Abstract
Folding of substrate proteins inside the sequestered and hydrophilic GroEL-GroES cis cavity favors production of the native state. Recent studies of GroEL molecules containing volume-occupying multiplications of the flexible C-terminal tail segments have been interpreted to indicate that close confinement of substrate proteins in the cavity optimizes the rate of folding: the rate of folding of a larger protein, Rubisco (51 kDa), was compromised by multiplication, whereas that of a smaller protein, rhodanese (33 kDa), was increased by tail duplication. Here, we report that this latter effect does not extend to the subunit of malate dehydrogenase (MDH), also 33 kDa. In addition, single-ring versions of tail-duplicated and triplicated molecules, comprising stable cis complexes, did not produce any acceleration of folding of rhodanese or MDH, nor did they show significant retardation of the folding of Rubisco. Tail quadruplication produced major reduction in recovery of native protein with both systems, the result of strongly reduced binding of all three substrates. When steady-state ATPase of the tail-multiplied double-ring GroELs was examined, it scaled directly with the number of tail segments, with more than double the normal ATPase rate upon tail triplication. As previously observed, disturbance of ATPase activity of the cycling double-ring system, and thus of “dwell time” for the folding protein in the cis cavity, produces effects on folding rates. We conclude that, within the limits of the ≈10% decrease of cavity volume produced by tail triplication, there does not appear to be an effect of “close confinement” on folding in the cis cavity.
Keywords: chaperonin, protein folding, single ring
The bacterial chaperonin GroEL is an essential double-ring assembly that assists folding of a large variety of newly translated proteins to native form, which it accomplishes through two major actions (1–4). One action involves binding of nonnative protein via exposed hydrophobic surfaces, preventing aggregation and potentially unfolding nonnative protein. As indicated by aggregation of a large collective of proteins in a severe conditional GroEL mutant strain, many newly translated proteins, including large ones, may be at least transiently bound by GroEL (ref. 5; see also refs. 6 and 7). For substrates that are more tightly bound, however, presumably via a greater amount of exposed hydrophobic surface (8), a subsequent step of ATP/GroES binding to the substrate-bound ring ejects the substrate off the cavity wall into a now GroES-encapsulated and hydrophilic so-called cis cavity, where folding commences (9–12). The reaction proceeds for ≈10 seconds before ATP hydrolysis in the cis ring followed by trans ring ATP binding ejects the cis ligands, including substrate polypeptide whether folded or not (11, 13, 14). For many such GroEL-GroES-dependent substrates, only a small fraction of GroEL-bound molecules reach the native state in any given round of this cycle, and further rounds of binding followed by attempted folding are required.
The nature of the folding reaction in the cis cavity is of considerable interest, particularly considering that for “stringent”, GroEL-GroES-dependent, substrates, unable to properly fold spontaneously in solution (15), the cis cavity is essential for reaching the native state (16). At least a dozen such bacterial proteins, many of them providing essential function, have been identified (17, 18). It seems clear that a major favorable feature of the cis cavity is sequestration, with solitary confinement of substrate in this space preventing multimolecular aggregation from occurring. The hydrophilic character of the cavity walls may also be supportive, potentially favoring burial of exposed hydrophobic surfaces and exposure of hydrophilic ones in the folding protein, properties of the native state (12). The benefit of this wall character has not, however, been formally demonstrated. Indeed, several experiments carrying out substitutions of charged, predominantly acidic, residues in the cis cavity wall to neutral character have shown no general effect in vitro (19) nor have such substitutions produced any effect on cell viability in vivo [e.g., see supporting information (SI) Fig. 6]. But are there other properties of the cis cavity that may generally facilitate productive folding?
