Abstract
We investigated the genetic diversity, extent of recombination, natural selection, and population divergence of Ralstonia solanacearum samples obtained from sources worldwide. This plant pathogen causes bacterial wilt in many crops and constitutes a serious threat to agricultural production due to its very wide host range and aggressiveness. Five housekeeping genes, dispersed around the chromosome, and three virulence-related genes, located on the megaplasmid, were sequenced from 58 strains belonging to the four major phylogenetic clusters (phylotypes). Whereas genetic variation is high and consistent for all housekeeping loci studied, virulence-related gene sequences are more diverse. Phylogenetic and statistical analyses suggest that this organism is a highly diverse bacterial species containing four major, deeply separated evolutionary lineages (phylotypes I to IV) and a weaker subdivision of phylotype II into two subgroups. Analysis of molecular variations showed that the geographic isolation and spatial distance have been the significant determinants of genetic variation between phylotypes. R. solanacearum displays high clonality for housekeeping genes in all phylotypes (except phylotype III) and significant levels of recombination for the virulence-related egl and hrpB genes, which are limited mainly to phylotype strains III and IV. Finally, genes essential for species survival are under purifying selection, and those directly involved in pathogenesis might be under diversifying selection.
Ralstonia solanacearum is one of the most destructive bacterial pathogens, able to cause disease on at least 200 different plant species (20). It affects a wide range of plants worldwide, including herbaceous plants, shrubs, and trees. Furthermore, R. solanacearum represents a major concern, given that it can seriously affect the production of ornamental plants and valuable crops such as tomato, potato, banana, peanut, eggplant, and others (19, 67). This gram-negative bacterium typically inhabits subtropical and tropical regions and recently has spread to the temperate regions of Europe (13).
R. solanacearum is a species complex composed of many genetic groups, some of which can be subdivided into clonal lines (14). Ralstonia syzygii and blood disease bacteria (BDB) belong to this species complex (7, 59). Attempts to organize the diverse genetic groups of R. solanacearum resulted in race classification based on host range (2). A phenotype-based structure also divides this population into biovars according to the ability of strains to use various sugar and alcohol carbohydrates (19, 21). Both classifications lack an exact concordance with the genetic background of the complex members. Recently, Fegan and Prior (7) analyzed the 16S-to-23S internal transcribed spacer region and mutS, hrpB, and egl gene sequences, together with amplified fragment length polymorphism/restriction fragment length polymorphism typing data (47, 48) and the 16S rRNA gene sequence (59) to develop a phylogeny-based scheme. This hierarchical classification is partitioned into four phylotypes (genetic groups), each of which is further subdivided into smaller groups named sequevars. Each phylotype reflects the geographic origin of strains: phylotype I and II are composed of Asian and American strains, respectively, whereas phylotype III members are African, and phylotype IV isolates, including R. syzygii and BDB, are from Indonesia, Japan, and Australia (7, 49).
The wide diversity of R. solanacearum is reflected in the bacterium's considerable variability in host range, aggressiveness (24), and the adaptation to different climates that is often influenced by host genotype, natural habitat, and agricultural practices (20). The fraction of phylotype II commonly known as race 3/biovar 2 (R3B2) infects tomato and common solanaceous weeds and causes brown rot, a serious disease of potato. This group is adapted to lower temperatures than other races; therefore, it constitutes a serious treat to agricultural production in temperate regions of the world (67).
R. solanacearum is organized into two large circular replicons called the chromosome (the larger replicon) and the megaplasmid (53). Both replicons contain essential and pathogenicity genes, the same dinucleotide relative abundances and codon usage, and similar distribution and composition of simple sequence repeats (4, 53). Thus, the two replicons in this bacterium have likely coevolved over a long time span. However, the evolutionary driving mechanism that shapes the chromosome and megaplasmid of R. solanacearum is still unclear.
The interaction between R. solanacearum and its plant hosts is strongly influenced by the ecological niche, the evolution of the bacterial genetic components, and the corresponding plant R proteins that might detect its presence on the host. Knowledge about the evolutionary driving forces acting upon this pathogen and its population structure can be tracked by focusing on housekeeping genes, which are less subject to horizontal gene transfer. However, high levels of discrimination can be achieved by combining analysis of housekeeping genes with that of more variable genes like those involved in virulence (42). Housekeeping genes, which represent the so-called “core genome” and typically encode proteins essential for the organism's survival, are present in all strains of a bacterial species and usually evolve slowly. Selective pressures that affect housekeeping genes reflect the selection forces working on the core genome. Conversely, the “flexible” or “accessory” genome contains genes that differ among strains, are dispensable, and usually improve bacterial fitness. The flexible genome includes genes associated with virulence, antibiotic resistance, and mobile elements (54). Genes of both genomes may experience different levels of selection depending mainly on the final function of the gene product.
Bacterial populations can be subject to different selective pressures that give distinct patterns of genetic diversity at the locus being examined. Diversifying (called also balancing) selection generates multiple alleles, some of which are retained over a long time period in the population, and the levels of polymorphism are appreciable. On the other hand, in the selective sweep mechanism (directional selection), an adaptive mutation that occurs in the target locus is preferentially fixed in the population as a result of positive selection, decreasing the number of alleles and the genetic diversity (50, 65). Purifying or stabilizing selection reduces the frequency of deleterious alleles (lower fitness) until they are eliminated from the population. Recombination could shape diversifying as well as directional selection and can provide high levels of variation even though selection is acting on an adaptive allele (9, 11). Geographic distance could also influence the genetic variation and structure of a population when spatially limited gene flow decreases the genetic similarity among populations and induces population divergence (41, 51).
In this study, we examined the evolutionary forces operating on R. solanacearum populations by analyzing sequences of representative genes from the core and flexible genomes, using samples obtained from worldwide sources. We were particularly interested in estimating the extent of recombination, selection, and geographic isolation that influenced this organism. We used multilocus sequence typing (MLST) (30) to analyze five housekeeping and three virulence-related genes. This approach uses sequences of internal gene fragments and assigns different allele numbers to the sequence at each locus, providing unique allelic profiles. We found R. solanacearum to be a diverse microorganism, showing high levels of nucleotide polymorphism and a number of unique alleles in the chromosome and in the megaplasmid. The core genome is essentially clonal (except in phylotype III) and is subject to purifying selection. On the other hand, virulence-related genes have two evolutionary profiles: hrpB and fliC behave like essential genes, whereas egl is undergoing diversifying selection together with high recombination mainly in phylotypes III and IV. We also found genetic evidence for the divergence of each phylotype due to geographic isolation, in which the spatial distance has had a key role. Finally, the population may have diverged a long time ago, as judged by the amount of fixed polymorphisms observed among phylotypes and by the relative abundance of single-locus and double-loci variants in the groups of clonal complexes.
MATERIALS AND METHODS
Sample collection and growth conditions.
Bolivian R. solanacearum samples were collected from wilted potato plants and diseased tubers harvested from naturally infested fields. A milk-like fluid was extracted and diluted in sterile distilled water. Bacterial suspensions were plated on Kelman medium (27) supplemented with 2,3,5-triphenyl tetrazolium chloride (50 μg·ml−1) and incubated for 48 h at 28°C. To identify each colony, we used a PCR-based approach that specifically detects the fliC gene (flagellin) of R. solanacearum (55). PCR was carried out in a total volume of 20 μl containing ExTaq DNA polymerase (Takara Bio Inc., Shiga, Japan), buffer, DNA template, the primer pair Rsol_fliC (55), bovine serum albumin (New England Biolabs, Ipswich, MA), and a mixture of deoxynucleotides. The genomic DNA template was extracted in Bolivia according to the hexadecyltrimethyl ammonium bromide method (68). Amplification conditions were 94°C (1 min) for denaturation and 63°C (1 min) and 72°C (1 min) for annealing and extension, respectively. A preceding denaturation step and a final extension step were carried out at 94°C and 72°C for 5 min each.
We used genomic DNA from 58 different strains, including that from the Bolivian samples and the DNA donated by members of the scientific community (Table 1). We included in our analysis Ralstonia solanacearum complex members BDB and R. syzygii (59). One strain of Ralstonia mannitolilytica (LMG6866T) was used as an outgroup, since this species is the closest relative of R. solanacearum and R. syzygii (63).
TABLE 1.
