conditions are the same as those described for Fig. 1. The traces in each panel have been normalized to the amplitude of the 20-μs phase to facilitate visual comparison of the kinetics. (A) D pathway mutants D124N and E278Q compared with WT enzyme (offset by −0.2 a.u., 0.2 ms and −0.4 a.u., 0.4 ms, respectively, for clarity). (B) K pathway mutant K354 M compared with WT enzyme.
K pathway mutant.
Fig. 2B shows the photoelectric response of the K354M mutant enzyme compared with the WT enzyme. In the case of K354M, again two phases can be observed. However, in contrast to the WT enzyme, the second phase is slowed down 5- to 6-fold (τ2 ≈ 900 μs). This result suggests that proton uptake associated with single-electron reduction of the oxidized enzyme occurs via the K pathway.
It has been proposed recently that the D pathway is better suited to the rapid uptake of protons than the K pathway, because the entrance of the D pathway is surrounded by a proton-collecting antenna (29, 30) and because “the D pathway site is rapidly protonated,” whereas “protonation of the K pathway is a long continuous process” (31). In contrast, our results support proton uptake via the K pathway during the O → E transition. This conclusion is also in agreement with other publications that stress the importance of the K pathway for the reductive part of the catalytic cycle (32, 33).
For the determination of the stoichiometry of charge translocation, it is possible to use the amplitude of the kinetically well resolved rapid phase (τ1) assigned to the CuA → heme a transition as an internal standard to calibrate other phases in the same trace (24). However, quantitation of the number of charges translocated in the cyanide-sensitive phases depends critically on the electrogenicity assigned to the CuA → heme a reaction (10). At present, the relative dielectric location of heme a (rdla), i.e., its dielectric distance from the extracellular surface of the membrane, is not known exactly. Quantitative interpretation of the results of time-resolved electrometric measurements should be treated carefully, because small changes in the rdla could have large effects on interpretation. Assuming that, coupled to the uptake of the first electron, one proton is transferred to the binuclear center from the inner side and that no electron backflow to CuA occurs on the same time scale, our data suggest a rdla of 0.6. This value would be in contrast to the value of 0.32 found recently (11, 12).
A rdla of 0.6 is additionally supported by the fact that it is able to explain the amplitudes of the electrogenic phases obtained for a different part of the catalytic cycle, namely the F → O transition in the Rhodobacter sphaeroides enzyme (ref. 17; these experiments could be reproduced in our lab with Paracoccus COX, yielding identical results; data not shown). In these experiments, three phases were observed in the electrical signal (with time constants τi and relative amplitudes αi) and assigned to the following processes: heme a reduction by one electron (τ1 = 15 μs and α1 = 0.3), uptake of one chemical proton (τ2 = 0.4 ms and α2 = 0.2), and pumping of one proton (τ3 = 1.5 ms and α3 = 0.5). The ratio of the potential amplitudes generated during uptake of a chemical proton and proton pumping yields the relative dielectric distance of heme a from the intracellular side (1-rdla) = α2/α3 = 0.4 or rdla = 0.6 as proposed above. A similar argument is valid for heme reduction and proton pumping, namely rdla = α1/α3, yielding again rdla = 0.6. (Note that here we have used the data of ref. 17. However, the interpretation we have adopted differs somewhat from that given in ref. 17.)
Double Reduction (O → R Transition).
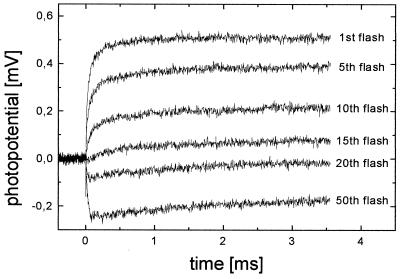
When the BLM-adsorbed COX vesicles are illuminated several times at intervals of 1 s in the absence of ferricyanide, a third phase with τ3 ≈ 1 ms appears in the electrical signal (Fig. 3). The relative amplitude of this phase increases with increasing flash number. The total amplitude of the signal, however, decreases and even eventually changes sign (see Fig. 3). In the following discussion, we propose that the additional phase is related to the uptake of the second electron and associated proton transfer processes. On the basis of this assignment, it is indeed possible to understand fully the behavior of the electrical signal under repeated illumination.
Figure 3.
