Abstract
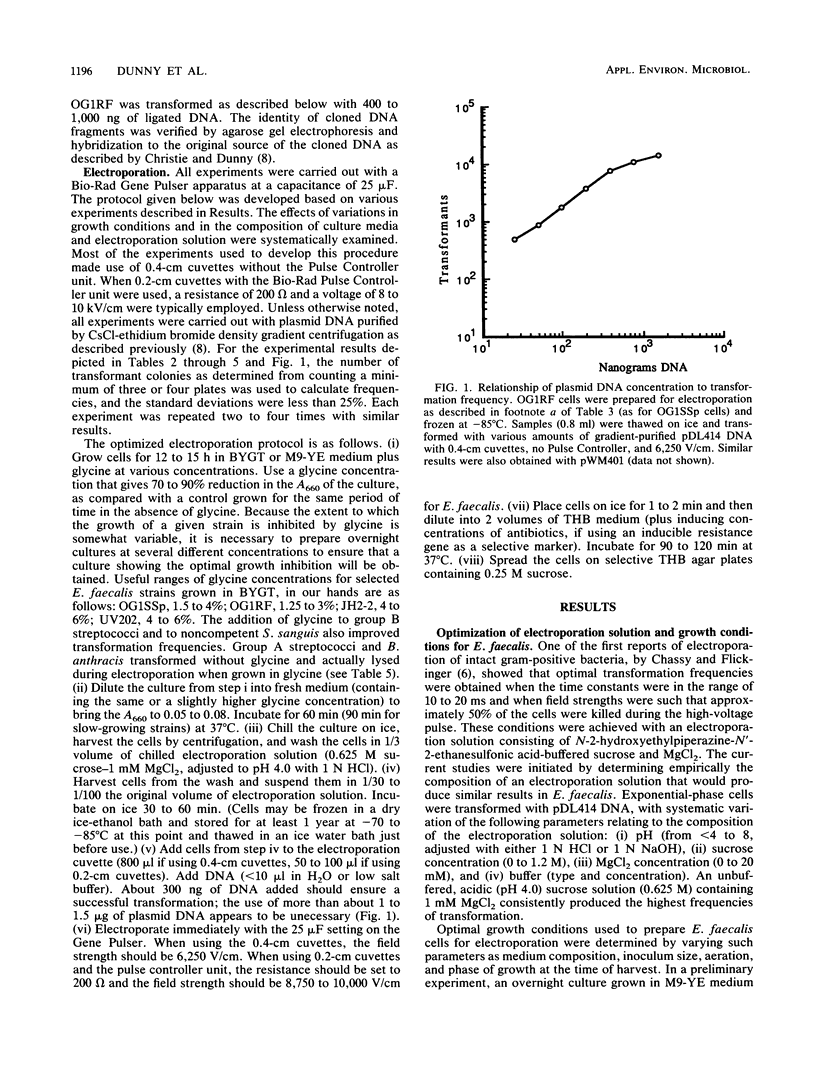
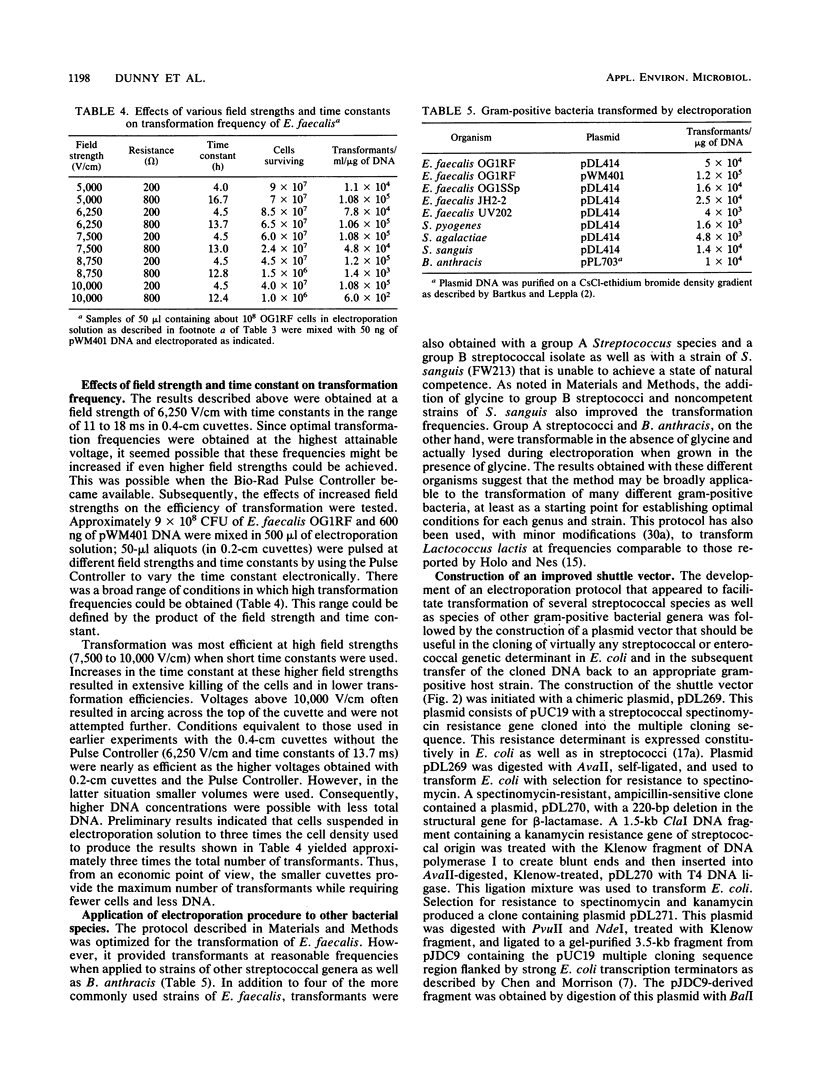
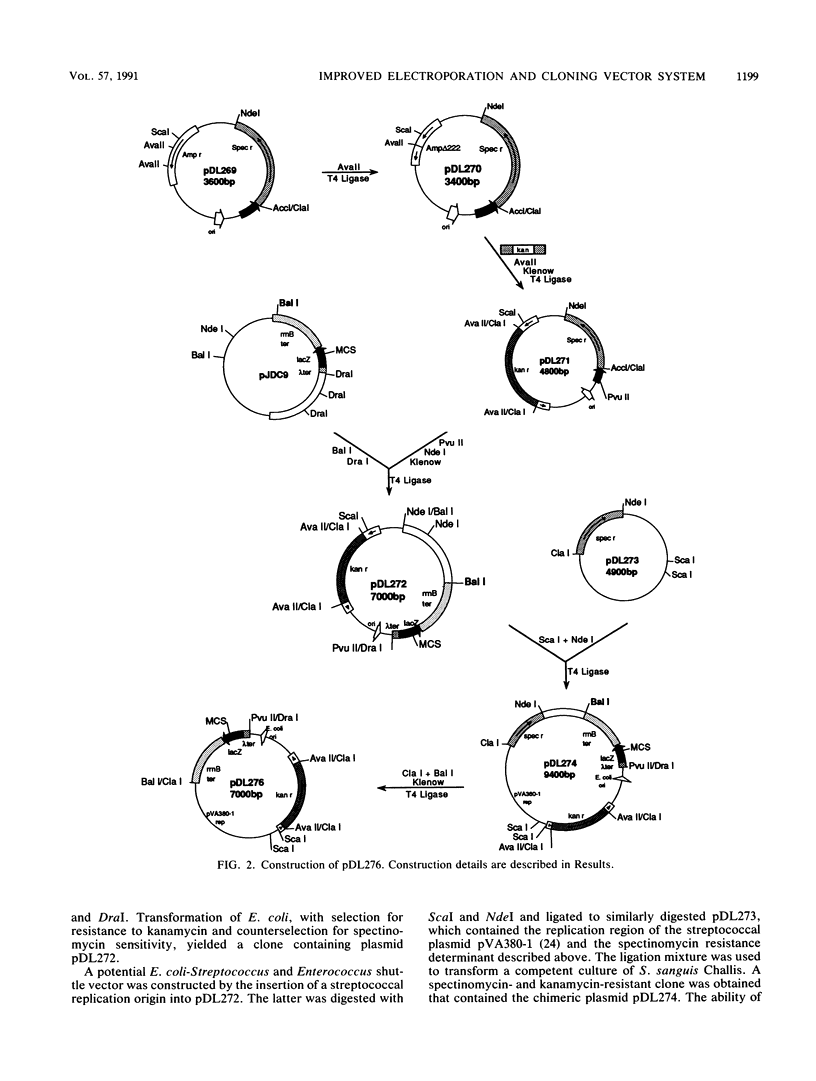
A protocol for transformation of intact Enterococcus faecalis cells by electroporation was developed through a systematic examination of the effects of changes in various parameters, including (i) growth conditions; (ii) composition of the electroporation solution; (iii) electroporation conditions, such as field strength and resistance; (iv) size, concentration, and purity of DNA used for transformation; and (v) conditions used to select for transformants. Key features of this protocol include the use of exponential-phase cells grown in inhibitory concentrations of glycine and the use of an acidic sucrose electroporation solution. Frequencies of greater than 2 x 10(5) transformants per microgram of plasmid DNA were obtained for E. faecalis cells, whereas various strains of streptococci and Bacillus anthracis were transformed at frequencies of 10(3) to 10(4) transformants per microgram of plasmid DNA with the same protocol. A novel Escherichia coli-Streptococcus and Enterococcus shuttle cloning vector, pDL276, was constructed for use in conjunction with the electroporation system. This vector features a multiple cloning site region flanked by E. coli transcription termination sequences, a relatively small size (less than 7 kb), and a kanamycin resistance determinant expressed in both gram-positive and gram-negative hosts. Various enterococcal and streptococcal DNA sequences were cloned in E. coli (including sequences that could not be cloned on other vectors) and were returned to the original host by electroporation. The vector and electroporation system was also used to clone directly into E. faecalis.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. G., McKay L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkus J. M., Leppla S. H. Transcriptional regulation of the protective antigen gene of Bacillus anthracis. Infect Immun. 1989 Aug;57(8):2295–2300. doi: 10.1128/iai.57.8.2295-2300.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke D. Plasmid transformation of Streptococcus sanguis (Challis) occurs by circular and linear molecules. Mol Gen Genet. 1981;182(3):490–497. doi: 10.1007/BF00293940. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassy B. M. A gentle method for the lysis of oral streptococci. Biochem Biophys Res Commun. 1976 Jan 26;68(2):603–608. doi: 10.1016/0006-291x(76)91188-8. [DOI] [PubMed] [Google Scholar]