DNA from R. solanacearum and BDB strains used in this study
| Strain name | Alternate name | Race | Biovar | Phylotype group | Host | Geographical origin | Sourcea | Year of sampling | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GMI1000 | JS753 | 1 | 3 | I | Tomato | French Guyana | 53 | ||||
| UW363 | TMPS2 | 3 | I | Tomato | China | C. Allen | |||||
| UW505 | CIP266 | 1 | 3 | I | Potato | Java, Indonesia | C. Allen | 1985 | |||
| BR629 | 629 | 1 | 1 | I | Pepper | Para, Brazil | N. Furuya | ||||
| UW298 | 148 BPI WP6 | 3 | I | Potato | Baguio, Philippines | C. Allen | 1978 | ||||
| UW51 | CMI-B2865 | 1 | 4 | I | Potato | Gorandiyantena, Ceylon | C. Allen | ||||
| UW296 | 133 | 3 | I | Potato | Sri Lanka | C. Allen | 1977 | ||||
| UW152 | S241 | 1 | 3 | I | Potato | Atherton, Australia | C. Allen | 1966 | |||
| UW503 | CIP 266 | 1 | 4 | I | Potato | Java, Indonesia | C. Allen | 1985 | |||
| MAFF301556 | 1 | 4 | I | Potato | Nagasaki, Japan | K. Tsuchiya, M. Horita | |||||
| R292 | CIP277 | 5 | 5 | I | Morus alba | China | K. Tsuchiya, M. Horita | ||||
| K | 1 | 3 | I | Eucalyptus sp. | South Africa | T. Coutinho | |||||
| 27B | 1 | 3 | I | Eucalyptus sp. | Uganda | T. Coutinho | |||||
| CK | 1 | 3 | I | Eucalyptus sp. | D. R. Congo | T. Coutinho | |||||
| CC | 1 | 3 | I | Eucalyptus sp. | D. R. Congo | T. Coutinho | |||||
| UW486 | CIP179 | 3 | I | Tomato | New Guinea | C. Allen | 1980 | ||||
| BS024 | 3 | 2 | II | Potato | Chuquisaca, Bolivia | This study | 2004 | ||||
| BS025 | 3 | 2 | II | Potato | Santa Cruz, Bolivia | This study | 2004 | ||||
| BS048 | 3 | 2 | II | Potato | Santa Cruz, Bolivia | This study | 2004 | ||||
| BS075 | 3 | 2 | II | Potato | Chuquisaca, Bolivia | This study | 2004 | ||||
| BS094 | 1 | 1 | II | Potato | Chuquisaca, Bolivia | This study | 2004 | ||||
| BS095 | 1 | 1 | II | Potato | Chuquisaca, Bolivia | This study | 2004 | ||||
| UW551 | 3 | 2 | II | Geranium | Naivasha, Kenya | C. Allen | 2003 | ||||
| UW477 | CIP10 | 3 | 2 | II | Potato | Peru | C. Allen | 1979 | |||
| UW72 | 3 | 2 | II | Potato | Greece | C. Allen | |||||
| UW134 | SS221 | 1 | 1 | II | Potato | Nairobi, Kenya | C. Allen | 1960 | |||
| UW224 | Harris 220 | 3 | 2 | II | Potato | Embu, Kenya | C. Allen | 1970 | |||
| UW276 | 3 | 2 | II | Potato | Mexico | C. Allen | 1977 | ||||
| UW420 | 0223A | 3 | 2 | II | Potato | Queensland, Australia | C. Allen | 1967 | |||
| UW469 | CIP239 | 1 | 1 | II | Potato | Brazil | C. Allen | 1983 | |||
| UW504 | CIP265 | 3 | 2 | II | Potato | East Java, Indonesia | C. Allen | 1985 | |||
| K60 | UW25 | 1 | 1 | II | Potato | N. Carolina, U.S. | C. Allen | 1975 | |||
| BR574 | 574 | 1 | 1 | II | Eucalyptus sp. | Para, Brazil | N. Furuya | ||||
| BR113 | 113 | 3 | 2 | II | Tomato | Parana, Brazil | N. Furuya | ||||
| UW448 | CIP258 | 3 | 2 | II | Potato | Burundi | C. Allen | 1984 | |||
| UW220 | 3 | 2 | II | Potato | Poona, India | C. Allen | 1971 | ||||
| UW365 | POUB | 3 | 2 | II | Potato | China | C. Allen | ||||
| UW344 | 10 1SC | 3 | 2 | II | Potato | Ituporanga, Brazil | C. Allen | 1981 | |||
| PD441 | 3 | 2 | II | Potato | Sweden | K. Tsuchiya, M. Horita | |||||
| PD1610 | 3 | 2 | II | Potato | Belgium | K. Tsuchiya, M. Horita | |||||
| PD1939 | 3 | 2 | II | Potato | Israel | K. Tsuchiya, M. Horita | |||||
| PD1100 | 3 | 2 | II | Potato | Egypt | K. Tsuchiya, M. Horita | |||||
| TK1-3-1 | 3 | 2 | II | Potato | Khan Kaen, Thailand | K. Tsuchiya, M. Horita | |||||
| BR855 | 855 | 3 | 2 | II | Potato | Brasilia, Brazil | N. Furuya | ||||
| Ant307 | 2 | 1 | II | Anthurium | French West Indies | C. Boucher | |||||
| UW386 | 0.983/1 | 1 | 1 | III | Tomato | Nigeria | C. Allen | 1983 | |||
| UW34 | K179 | 1 | III | Tobacco | Zambia | C. Allen | 1959 | ||||
| JT525 | GMI8237 | 1 | III | Geranium | Reunion island | C. Boucher | |||||
| NCPPB332 | JS949 | 1 | III | Potato | Zimbabwe | C. Boucher | |||||
| CFBP3059 | JS904 | 1 | III | Eggplant | Burkina Faso | C. Boucher | |||||
| UW519 | B9043 | 1 | 1 | IV | Clove | Indonesia | C. Allen | ||||
| MAFF211271 | 3 | N2 | IV | Potato | Shizuoka, Japan | K. Tsuchiya, M. Horita | |||||
| MAFF301558 | JS934 | 3 | N2 | IV | Potato | Nagasaki, Japan | K. Tsuchiya, M. Horita | ||||
| MAFF211402 | N2 | IV | Potato | Okinawa, Japan | K. Tsuchiya, M. Horita | ||||||
| WP20 | N2 | IV | Potato | Luzon, Philippines | K. Tsuchiya, M. Horita | ||||||
| UW443 | T334 | BBD | IV | Banana | West Java, Indonesia | C. Allen | 1987 | ||||
| UW445 | T394 | BBD | IV | Banana | N. Sulawisi, Indonesia | C. Allen | 1987 | ||||
| UW446 | T440 | BBD | IV | Banana | N. Sulawisi, Indonesia | C. Allen | 1987 | ||||
| Ralstonia mannitolilytica | LMG6866 (type strain) | Hospital infection | London, United Kingdom | M. Vaneechoutte | |||||||
C. Allen, Department of Plant Pathology, University of Wisconsin, Madison, WI; N. Furuya, Graduate School of Bioresource and Bioenvironmental Sciences, Kyushu University, Fukuoka, Japan; M. Horita, Hokkaido National Agricultural Experiment Station, Sapporo, Japan; T. Coutinho, Faculty of Natural and Agricultural Sciences, University of Pretoria, Republic of South Africa; C. Boucher, CNRS-INRA, Castanet-Tolosan, France; M. Vaneechoutte, University of Gent, Belgium.
Phylotype determination.
Multiplex PCR was performed as described previously (7) to determine the phylotype affiliation of all strains prior to starting sequence studies. Briefly, a PCR mixture containing phylotype-specific primers (Nmult:21:1F, Nmult:21:2F, Nmult:23:AF, Nmult:22:InF, and Nmult:22:RR) (Table 2) and R. solanacearum-specific primers (Table 2, 759 and 760) was prepared and subjected to thermocycling at the following temperatures: 94°C for 15 s, 58°C for 30 s, and 72°C for 30 s. PCR products were resolved using agarose 1.5% (wt/vol) gel electrophoresis.
TABLE 2.
Primers used for amplifying, sequencing, and phylotyping
| Gene | Primer designationa | Sequence of primer (5′-3′) | PCR conditions (°C)b | Reference |
|---|---|---|---|---|
| gdhA | GdhAF | GATGGATGACGGCCGCATCG | 63 | This study |
| GdhAR | TGAACGCCGCCGTCCGCAG | 63 | This study | |
| adk | AdkF | TCTGTTGGGCGCACCCGGC | 62 | This study |
| AdkR | CCCAGCCGGAGTAGTAGTCC | 62 | This study | |
| AdkNF | CGCCGGCAAAGGTACGCAAG | 63 | This study | |
| AdkNR | CGGGCGGGTCTGGTTCTCG | 63 | This study | |
| gyrB | GyrBF | AGGGCTTCGTCGAGTACATCAA | 62 | This study |
| GyrBR | GTTCCGCCGAGGCTCCACG | 62 | This study | |
| GyrBNF | GTGGAACGACGGCTTCAACGA | 63 | This study | |
| GyrBNR | GCGCGAGAACTGGTACTGCC | 63 | This study | |
| gapA | GapAF | ATGACCATCAAGATCGGCAT | 55 | This study |
| GapAR | GGGCCATTTCCAGCACCT | 55 | This study | |
| ppsA | PpsAF | CTGTACAACGACCGCGCTAT | 55 | This study |
| PpsAR | GTTGGTCAGGCCCATCTCTT | 55 | This study | |
| PpsANF | GGGCGTGATGTTCACGAT | 57 | This study | |
| PpsANR | CCAGCATGGGGTTCTCTTC | 57 | This study | |
| fliC | Rsol_fliCF | GAACGCCAACGGTGCGAACT | 63 | 55 |
| Rsol_fliCR | GGCGGCCTTCAGGGAGGTC | 63 | 55 | |
| hrpB | RShrpBF | TGCCATGCTGGGAAACATCT | 64 | 47 |
| RShrpBR | GGGGGCTTCGTTGAACTGC | 64 | 47 | |
| egl | EglF | AAATCCAGATATCGAATTGCCAA | 57 | This study |
| EglR | GCGTGCCGTACCAGTTCTG | 57 | This study | |
| Endo-Fc | ATGCATGCCGCTGGTCGCCGC | 70 | 47 | |
| Endo-Rc | GCGTTGCCCGGCACGAACACC | 70 | 47 | |
| Nmult:21:1F | CGTTGATGAGGCGCGCAATTT | 58 | 7 | |
| Nmult:21:2F | AAGTTATGGACGGTGGAAGTC | 58 | 7 | |
| Nmult:23:AF | ATTACSAGAGCAATCGAAAGATT | 58 | 7 | |
| Nmult:22:InF | ATTGCCAAGACGAGAGAAG TA | 58 | 7 | |
| Nmult:22:RR | TCGCTTGACCCTATAACGAGTA | 58 | 7 | |
| 759 | GTCGCCGTCAACTCACTTTCC | 58 | 7 | |
| 760 | GTCGCCGTCAGCAATGCGGAATCG | 58 | 7 |
F, forward primer; R, reverse primer; N, internal primer (nested).