Flash-induced membrane potential generation by WT COX. Anaerobic conditions as described for Fig. 1 are used, but no ferricyanide was added. The liposome-reconstituted COX is exposed to several laser pulses at a frequency of 1 Hz. The electrogenic response curves after the first and subsequent flashes are shown.
Repeated illumination opens up the possibility of double hits, i.e., some oxidase molecules receive a second electron during subsequent laser pulses. This fraction of COX molecules increases with increasing flash number, which explains the rising proportion of the additional third phase (τ3 ≈ 1 ms). After a large number of flashes, populations of two- and even three- and four-electron-reduced COX molecules build up. The fully reduced (four-electron-reduced) enzyme can no longer accept electrons, which leads to a decrease of the signal. Also, in the three-electron-reduced COX, the electrons probably partially reside on CuA. Also, these molecules contribute a smaller proportion to the signal.
Additionally, the excited ruthenium complex not only has reducing properties and the ability to transfer an electron to an oxidized protein [E(Ru(III)bpy/Ru(II)*bpy) = −0.86 V], but also can oxidize a reduced protein [E(Ru(II)*bpy/Ru(I)bpy) = 0.84 V] (34). In the three- and four-electron-reduced COX, the CuA center is partially and fully reduced, respectively. Therefore, the excited ruthenium complex can take up an electron from CuA, which subsequently leads to a backflow of electrons from heme a and the binuclear center to CuA, with similar kinetic properties as during reduction but with a negative amplitude. This feature explains the negative signal that is finally observed after many light flashes. The formed Ru(I)bpy is able to rereduce the enzyme in the dark [E(Ru(II)bpy/Ru(I)bpy) = −1.28 V] (34).
Indeed, dithionite-reduced COX shows the same electron backflow when illuminated in the presence of Rubpy (data not shown). This signal is not the result of light-induced structural changes in COX, because without Rubpy, no electrogenic events can be observed with dithionite-reduced enzyme after a laser pulse. Dithionite, together with Rubpy but without COX, does not show an electrical response on illumination either.
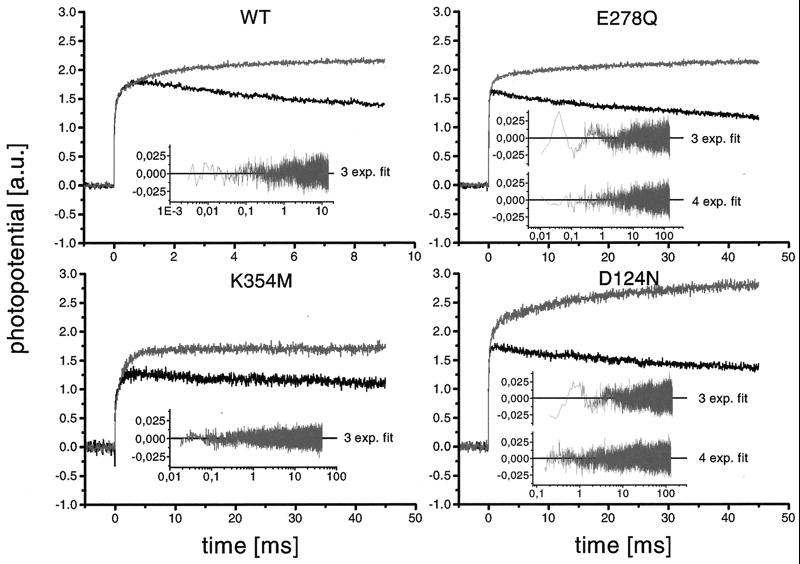
Fig. 4 compares the electrical signals after the first laser flash (O → E transition; black signals) to those after the fifth flash (O → R transition; gray signals). The results of WT, K pathway, and D pathway mutants are shown. Note that the traces in each panel have been normalized to the amplitude of the rapid phase (τ1 ≈ 20 μs) of the first flash to aid visual comparison of the kinetics.
Figure 4.
Effect of mutations on the membrane potential generation during O → R transition by P. denitrificans COX. Conditions are the same as those described for Fig. 3. In each panel, the photoelectric response after the fifth laser pulse (O → R; gray traces) is compared with that after the first flash (O → E; black traces). The curves have been normalized to the amplitude of the 20-μs phase for clarity. To illustrate the quality of the fitting of the gray curves, the Insets give the plots of the residuals.