- Chen J. D., Morrison D. A. Cloning of Streptococcus pneumoniae DNA fragments in Escherichia coli requires vectors protected by strong transcriptional terminators. Gene. 1987;55(2-3):179–187. doi: 10.1016/0378-1119(87)90278-2. [DOI] [PubMed] [Google Scholar]
- Christie P. J., Dunny G. M. Identification of regions of the Streptococcus faecalis plasmid pCF-10 that encode antibiotic resistance and pheromone response functions. Plasmid. 1986 May;15(3):230–241. doi: 10.1016/0147-619x(86)90041-7. [DOI] [PubMed] [Google Scholar]
- Christie P. J., Kao S. M., Adsit J. C., Dunny G. M. Cloning and expression of genes encoding pheromone-inducible antigens of Enterococcus (Streptococcus) faecalis. J Bacteriol. 1988 Nov;170(11):5161–5168. doi: 10.1128/jb.170.11.5161-5168.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao M. L., Ferretti J. J. Streptococcus-Escherichia coli shuttle vector pSA3 and its use in the cloning of streptococcal genes. Appl Environ Microbiol. 1985 Jan;49(1):115–119. doi: 10.1128/aem.49.1.115-119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny G. M., Clewell D. B. Transmissible toxin (hemolysin) plasmid in Streptococcus faecalis and its mobilization of a noninfectious drug resistance plasmid. J Bacteriol. 1975 Nov;124(2):784–790. doi: 10.1128/jb.124.2.784-790.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny G., Funk C., Adsit J. Direct stimulation of the transfer of antibiotic resistance by sex pheromones in Streptococcus faecalis. Plasmid. 1981 Nov;6(3):270–278. doi: 10.1016/0147-619x(81)90035-4. [DOI] [PubMed] [Google Scholar]
- Fachon-Kalweit S., Elder B. L., Fives-Taylor P. Antibodies that bind to fimbriae block adhesion of Streptococcus sanguis to saliva-coated hydroxyapatite. Infect Immun. 1985 Jun;48(3):617–624. doi: 10.1128/iai.48.3.617-624.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler S., Wirth R. Transformation of bacteria with plasmid DNA by electroporation. Anal Biochem. 1988 Apr;170(1):38–44. doi: 10.1016/0003-2697(88)90086-3. [DOI] [PubMed] [Google Scholar]