Ready-for-use annealing temperatures. All PCRs were preceded by a 5-min denaturation step at 96°C, followed by a final extension step of 5 min at 72°C.
Primer set used as internal primers.
PCR amplification and DNA sequencing.
Five chromosomal housekeeping genes (ppsA, phosphoenolpyruvate synthase; gyrB, DNA gyrase, subunit B; adk, adenylate kinase; gdhA, glutamate dehydrogenase oxidoreductase; and gapA, glyceraldehyde 3-phosphate dehydrogenase oxidoreductase) and three megaplasmid virulence-related genes (hrpB, regulatory transcription regulator; fliC, encoding flagellin protein; and egl, endoglucanase precursor) were analyzed. Sets of primers used to amplify internal fragments of these genes (Table 2) were also used as sequencing primers. Some genes required additional internal primers for sequencing. Primer design was carried out based on alignments of coding sequences of R. solanacearum GMI1000 (NCBI accession numbers NC_003295 and NC_003296) and close bacterial relatives, starting with the most conserved regions.
PCR amplification was carried out in a model PTC-100 thermocycler (MJ Research, Bio-Rad Laboratories, Inc., Hercules, CA) using ExTaq DNA polymerase, an enzyme with proofreading activity, and Q reagent (QIAGEN, Valencia, CA) to help the amplification of the GC-rich R. solanacearum DNA. Deoxyucleotide and primer concentrations were 350 nM each and 0.8 μM, respectively. Thirty cycles of amplification were performed with a denaturation temperature of 96°C for 5 min, the appropriate annealing temperature (Table 2), and an extension at 72°C for 1 to 1.5 min. All PCR products were cleaned with QIAquick PCR purification columns (QIAGEN) prior to sequencing.
DNA sequencing was performed in Applied Biosystems 3130 or 3730XL sequencers using forward and reverse primers. Raw sequences from both strands were assembled with Sequencher 4.1.2 (GeneCodes Corp., Ann Arbor, MI). Sequences were edited with BioEdit 7.0.5.1 (16) and aligned using Clustal W (62). All ambiguous and terminal sequences were trimmed before data analysis. Inconsistencies were solved by resequencing. We were not able to amplify the fliC gene from strains BR113, UW445, UW446, UW344, and LMG6866. As a control, we tried to amplify the fliD gene that encodes the flagellar cap (23), using two different sets of primers from the same strains without success. This finding suggests that these strains do not have flagella, consistent with the observation that some strains of R. solanacearum (3) and R. mannitolilytica (5) are nonmotile. We found a six-base deletion in some egl sequences belonging mainly to phylotype I. We could not amplify the egl gene from R. mannitolilytica, since this species is a human pathogen that lacks the gene.
Data analysis.
Each gene was analyzed independently as well as collectively. From two concatenated sequence sets we created one set corresponding to the chromosome and a second set corresponding to the megaplasmid. In addition, we created a third concatenated sequence using the chromosome and megaplasmid sequences that we named sequence “C+M.” We differentiated each sequence type (ST) using the NRDB program, available at www.mlst.net. STs were grouped into clonal complexes using the eBURST program (10).
We retrieved sequences of R. solanacearum egl and hrpB genes (56 sequences each) from the GenBank database. These sequences were added to those obtained from this work to achieve a pool of 114 sequences for each gene. The new sequence groups were designated egl-114 and hrpB-114 (for a list of GenBank accession numbers see reference 64).
The data analysis was started by assessing models of nucleotide substitution using the maximum likelihood (ML) approach as implemented in the Modeltest 3.7 (44) program. First, neighbor-joining (NJ) trees were obtained using PAUP* 4.0b10 (58) to get initial likelihood scores, and then the best-fit nucleotide substitution model was selected for a set of aligned sequences. Model selection was conducted on the basis of hierarchical likelihood ratio tests (hLRT) instead of the Akaike or Bayesian information criterion (45). Phylogenetic trees for each sequence fragment or for concatenated data were inferred using the NJ, parsimony (MP), and ML methods by using PAUP* and MEGA 3.1 (28). Heuristic search methods using the tree bisection reconnection branch-swapping algorithm, with the random addition of taxa and 10 replicates, were applied to create ML trees. The Tamura-Nei model with gamma correction and 1,000 bootstrapping replicates were used to generate NJ trees. ML trees were visualized with TreeView 1.6.6 (40).
The RDP 2.0 (33) program was used to detect recombination. This package compiles tools for performing a comparable search for recombination evidence from sequence alignments. We estimated the population recombination rate using the RDP, MaxChi, Chimaera, and GENECONV methods. The analysis options of the RDP program were adjusted to the following general settings: sequences were considered linear, the P value cutoff was set to 0.05, the Bonferroni correction was applied, consensus daughter sequences were found, breakpoints were polished, and only recombination events detected by at least two programs were listed. Each program was run on a single gene alignment using different window sizes (i.e., 10, 20, 30, or 40) depending on the number of variable sites. Specific settings for each program were as follows. For the RDP method, no reference sequence was selected and the percentage of identity between recombinant sequences was set from 0 to 100. MaxChi was adjusted to include gaps and generate 1,000 permutations. For Chimaera, 1,000 permutations were performed; and for GENECONV, we scanned sequence triplets, treating each indel as a polymorphism and setting the g scale to 1.
The “standardized” index of association, IAs, was estimated using LIAN 3.5 (17, 18), a Web tool available at the MLST site http://pubmlst.org. P values, calculated using the parametric method, were used to discriminate the significance of IAs. The homoplasy ratio (H) was carried out with START 1.0.5 (25), adjusting the homoplasy settings to 1.00 × the number of sites. The START 1.0.5 (25) package was also used to calculate nonsynonymous (N) to synonymous (S) rate (dN/dS) ratios, loading data of allelic profiles and allele sequences at the same time. This program uses the Nei and Gojobori method for the estimation of synonymous and nonsynonymous substitutions. Tajima's D, genetic diversity, and subpopulation divergence estimations were computed with DnaSP 4.10 (52). Spatial patterns of isolation by distance were calculated using the Mantel test as implemented in Arlequin 3.01 (6).
Nucleotide sequence accession numbers.
Sequence data from this article have been deposited in the GenBank data library under accession numbers DQ657359 to DQ657824.
RESULTS
Sequence analysis and DNA polymorphism.
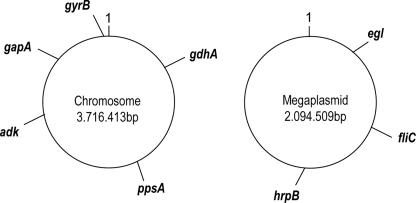
About 480 sequences were generated from different strains that represent the four R. solanacearum phylotypes proposed by Fegan and Prior (7) that include a variety of races and biovars (Table 1). The selected genes (five housekeeping genes and three genes involved with virulence) were distributed as much as possible across both replicons, as shown in Fig. 1. Only a single copy of each gene studied in this work was found in the genome of R. solanacearum strain GMI1000 (53) and that of strain UW551 (12). The selection of these genes was based on their use in an MLST scheme of other bacterial species and the availability of some sequence data from the virulence-related egl and hrpB genes in databases. The virulence-related genes are implicated directly (egl) or indirectly (hrpB and fliC) in disease-causing processes. The egl gene encodes an endoglucanase that likely acts at the front line of host invasion by partially degrading host cell walls (29). hrpB encodes an araC (1-β-d-arabinofuranosylcytosine) type transcriptional regulatory protein that governs multiple virulence pathways (38). Flagellin, encoded by the fliC gene, is the essential subunit of the flagellar filament that is needed for invasive virulence (61). In R. solanacearum, flagellin is not a major elicitor of host defenses (43).
FIG. 1.
Schematic representation of locus positions studied in this work. The locations are based on the sequence of R. solanacearum GMI1000 (53). ppsA, phosphoenolpyruvate synthase; gyrB, DNA gyrase (subunit B); adk, adenylate kinase (ATP-AMP transphosphorylase); gdhA, glutamate dehydrogenase [NAD(P)+] oxidoreductase; gapA, glyceraldehyde 3-phosphate dehydrogenase oxidoreductase; hrpB, regulatory transcription regulator; fliC, flagellin protein; egl, endoglucanase precursor (endo-1,4-beta-glucanase).
We found similar levels of genetic variation among the 58 R. solanacearum isolates for all housekeeping genes studied (Table 3, polymorphic sites and nucleotide diversity [π]), except for ppsA, which showed greater genetic variation than the others. The genetic variation was consistent for all chromosomal loci studied, with a mean number of 77.4 polymorphic sites (a mean percentage of 10.74), suggesting that the rates of evolution are similar except for the ppsA gene. There was a high level of genetic variation for chromosomal concatenated data: 387 polymorphic sites in 3,558 bp of sequence and a π value for synonymous sites equal to 9.012% (calculated with the Jukes-Cantor correction). The genetic variation in the megaplasmid was amply influenced by two genes (egl and hrpB) that exhibited higher genetic variation than fliC. The number of polymorphic sites for concatenated data for the megaplasmid sequences was 113 of 1,497 bp with a π value for synonymous sites of 14.3%. Thus, the megaplasmid showed greater diversity than the chromosome.