In all four cases, the first two time constants (τ1 and τ2) remain the same after several flashes, but additional slow phases appear (see Table 1). In the case of the WT enzyme, a third phase of τ3 ≈ 1 ms is obtained; the K354 M mutant yields an additional time constant of τ3 ≈ 4 ms. The signals of the D pathway mutants E278Q and D124N cannot be fitted properly with three exponential functions, and the best fit yields four phases (see Fig. 4 Insets and Table 1) with two additional time constants of τ3 ≈ 2 ms and τ4 ≈ 15 ms in both cases. Addition of ferricyanide to the WT and mutant enzymes leads to a loss of the slower phases τ3 and τ4, again confirming that these phases result from the reduction of the one-electron-reduced state. Thus, our results imply that both K and D pathways are involved in proton uptake associated with the transfer of the second electron. However, because the redox state after multiple flashes is not known exactly, it could not be ruled out that the fourth phase (τ4 = 15 ms) observed for the D pathway mutants results from proton uptake coupled to the transfer of the third electron. To clarify this possibility, single-electron reduction measurements starting from a defined one-electron-reduced state will be necessary.
Conclusions
On single-electron reduction of the oxidized state of Paracoccus denitrificans COX, an electron is transferred from CuA to heme a with a time constant of ≈20 μs (pH 7.4; T = 25°C). This electron transport is probably coupled to the uptake of a proton via the K pathway with a time constant of ≈175 μs. The results of the multiflash experiments suggest that the D pathway may be involved in proton uptake coupled to the transfer of the second electron.
Acknowledgments
We are grateful to Hannelore Müller for excellent technical assistance and Ute Pfitzner for initially supplying the COX enzyme. This work was supported by Deutsche Forschungsgemeinschaft Grant SFB 472.
Abbreviations
- BLM
black lipid membrane
- COX
cytochrome oxidase
- Rubpy
[RuII2,2′-bipyridyl3]2+
- WT
wild type
- rdla
relative dielectric location of heme a
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080079097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080079097
References
- 1.Michel H, Behr J, Harrenga A, Kannt A. Annu Rev Biophys Biomol Struct. 1998;27:329–356. doi: 10.1146/annurev.biophys.27.1.329. [DOI] [PubMed] [Google Scholar]
- 2.Gennis R B. Biochim Biophys Acta. 1998;1365:241–248. doi: 10.1016/s0005-2728(98)00095-4. [DOI] [PubMed] [Google Scholar]
- 3.Konstantinov A A. J Bioenerg Biomembr. 1998;30:121–130. doi: 10.1023/a:1020571930850. [DOI] [PubMed] [Google Scholar]
- 4.Einarsdottir O. Biochim Biophys Acta. 1995;2:129–147. doi: 10.1016/0005-2728(94)00196-c. [DOI] [PubMed] [Google Scholar]
- 5.Casey R P, Chappell J B, Azzi A. Biochem J. 1979;182:149–156. doi: 10.1042/bj1820149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwata S, Ostermeier C, Ludwig B, Michel H. Nature (London) 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 7.Ostermeier C, Harrenga A, Ermler U, Michel H. Proc Natl Acad Sci USA. 1997;94:10547–10553. doi: 10.1073/pnas.94.20.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wikström M. Nature (London) 1977;266:271–273. doi: 10.1038/266271a0. [DOI] [PubMed] [Google Scholar]
- 9.Wikström M. Nature (London) 1989;338:776–778. doi: 10.1038/338776a0. [DOI] [PubMed] [Google Scholar]
- 10.Michel H. Proc Natl Acad Sci USA. 1998;95:12819–12824. doi: 10.1073/pnas.95.22.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michel H. Biochemistry. 1999;38:15129–15140. doi: 10.1021/bi9910934. [DOI] [PubMed] [Google Scholar]