- Holo H., Nes I. F. High-Frequency Transformation, by Electroporation, of Lactococcus lactis subsp. cremoris Grown with Glycine in Osmotically Stabilized Media. Appl Environ Microbiol. 1989 Dec;55(12):3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc D. J., Inamine J. M., Lee L. N. Broad geographical distribution of homologous erythromycin, kanamycin, and streptomycin resistance determinants among group D streptococci of human and animal origin. Antimicrob Agents Chemother. 1986 Apr;29(4):549–555. doi: 10.1128/aac.29.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc D. J., Lee L. N. Characterization of two tetracycline resistance determinants in Streptococcus faecalis JH1. J Bacteriol. 1982 May;150(2):835–843. doi: 10.1128/jb.150.2.835-843.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindler L. E., Macrina F. L. Characterization of genetic transformation in Streptococcus mutans by using a novel high-efficiency plasmid marker rescue system. J Bacteriol. 1986 May;166(2):658–665. doi: 10.1128/jb.166.2.658-665.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchansky J. B., Muriana P. M., Klaenhammer T. R. Application of electroporation for transfer of plasmid DNA to Lactobacillus, Lactococcus, Leuconostoc, Listeria, Pediococcus, Bacillus, Staphylococcus, Enterococcus and Propionibacterium. Mol Microbiol. 1988 Sep;2(5):637–646. doi: 10.1111/j.1365-2958.1988.tb00072.x. [DOI] [PubMed] [Google Scholar]
- Macrina F. L., Evans R. P., Tobian J. A., Hartley D. L., Clewell D. B., Jones K. R. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene. 1983 Nov;25(1):145–150. doi: 10.1016/0378-1119(83)90176-2. [DOI] [PubMed] [Google Scholar]
- Macrina F. L., Jones K. R., Wood P. H. Chimeric streptococcal plasmids and their use as molecular cloning vehicles in Streptococcus sanguis (Challis). J Bacteriol. 1980 Sep;143(3):1425–1435. doi: 10.1128/jb.143.3.1425-1435.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrina F. L., Wood P. H., Jones K. R. Genetic transformation of Streptococcus sanguis (Challis) with cryptic plasmids from Streptococcus ferus. Infect Immun. 