TABLE 3.
Genetic diversity and selection analyses
| Gene | Length (bp) | No. of ST | No. of polymorphic sites | % of polymorphic sites | Θa | πb
|
Tajima's Dc | dN/dS ratio | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Synonymous | Nonsynonymous | ||||||||
| gyrB | 873 | 24 | 73 | 8.4 | 0.018 | 0.020 | 0.071 | 0.004 | 0.222 | 0.044 |
| adk | 420 | 18 | 32 | 7.6 | 0.016 | 0.023 | 0.077 | 0.007 | 0.626 | 0.093 |
| gdhA | 774 | 22 | 85 | 11.0 | 0.024 | 0.022 | 0.065 | 0.008 | −0.468 | 0.113 |
| gapA | 774 | 22 | 78 | 10.1 | 0.022 | 0.028 | 0.093 | 0.008 | 0.626 | 0.074 |
| ppsA | 717 | 23 | 119 | 16.6 | 0.036 | 0.041 | 0.156 | 0.008 | 0.038 | 0.042 |
| hrpB | 810 | 25 | 150 | 18.5 | 0.040 | 0.052 | 0.164 | 0.019 | 0.389 | 0.118 |
| fliC | 318 | 18 | 40 | 12.6 | 0.028 | 0.024 | 0.068 | 0.010 | −0.647 | 0.250 |
| egl | 686 | 24 | 149 | 21.7 | 0.047 | 0.056 | 0.123 | 0.034 | 0.195 | 1.378 |
| hrpB-114 | 810 | 55 | 176 | 21.7 | 0.041 | 0.056 | 0.171 | 0.021 | 0.427 | 0.135 |
| egl-114 | 668 | 45 | 161 | 24.1 | 0.045 | 0.059 | 0.029 | 0.071 | 0.524 | 2.705 |
Theta value per site (Watterson estimator).
Nucleotide diversity calculated with the Jukes-Cantor correction.
Not significant Tajima D values at a P value of >0.1 (see reference 60 for confidence limits).
Phylogenetic reconstruction.
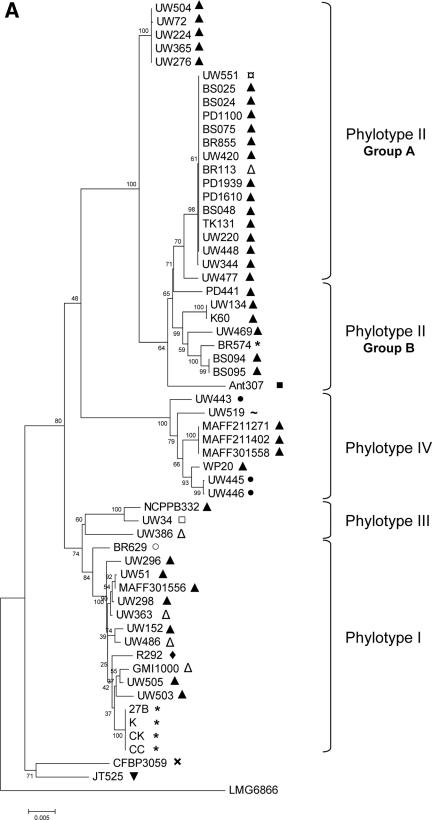
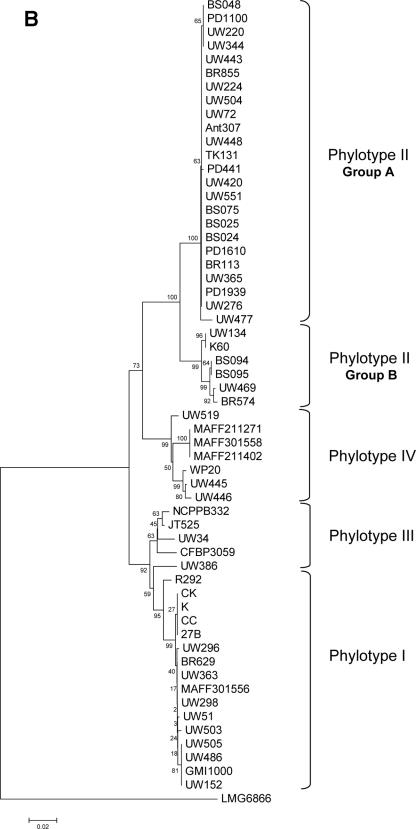
ML, MP, and NJ phylogenetic trees were constructed using single sequences of each locus and concatenated data (chromosome, megaplasmid, and “C+M” sequences). The best-fit nucleotide substitution model used to infer ML trees varied according to each locus, but the majority of them fit in the GTR+I+Γ (general time-reversible with invariant sites and a gamma rate distribution) or HKY+I+Γ (Hasegawa, Kishino, Yano with invariant sites and a gamma rate distribution) models. hLRT was used to select the best model, although in most cases, hLRT and the Akaike information criterion found the same models. We validated our sample size used in the phylogenetic reconstruction of R. solanacearum genes by comparing egl and hrpB trees generated using sequences obtained in this work against trees inferred using sequences retrieved from databases (egl-114 and hrpB-114). Trees constructed with both sets of sequences are in good agreement (e.g., egl-114 [Fig. 2C]).
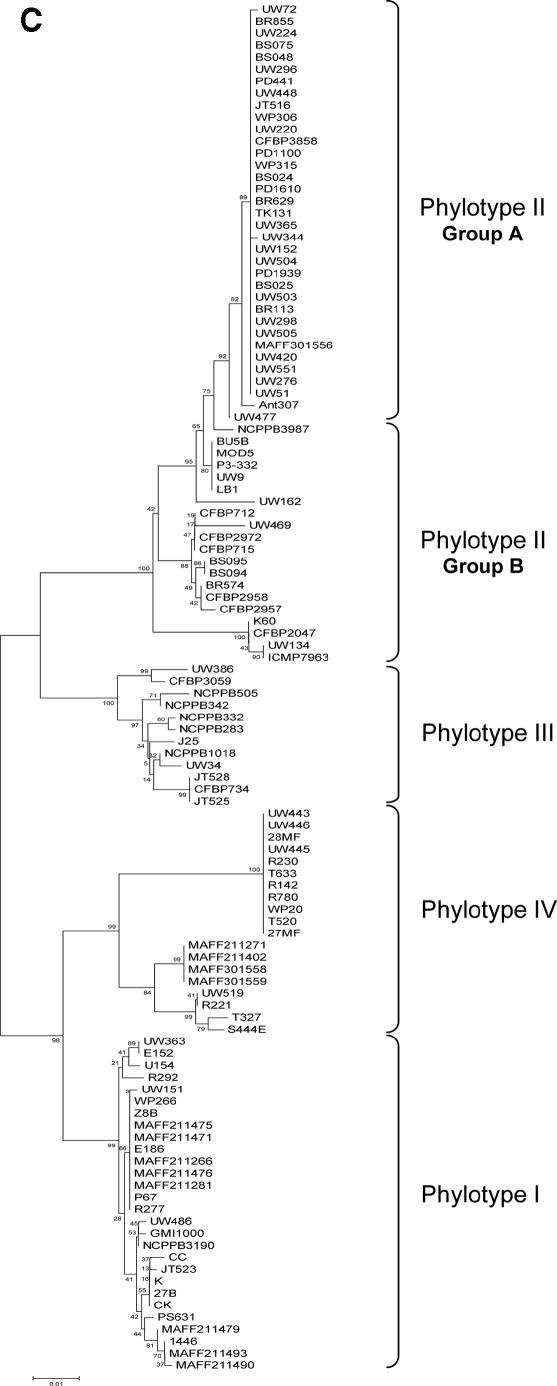
FIG. 2.
NJ phylogenetic trees based on (A) chromosome concatenated, (B) megaplasmid concatenated, and (C) egl-114 data sets. The four major phylogenetic groups are highlighted, including the subdivision of phylotype II. Bootstrap support values are shown next to the nodes. Plant hosts: ▴, potato; ¤, geranium; ▵, tomato; *, Eucalyptus sp.; ▪, Anthurium sp.; •, banana; ∼, clove; □, tobacco; ○, Capsicum annuum; ⧫, Morus alba; ×, Solanum melongena; ▾, Pelargonium asperum. (A) Chromosome concatenated data; (B) megaplasmid concatenated data; (C) egl-114 data.
Trees constructed using different methods showed a similar branching pattern. We clearly distinguished four phylogenetic clusters in the R. solanacearum population (Fig. 2). Phylotype I comprises mainly strains of race 1/biovar 3 samples from Asia. Phylotype II contains primarily strains from America and around the world. Trees inferred in this work showed a further division in phylotype II, which was separated into two subgroups: the first one (referred to as IIa, see Fig. 2), composed exclusively of R3B2 isolates, is more compact relative to those of other groups. The second subgroup (IIb) showed more diversity than IIa and included strains isolated from the tropical Atlantic coast of South America and Central America, with some exceptions (CFPB715/712 and CMP7963) that come from Africa. The IIb group is composed mainly of biovar 1 isolates that are pathogens of potato, tomato, banana, and others. Phylotype III, composed of African isolates, was the most diverse cluster at the sequence level and in terms of host diversity (potato, tomato, tobacco, Symphytum sp., Solanum panduriforme, S. melongena, etc.). The fourth phylotype was also a very diverse cluster, composed of isolates from Indonesia, belonging to races 1, 2, and 3 and biovars 1, 2, and 2T.