- 12.Verkhovsky M I, Jasaitis A, Verkhovskaya M L, Morgan J E, Wikström M. Nature (London) 1999;400:480–483. doi: 10.1038/22813. [DOI] [PubMed] [Google Scholar]
- 13.Michel H. Nature (London) 1999;402:602–603. doi: 10.1038/45133. [DOI] [PubMed] [Google Scholar]
- 14.Morgan J E, Verkhovsky M I, Wikström M. J Bioenerg Biomembr. 1994;26:599–608. doi: 10.1007/BF00831534. [DOI] [PubMed] [Google Scholar]
- 15.Thomas J W, Puustinen A, Alben J O, Gennis R B, Wikström M. Biochemistry. 1993;32:10923–10928. doi: 10.1021/bi00091a048. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Horsman J A, Puustinen A, Gennis R B, Wikström M. Biochemistry. 1995;34:4428–4433. doi: 10.1021/bi00013a035. [DOI] [PubMed] [Google Scholar]
- 17.Konstantinov A A, Siletsky S, Mitchell D, Kaulen A, Gennis R B. Proc Natl Acad Sci USA. 1997;94:9085–9090. doi: 10.1073/pnas.94.17.9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfitzner U, Odenwald A, Ostermann T, Weingard L, Ludwig B, Richter O M H. J Bioenerg Biomembr. 1998;30:89–97. doi: 10.1023/a:1020515713103. [DOI] [PubMed] [Google Scholar]
- 19.Gerhus E, Steinrücke P, Ludwig B. J Bacteriol. 1990;172:2392–2400. doi: 10.1128/jb.172.5.2392-2400.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleymann G, Ostermeier C, Ludwig B, Skerra A, Michel H. Biotechnology. 1995;13:155–160. doi: 10.1038/nbt0295-155. [DOI] [PubMed] [Google Scholar]
- 21.Gropp T, Cornelius F, Fendler K. Biochim Biophys Acta. 1998;1368:184–200. doi: 10.1016/s0005-2736(97)00162-4. [DOI] [PubMed] [Google Scholar]
- 22.Kannt A, Pfitzner U, Ruitenberg M, Hellwig P, Ludwig B, Mäntele W, Fendler K, Michel H. J Biol Chem. 1999;274:37974–37981. doi: 10.1074/jbc.274.53.37974. [DOI] [PubMed] [Google Scholar]
- 23.Nilsson T. Proc Natl Acad Sci USA. 1992;89:6497–6501. doi: 10.1073/pnas.89.14.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaslavsky D, Kaulen A D, Smirnova I A, Vygodina T, Konstantinov A A. FEBS Lett. 1993;336:389–393. doi: 10.1016/0014-5793(93)80843-j. [DOI] [PubMed] [Google Scholar]
- 25.Hallen S, Brzezinski P, Malmström B G. Biochemistry. 1994;33:1467–1472. doi: 10.1021/bi00172a024. [DOI] [PubMed] [Google Scholar]
- 26.Siletsky S, Kaulen A D, Konstantinov A A. Biochemistry. 1999;38:4853–4861. doi: 10.1021/bi982614a. [DOI] [PubMed] [Google Scholar]
- 27.Zaslavsky D L, Smirnova I A, Siletsky S A, Kaulen A D, Millet F, Konstantinov A A. FEBS Lett. 1995;359:27–30. doi: 10.1016/0014-5793(94)01443-5. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi K, Une H, Hayashi K. J Biol Chem. 1989;264:7976–7980. [PubMed] [Google Scholar]
- 29.Marantz Y, Nachliel E, Aagaard A, Brzezinski P, Gutman M. Proc Natl Acad Sci USA. 1998;95:8590–8595. doi: 10.1073/pnas.95.15.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karpefors M, Ädelroth P, Aagaard A, Sigurdson H, Svensson Ek M, Brzezinski P. Biochim Biophys Acta. 1998;1365:159–169. doi: 10.1016/s0005-2728(98)00058-9. [DOI] [PubMed] [Google Scholar]
- 31.Sacks V, Marantz Y, Aagaard A, Checover S, Nachliel E, Gutman M. Biochim Biophys Acta. 1998;1365:232–240. [Google Scholar]
- 32.Vygodina T V, Pecoraro C, Mitchell D, Gennis R, Konstantinov A A. Biochemistry. 1998;37:3053–3061. doi: 10.1021/bi971876u. [DOI] [PubMed] [Google Scholar]
- 33.Ädelroth P, Gennis R B, Brzezinski P. Biochemistry. 1998;37:2470–2476. doi: 10.1021/bi971813b. [DOI] [PubMed] [Google Scholar]
- 34.Roundhill D M. In: Modern Inorganic Chemistry. Fackler J P, editor. New York: Plenum; 1994. pp. 165–215. [Google Scholar]