1980 Jun;28(3):692–699. doi: 10.1128/iai.28.3.692-699.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrina F. L., Wood P. H., Jones K. R. Simple method for demonstrating small plasmid deoxyribonucleic acid molecules in oral streptococci. Appl Environ Microbiol. 1980 May;39(5):1070–1073. doi: 10.1128/aem.39.5.1070-1073.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre D. A., Harlander S. K. Genetic transformation of intact Lactococcus lactis subsp. lactis by high-voltage electroporation. Appl Environ Microbiol. 1989 Mar;55(3):604–610. doi: 10.1128/aem.55.3.604-610.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsuk S., Chiang Y. W., Reynolds R. B., Lovett P. S. Restriction fragments that exert promoter activity during postexponential growth of Bacillus subtilis. J Bacteriol. 1983 Sep;155(3):1399–1406. doi: 10.1128/jb.155.3.1399-1406.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecenka V., Dvorák M., Trávnícek M. Simple and efficient method for cloning of large DNA fragments with identical ends into plasmid vectors. Nucleic Acids Res. 1988 May 11;16(9):4179–4179. doi: 10.1093/nar/16.9.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell Ian B., Achen Marc G., Hillier Alan J., Davidson Barrie E. A Simple and Rapid Method for Genetic Transformation of Lactic Streptococci by Electroporation. Appl Environ Microbiol. 1988 Mar;54(3):655–660. doi: 10.1128/aem.54.3.655-660.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stassi D. L., Lopez P., Espinosa M., Lacks S. A. Cloning of chromosomal genes in Streptococcus pneumoniae. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7028–7032. doi: 10.1073/pnas.78.11.7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth R., An F. Y., Clewell D. B. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J Bacteriol. 1986 Mar;165(3):831–836. doi: 10.1128/jb.165.3.831-836.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y., Clewell D. B. Recombination-deficient mutant of Streptococcus faecalis. J Bacteriol. 1980 Aug;143(2):966–970. doi: 10.1128/jb.143.2.966-970.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]