MLST scheme.
We used the MLST scheme to assign different allele numbers to the sequence at each locus, getting unique allelic profiles, called STs. The R. solanacearum population has a number of STs that varies from 18 to 24 among housekeeping loci and from 18 to 25 among virulence-related loci. The total numbers of STs of the chromosome and the megaplasmid concatenated data are 35 in both cases and 41 for “C+M” data.
We used the eBURST program to define clonal complexes of related isolates derived from a common ancestor. This program assigned five clonal complexes to the R. solanacearum chromosome, where a clonal complex was defined as groups of STs that shared alleles at four out of the five housekeeping loci with at least one other member of the complex. For the megaplasmid, we also found five clonal complexes (two out of three loci) that concur with those from the chromosome. Clonal complexes using chromosome data are arranged in the following way: clonal complex 1 sorts STs represented by strains GMI1000 and UW505 (phylotype I); clonal complex 2 is represented by strains UW551, UW72, BS048, BR113, UW224, UW334, and PD441 (phylotype IIa); clonal complex 3 is represented by strains UW51, MAFF301556, and BR629 (phylotype I); clonal complex 4 is represented by strains UW298 and UW363 (phylotype I); and clonal complex 5 is represented by strains UW445 and UW446 (phylotype IV). No clonal complex was detected in phylotype III, whereas an appreciable number of singletons were found. eBURST did not find a clear common ancestor in clonal complexes 1, 4, and 5, perhaps due to the small number of isolates. However, clonal complexes 2 and 3 each contained common ancestors (UW551 and UW51, respectively) and a number of single-locus variants (SLVs, i.e., differing from the ancestral genotype at one locus) and double-locus variants (DLVs, i.e., differing from the ancestral genotype at two loci), suggesting that they are moderately old complexes. The abundance of SLVs and DLVs reflects the relative age of clonal complexes (57). Clonal complex 2 is one of the largest and most well-defined clonal groups that encompasses members of R3B2, sharing identical sequences.
Selection.
We used a series of tests to identify the selective pressures working on housekeeping and virulence-related genes of R. solanacearum. First, we determined the dN/dS ratios. Values of dN/dS of 1, dN/dS of >1, and dN/dS of <1 indicate neutrality, diversifying selection, and purifying selection, respectively. hrpB, fliC, and all housekeeping genes showed low levels of dN/dS (Table 3), indicating that these loci are under strong purifying selection conditions. In contrast, egl acquired values over 1, a dN/dS ratio of 1.378 for egl (alleles sequenced in this work) and a dN/dS ratio of 2.705 for egl-114. This suggests that diversifying selection is determining the evolution of the egl gene (Table 3).
We also performed the Tajima D test (60), a statistic that estimates whether the number of segregating/polymorphic sites and the average number of nucleotide differences are correlated. If the value of D is too large or too small, the neutral “null” hypothesis is rejected. Thus, D is negative for selective sweep and population growth and positive for diversifying selection. No loci showed significant deviation from neutral evolution for either individual housekeeping or virulence-related genes (Table 3). Additionally, we analyzed each phylotype using concatenated sequence data for the chromosome and, separately, for the megaplasmid. No sequence from any phylotype revealed significant deviation from the neutral model for chromosomal or megaplasmid data with one exception: phylotype I of megaplasmid concatenated data acquired a positive value with low significance (P < 0.1). This indicates that the nucleotide diversity is more dependent on alleles with higher frequencies, a signature of diversifying selection. We also calculated the Tajima D for each gene of each phylotype. The data set of egl-114 sequences obtained exclusively from phylotype I and IV strains showed a weak positive value for D (1.26 and 1.84; P < 0.1; respectively). In contrast, strains of phylotype I might be constrained by a selective sweep at the hrpB locus, since we found a significant negative value (−2.105; P < 0.05) for D upon analysis of hrpB-114 sequences. The rest of the other loci showed no significant values for Tajima's D for every phylotype analyzed.
Recombination.
To test the possible role of homologous recombination in the R. solanacearum population, we assessed the levels of linkage disequilibrium in our worldwide samples. If the alleles of different loci are under linkage equilibrium, high rates of recombination are operating in the population. The index of association (37) measures the extent of linkage. In our analysis, we used the “standardized” IA (IAs [18]) to test the statistical independence of alleles at all loci. The statistic IAs is equal to zero when the population is experiencing free recombination, whereas a population significantly different from zero is considered to be clonal (linkage disequilibrium). We calculated the IAs value using the allelic profile (grouped STs) of chromosome and “C+M” concatenated data. We also compared the “simulate critical values” LMC (Monte Carlo) (or calculated critical value Lpara [parametric]) with the variance VD, as suggested by Haubold and collaborators (18). If LMC was <VD, the population would be under linkage disequilibrium. After 1,000 random resamplings of the input data, IAs gave a value that was statistically significantly different from zero (P, <0.0001) for the chromosomal and for the “C+M” ST series (IAs = 0.3707 and 0.3736, respectively [Table 4]), suggesting a clonal population structure. When we calculated the IAs value for each phylotype, we found some differences in population structure. All phylotypes were essentially clonal, showing IAs values that were statistically significantly different from zero and VD values that exceeded LMC values (Table 4). However, phylotype III could be experiencing higher levels of recombination, since the IAs values were low for the chromosome (IAs = 0.0031) and for the “C+M” (IAs = 0.0982 [Table 4]).
TABLE 4.
Measures of association (IAs) between loci in R. solanacearum
| Group | Subgroup | No. of STs | IAs | Pparaa | PMCb | Lparaa | LMCb | VD |
|---|---|---|---|---|---|---|---|---|
| All phylotypes | “C+M” | 41 | 0.3736 | 0.0000 | 0.0010 | 0.6477 | 0.6513 | 2.1214 |
| Chromosome | 35 | 0.3707 | 0.0000 | 0.0010 | 0.2448 | 0.2451 | 0.5582 | |
| Phylotype I | “C+M” | 13 | 0.1694 | 0.0000 | 0.0010 | 1.5594 | 1.5829 | 2.6940 |
| Chromosome | 14 | 0.2194 | 0.0000 | 0.0010 | 0.8127 | 0.8420 | 1.2316 | |
| Phylotype II | “C+M” | 16 | 0.5371 | 0.0000 | 0.0010 | 1.9542 | 2.0078 | 6.9321 |
| Chromosome | 11 | 0.5441 | 0.0000 | 0.0010 | 0.9847 | 1.0249 | 2.2471 | |
| Phylotype III | “C+M” | 5 | 0.0982 | 0.0116 | 0.2840 | 0.4410 | 0.4556 | 0.4556 |
| Chromosome | 5 | 0.0031 | 1.0000 | 1.0000 | 0.2787 | 0.4000 | 0.1778 | |
| Phylotype IV | “C+M” | 6 | 0.5904 | 0.0000 | 0.0010 | 1.2340 | 1.4952 | 3.9238 |
| Chromosome | 6 | 0.6502 | 0.0000 | 0.0020 | 0.8601 | 0.9524 | 1.9524 |
We sought additional evidence for recombination by analyzing each R. solanacearum locus independently. The nonparametric, phylogenetically based RDP method (32, 33) applies a pair-wise scanning approach to detect informative sites (triplets) within an unweighted-pair group method with arithmetic means (UPGMA) dendrogram to find potential recombinant regions by comparing each site with the other. Once a potentially recombinant region has been detected, the method determines which of the three sequences is the recombinant and which are the “parentals.” The RDP method did not find significant evidence for recombination in the data analyzed, suggesting that this population is highly clonal (Table 5).
TABLE 5.
Tests for recombination for single genes
| Locus | Homoplasy H ratio | RDP | MaxChi | Chimaera | GENECOV |
|---|---|---|---|---|---|
| gyrB | 0.141 | No | No | No | No |
| adk | −0.017 | No | No | No | No |
| gdhA | −0.001 | No | No | No | No |
| gapA | −0.049 | No | No | No | No |
| ppsA | −0.016 | No | No | No | No |
| fliC | 0.339 | No | No | No | No |
| egl-114a | No | Yes | Yes | No | |
| hrpB-114 | 0.001 | No | Yes | Yes | Yes |
These sequences had too few informative sites for H ratio estimation.
The homoplasy test (36) is useful for detecting recombination in sequences of closely related organisms that differ only by 1 to 5% of nucleotides, whereas GENECONV (39), MaxChi, and Chimaera (34, 46) are useful for sequences that might be more diverse. The homoplasy test compares the observed numbers of homoplasy in a maximum-parsimony tree of the sequences with the number expected in the absence of recombination. The homoplasy ratio, H, is an indicator of the frequency of recombination. H values vary from 0 (clonal) to 1.0 (complete linkage equilibrium, indicating free recombination). The homoplasy test had a sufficient number of informative sites from which to give interpretable results with seven out of the eight loci studied. The H ratio for all loci was low, except for fliC, indicating that these loci are close to being clonal (Table 5). Some loci showed negative H ratio values due to the estimation of expected numbers of homoplasy if being clonal is slightly superior to the true homoplasy, affected in its turn by a possible misestimation of the effective number of sites.
The MaxChi and Chimaera functions are based on the maximum chi-square method for detecting recombination breakpoints using a sliding window. GENECONV recognizes distinct recombination breakpoints in sequence alignments by searching for unusually long fragments that are identical or nearly identical in pairs of aligned sequences. Table 5 summarizes the results from the single data sets given by these three tests. We used different window sizes with concordant results within each test; however, results given by the three recombination detection programs were only partially concordant. In a previous evaluation of their performance using simulated and empirical data, Posada and Crandall (46) found MaxChi to be the most powerful test, followed by Chimaera and GENECONV. Thus, we principally utilized the information on recombination inferred by MaxChi and Chimaera. Housekeeping genes showed no evidence of recombination. In contrast, two out of three virulence-related genes (egl and hrpB) appeared to have undergone significant levels of recombination. The most remarkable finding was that recombination events were not distributed homogeneously in the population but mainly in phylotypes III and IV. Thus, RPD programs helped to infer which strains have been participating in the recombination process, including the parent (donor) and the daughter strains. For egl, strains that belong to phylotype III (NCPPB332, CFBP734, NCPPB1018, NCPPB283, JT525, UW34, JT528, and UW386) and IV (WP20, S444E, and UW519) have been participating in recombination events. Similarly, phylotype IV strains that showed evidence of recombination in the hrpB gene were UW445, UW446, WP20, 28MF, 27MF, R780, R230, R142, T520, T633, and R780.
Divergence between populations and isolation by distance.
R. solanacearum classification based on phylogeny may reflect the geographical basis for each group (7). To test this, we applied statistical tests to R. solanacearum subpopulations under the null hypothesis that they are not genetically different but come from the same geographic origin. We measured the nucleotide diversity within each subpopulation (the π value) and between populations (Dxy value). As expected, the mean divergence between groups (Dxy) in all cases was higher than the mean divergence within subpopulations (Table 6), which agrees well with phylogenetic inference. The mean divergence value between subpopulations (Dxy = 0.056) was lower than that between R. solanacearum and R. mannitolilytica (Dxy = 0.120).
TABLE 6.
Divergence within and between R. solanacearum phylotypes and analysis of geographic isolation
| Locus |
|
Dxya | No. of fixed differences | No. of shared mutations | Statistical test valuesb
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Phylotype group | π values within group | χ2 | KS* | FST | HT | ||||
| Chromosome (concatenated) | I vs II II vs III | 0.00525 vs 0.00894 0.00894 vs 0.02260 | 0.03425 0.04144 | 26 30 | 19 45 | 0.0040** 0.0029** | 2.73915*** 2.81535*** | 0.78946 0.61466 | 0.89394 0.81250 |
| II vs IV | 0.00894 vs 0.00683 | 0.04193 | 39 | 15 | 0.0021** | 2.73999*** | 0.80797 | 0.84685 | |
| IIa vs IIb | 0.00536 vs 0.01102 | 0.01403 | 5 | 12 | 0.0012** | 2.16003*** | 0.41568 | 0.75616 | |
| III vs IV | 0.02290 vs 0.00683 | 0.04108 | 60 | 11 | 0.2237 NS | 3.35492*** | 0.63321 | 0.96154 | |
| hrpB | I vs II | 0.00385 vs 0.02046 | 0.08336 | 35 | 9 | 0.0000*** | 1.73260*** | 0.84913 | 0.90275 |
| II vs III | 0.02046 vs 0.01717 | 0.07806 | 28 | 12 | 0.0019** | 2.27145*** | 0.75132 | 0.88475 | |
| II vs IV | 0.02046 vs 0.01793 | 0.07099 | 12 | 11 | 0.0001*** | 2.24644*** | 0.72261 | 0.89744 | |
| IIa vs IIb | 0.00437 vs 0.01910 | 0.03790 | 1 | 6 | 0.0004*** | 1.49855*** | 0.68691 | 0.81915 | |
| III vs IV | 0.01717 vs 0.01793 | 0.07456 | 22 | 11 | 0.0699 NS | 2.4071*** | 0.75711 | 0.96322 | |
| egl | I vs II | 0.00674 vs 0.02317 | 0.08051 | 0 | 10 | 0.0000*** | 1.8306*** | 0.8093 | 0.84739 |
| II vs III | 0.02235 vs 0.01538 | 0.06902 | 6 | 8 | 0.0000*** | 2.01279*** | 0.7266 | 0.79917 | |
| II vs IV | 0.02317 vs 0.02733 | 0.09685 | 0 | 25 | 0.0000*** | 2.00376*** | 0.73068 | 0.80912 | |
| IIa vs IIb | 0.01478 vs 0.02205 | 0.03067 | 0 | 18 | 0.0000*** | 1.35614*** | 0.40338 | 0.70841 | |
| III vs IV | 0.01558 vs 0.02733 | 0.09583 | 32 | 1 | 0.0076** | 2.19117*** | 0.76702 | 0.87356 | |
| All loci (mean) | All phylotypes | 0.0144 | 0.056 | 19.7 | 14.2 | NA | NA | 0.7 | 0.852 |
Dxy, average number of nucleotide substitutions per site between populations, calculated using the Jukes-Cantor correction.
Significance obtained by the permutation test with 10,000 replicates: NS, not significant; **, 0.001 < P < 0.01; ***, P < 0.001. HT, haplotype diversity. NA, not applicable.
We used the χ2, KS*, KST, ZS, and ZS* statistical tests to measure the extent and significance of genetic differentiation (22). When haplotype diversity (HT) is below a critical value, χ2 is the best test, whereas, if HT is above the critical value, KS* is usually best (22). In our situation, the critical value for the megaplasmid locus egl and hrpB locus appears to be ∼0.97, whereas for chromosomal loci, it is ∼0.91. Table 6 shows comparisons of some phylotypes. The statistical tests suggest that housekeeping and virulence-related gene sequences from different phylotypes have evolved with significant geographic isolation (P < 0.01 and P < 0.001 for χ2 and KS* test statistics, respectively) and have undergone a high level of genetic differentiation (average level of gene flow [FST] = 0.7). Surprisingly, phylotype II also showed genetic differentiation in two subpopulations (i.e., IIa and IIb) for both egl and hrpB, as well as for housekeeping genes. These results suggest that each phylotype of the R. solanacearum population is genetically different and has evolved in geographic isolation. This conclusion is highly robust considering that the power of the KS* test is substantial with a sample size of ≥50 and with genes that are experiencing recombination (22), as occurs in our case (i.e., egl and hrpB).
We sought evidence to determine if spatial distance between groups was a critical factor in generating the geographically independent evolution of each phylotype. The genetic similarity between populations decreases exponentially as the geographic distance between them increases, because of the limiting effect of geographic distance on rates of gene flow (51). We used the Mantel test to assess the correlation between genetic differentiation and geographic distance, comparing the FST values of the four phylotypes. After running 1,000 permutations, the Mantel test revealed a highly significant association between geographic and genetic distances for both housekeeping (r = 0.988; P = 0.042) and virulence-related (hrpB-114, r = 0.837, P = 0.072; and egl-114, r = 0.921, P = 0.085) genes. This indicates that spatial distance explains the observed level of genetic and geographic structuring in the R. solanacearum population.
Analyses of fixed polymorphisms (the number of sites at which all of the sequences in one sample are different from all of the sequences in a second sample) and of shared polymorphisms are useful for inferring times of population divergence (26). The distribution of these polymorphisms in our R. solanacearum population was different for chromosomal and hrpB loci compared to those of egl (Table 6). The number of fixed differences among major clusters (phylotype I versus II, II versus III, and II versus IV) was higher than shared polymorphisms (varying from 26 to 60 for chromosome loci and from 12 to 35 for hrpB). This suggests a medium to long time period of separation from the common ancestor. In the case of phylotype II, comparisons within the subgroup (IIa versus IIb) showed a lower number of fixed polymorphisms, indicating that these subpopulations could be younger than the main clusters. In contrast, almost all comparisons among phylotypes in the egl locus showed a lower number of fixed differences. This suggests that either the subpopulations diverged very recently or that recombination between subpopulations had hidden population differentiation. Considering that the egl gene is experiencing higher levels of recombination, the last explanation seems the most appropriate.
DISCUSSION
We sequenced internal fragments of five housekeeping and three virulence-related genes for 58 R. solanacearum isolates representing a worldwide sample. Phylogenetic and statistical analyses showed that this organism (i) is a highly diverse bacterial species that contains four major, deeply separated evolutionary lineages (phylotypes I to IV) as well as a weaker subdivision of phylotype II consisting of two subgroups; (ii) it evolved in geographic isolation where the spatial distance has played an important role; (iii) it displays high clonality for housekeeping genes in all phylotypes except phylotype III and a significant level of recombination for egl and hrpB genes, which is limited mainly to phylotype III strains, but also to phylotype IV; and (iv) it is under different selection forces depending on gene products: those essential for species survival are under a purifying selection and those directly involved in pathogenesis (like egl) might be under diversifying selection.
Our phylogenetic analysis allowed the generation of trees that agreed with those inferred by Fegan and Prior (7, 49). All of our trees showed four separate phylogenetic clusters as well as a subdivision of the phylotype II into two subpopulations (IIa and IIb); however, this subdivision may also depend on the gene used to infer the phylogeny. Phylotype II subgroups showed a lower number of fixed polymorphisms than main clusters, suggesting that they split recently and only group IIa is arranged in clonal complexes in contrast to group IIb.
Phylogenetic inference and statistical tests applied to find evidence for population subdivision suggest that each phylotype is genetically different and this variation resulted from a diverging evolution. Values similar to those of the R. solanacearum average nucleotide divergence (Dxy) were obtained among two clades of Pseudomonas viridiflava (15) and three P. syringae pathovars (54). The R. solanacearum population divergence is explained by geographically restricted gene flow, where the geographic isolation has played a crucial role. Usually, it is uncommon that geographic isolation could shape the population structure of bacteria, since population isolation events in nature have rarely been observed (41). However, the data assessed in this work suggest that the physical isolation and spatial distance have been the significant and primary determinants of genetic variation between phylotypes, which co-occurred together with other evolutionary forces to structure the genetic variation of the R. solanacearum population.
It is difficult to state when the divergence of R. solanacearum phylotypes from their ancestor occurred, but the remarkable accumulation of fixed polymorphisms, the occurrence of SLV and DLV, and the restriction of gene flow indicate that R. solanacearum is an ancient pathogen. Furthermore, genome analysis suggests that the chromosome and megaplasmid coevolved long enough ago for the megaplasmid to acquire several duplications of housekeeping genes and a few RNAs and for the base composition (GC content, 65 to 67%) of both replicons to become homogenized (13).
The genetic diversity of R. solanacearum is striking. The substantial pathogenic variability in host range and aggressiveness and the ample adaptation to diverse ecological niches, together with genetic estimations like the number of alleles per loci, the nucleotide diversity, and the abundance of singletons indicate that R. solanacearum is a bacterium that is significantly diverse. We would expect high levels of recombination acting on this organism, since recombination is one of the most important cellular processes that increase the genetic variability in bacterial genomes (56). However, data analyzed in this work demonstrate that R. solanacearum is essentially clonal. How is it possible to see such diversity within a clonal population? We can consider some explanations for such patterns that seem mutually incompatible. This organism has evolved in geographical isolation, and it can occupy different niches, suggesting the possibility of a notable geographical structure. Frequent recombination could have taken place within each geographically isolated subpopulation, between which gene flow is limited (35). For instance, we saw higher recombination within phylotype III and IV strains for egl and hrpB loci, respectively, but there was no evidence that these two genes are recombining with their counterparts from other phylotypes. An alternative or perhaps complementary scenario could be that subpopulations have been composed of a large number of relatively rare genotypes that are recombining at high frequency. These highly recombinant entities would be the “founding genotypes” for the whole population. Some genotypes that have acquired selective advantages have arisen from the founding genotypes as clones (very closely related genotypes). After emerging, the clonal complexes could be subject to strong selective pressures or to repeated bouts of periodic selection and usually compete against preexisting subpopulations to occupy the same geographical niche (8). According to our results, phylotypes III and IV are the most diverse groups, suggesting that they could be the founding genotypes or preceding groups. Phylotype III has no clonal complexes but instead is composed of many singletons. It has been geographically restricted mainly to Africa and may have evolved a long time ago. Additionally, some of its genes are experiencing high levels of recombination and positive selection. Conversely, phylotypes I and II are arranged in clonal complexes and have spread successfully worldwide. This may be due to the cold tolerance of R3B2 strains (phylotype IIa). In fact, currently there are some reports about the emergence of new strains that may eventually overrun the former population of R. solanacearum (strains from phylotype II in Martinique [66], race 1 in Egypt [1], and biovar 2 in Peru [31]).
The finding that R. solanacearum is an essentially clonal organism agrees with current knowledge about the genome and biology of this bacterium. The genome of R. solanacearum contains regions of close similarity interspersed with divergent regions (a mosaic structure). This structure is evidenced by regions with different GC content disseminated over the two replicons and often associated with mobile genetic elements (insertion sequences and bacteriophage [53]). Thus, these regions have arisen through lateral gene transfer. The mosaic structure is possible to detect only if recombination events have not been frequent enough to mix the different gene sequences within the population, obscuring the signal of GC-biases in the sequence (35).
Usually, genes encoding vital metabolic enzymes are subject to strong levels of purifying selection, while other gene loci that may elicit host resistance response are under diversifying selection (11). We found this phenomenon in our data. Our data suggest that the housekeeping genes fliC and hrpB have been subjected to purifying selective pressures in all phylotypes. The high degree of clonality we observed in some groups may be indicative of a recent population selective sweep or bottleneck. Similarly, purifying pressures have constrained egl in most phylotypes but not in all: this locus is under diversifying selection in phylotypes I and IV. Since the egl gene product, a plant cell wall-degrading exported enzyme, is directly involved in pathogenesis (29), diversifying selection may help to accelerate the accumulation of new divergent alleles to evade the host surveillance system. Successful alleles may then be fixed in the population by a positive selection mechanism.
Acknowledgments
We are grateful to all the scientists from the international community (C. Allen, N. Furuya, M. Horita, T. Coutinho, C. Boucher, and M. Vaneechoutte) who provided DNA for this study. We thank Carlos Medina, Gail Teitzel, Erica Goss, and especially David Guttman for critical comments on the manuscript.
This work was supported by a supplement to a subcontract from National Science Foundation grant 00RA6325-DBI 0211923 from the Plant Genome Program.
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Aly, M. M., and N. Y. A. El Ghafar. 2000. Bacterial wilt of artichoke caused by Ralstonia solanacearum in Egypt. Plant Pathol. 49:807. [Google Scholar]
- 2.Buddenhagen, I., L. Sequeira, and A. Kelman. 1962. Designation of races in Pseudomonas solanacearum. Phytopathology 52:726. [Google Scholar]
- 3.Coenye, T., E. Falsen, M. Vancanneyt, B. Hoste, J. R. W. Govan, K. Kersters, and P. Vandamme. 1999. Classification of Alcaligenes faecalis-like isolates from the environment and human clinical samples as Ralstonia gilardii sp. nov. Int. J. Syst. Bacteriol. 49:405-413. [DOI] [PubMed] [Google Scholar]
- 4.Coenye, T., and P. Vandamme. 2003. Simple sequence repeats and compositional bias in the bipartite Ralstonia solanacearum GMI1000 genome. BMC Genomics 4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Baere, T., S. Steyaert, G. Wauters, P. De Vos, J. Goris, T. Coenye, T. Suyama, G. Verschraegen, and M. Vaneechoutte. 2001. Classification of Ralstonia pickettii biovar 3/′Thomasii' strains (Pickett, 1994) and of new isolates related to nosocomial recurrent meningitis as Ralstonia mannitolylitica sp. nov. Int. J. Syst. Evol. Microbiol. 51:547-558. [DOI] [PubMed] [Google Scholar]
- 6.Excoffier, L., G. Laval, and S. Schneider. 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. 1:47-50. [PMC free article] [PubMed] [Google Scholar]
- 7.Fegan, M., and P. Prior. 2005. How complex is the Ralstonia solanacearum species complex? p. 449-461. In C. Allen, P. Prior, and A. C. Hayward (ed.), Bacterial wilt disease and the Ralstonia solanacearum species complex. American Phytopathological Society Press, St. Paul, MN.
- 8.Feil, E. J. 2004. Small change: keeping pace with microevolution. Nat. Rev. Microbiol. 2:483-495. [DOI] [PubMed] [Google Scholar]
- 9.Feil, E. J., E. C. Holmes, D. E. Bessen, M.-S. Chan, N. P. J. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feil, E. J., and B. G. Spratt. 2001. Recombination and the population structures of bacterial pathogens. Annu. Rev. Microbiol. 55:561-590. [DOI] [PubMed] [Google Scholar]
- 12.Gabriel, D., C. Allen, M. Schell, T. P. Denny, J. T. Greenberg, Y. P. Duan, Z. Flores-Cruz, Q. Huang, J. M. Clifford, G. Presting, E. T. González, J. Reddy, J. Elphinstone, J. Swanson, J. Yao, V. Mulholland, L. Liu, W. Farmerie, M. Patnaikuni, B. Balogh, D. Norman, A. Alvarez, J. A. Castillo, J. Jones, G. Saddler, T. Walunas, A. Zhukov, and N. Mikhailova. 2006. Identification of open reading frames unique to a select agent: Ralstonia solanacearum race 3 biovar 2. Mol. Plant-Microbe Interact. 19:69-79. [DOI] [PubMed] [Google Scholar]
- 13.Genin, S., and C. Boucher. 2004. Lessons learned from the genome analysis of Ralstonia solanacearum. Annu. Rev. Phytopathol. 42:107-134. [DOI] [PubMed] [Google Scholar]
- 14.Gillings, M. R., and P. Fahy. 1994. Genomic fingerprinting: towards a unified view of the Pseudomonas solanacearum species complex, p. 95-112. In A. C. Hayward and G. L. Hartman (ed.), Bacterial wilt: the disease and its causative agent, Pseudomonas solanacearum. CAB International, Wallingford, United Kingdom.
- 15.Goss, E. M., M. Kreitman, and J. Bergelson. 2005. Genetic diversity, recombination and cryptic clades in Pseudomonas viridiflava infecting natural populations of Arabidopsis thaliana. Genetics 169:21-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 17.Haubold, B., and R. R. Hudson. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16:847-848. [DOI] [PubMed] [Google Scholar]
- 18.Haubold, B., M. Travisano, P. B. Rainey, and R. R. Hudson. 1998. Detecting linkage disequilibrium in bacterial populations. Genetics 150:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayward, A. C. 1964. Characteristics of Pseudomonas solanacearum. J. Appl. Bacteriol. 27:265-277. [Google Scholar]
- 20.Hayward, A. C. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29:65-87. [DOI] [PubMed] [Google Scholar]
- 21.Hayward, A. C. 1994. Systematics and phylogeny of Pseudomonas solanacearum and related bacteria, p. 123-135. In A. C. Hayward and G. L. Hartman (ed.), Bacterial wilt: the disease and its causative agent, Pseudomonas solanacearum. CAB International, Wallingford, United Kingdom.
- 22.Hudson, R. R., D. D. Boos, and N. L. Kaplan. 1992. A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 9:138-151. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda, T., K. Oosawa, and H. Hotani. 1996. Self-assembly of the filament capping protein, FLID, of bacterial flagella into an annular structure. J. Mol. Biol. 259:679-686. [DOI] [PubMed] [Google Scholar]
- 24.Jaunet, T. X., and J.-F. Wang. 1999. Variation in genotype and aggressiveness of Ralstonia solanacearum race 1 isolated from tomato in Taiwan. Phytopathology 89:320-327. [DOI] [PubMed] [Google Scholar]
- 25.Jolley, K. A., E. J. Feil, M.-S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 26.Kalia, A., A. K. Mukhopadhyay, G. Dailide, Y. Ito, T. Azuma, B. C. Y. Wong, and D. E. Berg. 2004. Evolutionary dynamics of insertion sequences in Helicobacter pylori. J. Bacteriol. 186:7508-7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelman, A. 1954. The relationship of pathogenicity in Pseudomonas solanacearum to colony appearance on a tetrazolium medium. Phytopathology 44:693-695. [Google Scholar]
- 28.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 29.Liu, H., S. Zhang, M. A. Schell, and T. P. Denny. 2005. Pyramiding unmarked deletions in Ralstonia solanacearum shows that secreted proteins in addition to plant cell-wall-degrading enzymes contribute to virulence. Mol. Plant-Microbe Interact. 18:1296-1305. [DOI] [PubMed] [Google Scholar]
- 30.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. S. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marin, J. E., and H. M. El-Nashaar. 1993. Pathogenicity and new phenotypes of Pseudomonas solanacearum from Peru. ACIAR Proc. 45:78-84. [Google Scholar]
- 32.Martin, D., and E. Rybicki. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562-563. [DOI] [PubMed] [Google Scholar]
- 33.Martin, D., C. Williamson, and D. Posada. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260-262. [DOI] [PubMed] [Google Scholar]
- 34.Maynard Smith, J. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126-129. [DOI] [PubMed] [Google Scholar]
- 35.Maynard Smith, J., E. J. Feil, and N. H. Smith. 2000. Population structure and evolutionary dynamics of pathogenic bacteria. BioEssays 22:1115-1122. [DOI] [PubMed] [Google Scholar]
- 36.Maynard Smith, J., and H. H. Smith. 1998. Detecting recombination from gene trees. Mol. Biol. Evol. 15:590-599. [DOI] [PubMed] [Google Scholar]
- 37.Maynard Smith, J., N. H. Smith, M. O’Rourke, and B. S. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Occhialini, A., S. Cunnac, N. Reymond, S. Genin, and C. Boucher. 2005. Genome-wide analysis of gene expression in Ralstonia solanacearum reveals that the hrpB gene acts as a regulatory switch controlling multiple virulence pathways. Mol. Plant-Microbe Interact. 18:938-949. [DOI] [PubMed] [Google Scholar]
- 39.Padidam, M., S. Sawyer, and C. M. Fauquet. 1999. Possible emergence of new geminiviruses by frequent recombination. Virology 265:218-225. [DOI] [PubMed] [Google Scholar]
- 40.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 41.Papke, T. R., and D. M. Ward. 2004. The importance of physical isolation to microbial diversification. FEMS Microbiol. Ecol. 48:293-303. [DOI] [PubMed] [Google Scholar]
- 42.Perez-Losada, M., R. P. Viscidi, J. C. Demma, J. Zenilman, and K. A. Crandall. 2005. Population genetics of Neisseria gonorrhoeae in a high-prevalence community using a hypervariable outer membrane porB and 13 slowly evolving housekeeping genes. Mol. Biol. Evol. 22:1887-1902. [DOI] [PubMed] [Google Scholar]
- 43.Pfund, C., J. Tans-Kersten, F. M. Dunning, J. M. Alonso, J. R. Ecker, C. Allen, and A. F. Bent. 2004. Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana. Mol. Plant Microbe Interact. 17:696-706. [DOI] [PubMed] [Google Scholar]
- 44.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 45.Posada, D., and K. A. Crandall. 2001. Selecting the best-fit model of nucleotide substitution. Syst. Biol. 50:580-601. [PubMed] [Google Scholar]
- 46.Posada, D., and K. A. Crandall. 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. USA 98:13757-13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poussier, S., P. Prior, J. Luisetti, C. Hayward, and M. Fegan. 2000. Partial sequencing of the hrpB and endoglucanase genes confirms and expands the known diversity within the Ralstonia solanacearum species complex. Syst. Appl. Microbiol. 23:479-486. [DOI] [PubMed] [Google Scholar]
- 48.Poussier, S., D. Trigalet-Demery, P. Vandewalle, B. Goffinet, J. Luisetti, and A. Trigalet. 2000. Genetic diversity of Ralstonia solanacearum as assessed by PCR-RFLP of the hrp gene region, AFLP and 16S rRNA sequence analysis and identification of an African subdivision. Microbiology 146:1679-1692. [DOI] [PubMed] [Google Scholar]
- 49.Prior, P., and M. Fegan. 2005. Recent developments in the phylogeny and classification of Ralstonia solanacearum. Acta Hortic. 695:127-136. [Google Scholar]
- 50.Rand, D. M. 1996. Neutrality tests of molecular markers and the connection between DNA polymorphism, demography, and conservation biology. Conserv. Biol. 10:665-671. [Google Scholar]
- 51.Relethford, J. H. 2004. Global patterns of isolation by distance based on genetic and morphological data. Hum. Biol. 76:499-513. [DOI] [PubMed] [Google Scholar]
- 52.Rozas, J., J. C. Sánchez-Delbarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 53.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangeno, M. Arlat, A. Billault, P. Brottier, J. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demang, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schie, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 54.Sarkar, S. F., and D. S. Guttman. 2004. Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl. Environ. Microbiol. 70:1999-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schönfeld, J., H. Heuer, J. D. van Elsas, and K. Smalla. 2003. Specific and sensitive detection of Ralstonia solanacearum in soil on the basis of PCR amplification of fliC fragments. Appl. Environ. Microbiol. 69:7248-7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spratt, B. G., W. P. Hanage, and E. J. Feil. 2001. The relative contributions of recombination and point mutation to the diversification of bacterial clones Curr. Opin. Microbiol. 4:602-606. [DOI] [PubMed] [Google Scholar]
- 57.Spratt, B. G., W. P. Hanage, B. C. Li, D. M. Aanensen, and E. J. Feil. 2004. Displaying the relatedness among isolates of bacterial species—the eBURST approach. FEMS Microbiol. Lett. 241:129-134. [DOI] [PubMed] [Google Scholar]
- 58.Swofford, D. L. 2003. PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4.0. Sinauer Associates, Sunderland, MA.
- 59.Taghavi, M., C. Hayward, L. I. Sly, and M. Fegan. 1996. Analysis of the phylogenetic relationships of strains of Burkholderia solanacearum, Pseudomonas syzygii, and the blood disease bacterium of banana based on 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 46:10-15. [DOI] [PubMed] [Google Scholar]
- 60.Tajima, F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tans-Kersten, J., H. Huang, and C. Allen. 2001. Ralstonia solanacearum needs motility for invasive virulence on tomato. J. Bacteriol. 183:3597-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vaneechoutte, M., P. Kämpfer, T. De Baere, E. Falsen, and G. Verschraegen. 2004. Wautersia gen. nov., a novel genus accommodating the phylogenetic lineage including Ralstonia eutropha and related species, and proposal of Ralstonia [Pseudomonas] syzygii (Roberts et al. 1990) comb. nov. Int. J. Syst. Evol. Microbiol. 54:317-327. [DOI] [PubMed] [Google Scholar]
- 64.Villa, J. E., K. Tsuchiya, M. Horita, N. Opina, and M. Hyakumachi. 2005. Phylogenetic relationships of Ralstonia solanacearum species complex strains from Asia and other continents based on 16S rDNA, endoglucanase, and hrpB gene sequences. J. Gen. Plant Pathol. 71:39-46. [Google Scholar]
- 65.Wayne, M. L., and K. L. Simonsen. 1998. Statistical tests of neutrality in the age of weak selection. Trends Ecol. Evol. 13:236-240. [DOI] [PubMed] [Google Scholar]
- 66.Wicker, E., L. Grassart, R. Coranson-Beaudu, D. Mian, C. Guilbaud, and P. Prior. 2005. Emerging strains of Ralstonia solanacearum in Martinique (French West Indies): a case study for epidemiology of bacterial wilt. Acta Hortic. 695:145-152. [Google Scholar]
- 67.Williamson, L., B. D. Hudelson, and C. Allen. 2002. Ralstonia solanacearum strains isolated from geranium belong to race 3 and are pathogenic on potato. Plant Dis. 86:987-991. [DOI] [PubMed] [Google Scholar]
- 68.Wilson, K. 1994. Preparation of genomic DNA from bacteria, p. 2.4.1-2.4.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, New York, NY. [DOI] [PubMed]