A recent study has suggested that “close confinement” of nonnative substrate protein in the cis cavity is critical to producing an optimum rate of folding to the native state in the enclosed space (19). It was postulated that the volume of the cis cavity (≈160,000 Å3) would provide optimum confinement for “larger” substrate proteins of ≈40–50 kDa, e.g., Rubisco from Rhodospirillum rubrum (51 kDa), and that any decrease of cavity volume would lead to physical restriction of the folding protein and thus to a decrease in rate of folding. Moreover, for such proteins, an increase of volume would also be expected to reduce the rate of cis folding due to reduced, nonoptimum confinement. Conversely, it was hypothesized that relatively “small” proteins of ≈30 kDa, such as the well studied substrate bovine rhodanese (33 kDa), would experience an acceleration of folding if the volume of the cavity was decreased, i.e., if these substrates were more closely confined. With progressive decrease of the cavity volume, an optimum confinement would, of course, be reached, beyond which there would be physical restriction of space needed for folding, and the rate of reaching native form would then be reduced. Experimentally, the volume of the cavity was manipulated by multiplying or removing the flexible C-terminal tails of the GroEL subunits, which extend into the cavity from each equatorial domain as a GGM × 4 repeat from residue Ala-535 to the terminal residue, Met-548. The C-terminal tails are not crystallographically ordered, but the collective within a ring (a net molecular mass of ≈10 kDa) is visualized by EM studies as a mass in the central cavity at the level of the equatorial domains (20). Each increment of tail multiplication was predicted to reduce the volume of the cis cavity by ≈4%. Data supporting the foregoing models of folding rate behavior were presented, comparing rates of reaching the native state for the well-studied GroEL-GroES-dependent substrates, rhodanese and Rubisco, as well as for two other proteins. Rates of folding of these substrates were reported for cycling reactions, carried out with various tail-multiplied versions of double-ring GroEL, GroES, and ATP. Here, we test the conclusions drawn from that study, using reactions carried out with tail-multiplied versions of SR1, a single-ring version of GroEL that reports specifically on the cis folding phase of the chaperonin reaction, where the proposed effects on cis folding would be expected to be observed.
Results
Effects of Volume Reduction of the cis Cavity on Folding Rate, Using Tail-Multiplied Versions of SR1.
Cycling chaperonin reactions with tail-multiplied double-ring GroELs were reported on in ref. 19. In addition to cis folding, such reactions have ongoing phases of substrate protein release and rebinding. The proposed effects of close physical confinement by C-terminal tail multiplication, however, would be exerted specifically during the cis folding phase of the reaction cycle, during which the GroES “lid” structure encapsulates the folding substrate protein. If this phase of the reaction is the specific site of action, then it would be predicted that the effects observed with the double-ring system should be reproduced by the single-ring (SR1) system. Here, upon the addition of ATP and GroES, the reaction is confined strictly to the cis phase of the reaction cycle, with polypeptide folding proceeding to completion inside a stable, long-lived, cis SR1-GroES folding chamber (9). This chamber, formed after the addition of ATP and GroES to a substrate-SR1 binary complex, is stable because of the absence of the opposite GroEL ring, which normally provides the allosteric GroES ejection signal upon ATP binding (11, 21). The kinetics of folding to native form of stringent (GroEL-GroES-dependent) proteins such as rhodanese, malate dehydrogenase (MDH), and Rubisco in the stable SR1–GroES complexes have been observed to be virtually identical to those in the cycling GroEL-GroES reaction (9, 11), in the latter of which the ongoing nucleotide cycle drives regular departure of GroES and substrate protein into the bulk solution at ≈10 sec intervals (13). Thus, if cavity volume alterations exerted by changes of GroEL tail length are specifically affecting the rate of cis folding in the GroEL-GroES reaction, they would be expected to likewise affect the rates of folding inside SR1–GroES complexes. To test this hypothesis, we prepared versions of SR1 containing the various multiples of GroEL C-terminal tails that had been previously reported (designated here by T and the total number of tails).
No Acceleration of cis Folding of Rhodanese or MDH Mediated by Tail-Multiplied SR1 and GroES.
Binary complexes were formed between the various tail-multiplied SR1 molecules and three different stringent substrates, rhodanese, MDH, and Rubisco, and refolding was carried out by adding ATP and GroES. Recovery of enzymatic activity was measured at various times after initiating the reaction. For MDH and Rubisco, a brief cold temperature exposure was required at each of these times, as in previous kinetic studies with SR1 (11), to release GroES and substrate from the SR-GroES complexes and allow assembly of the folded subunits into the respective native homodimers. Figs. 1–3 show representative experiments for each of the three substrates. Each experiment was conducted at least three times, with rates and extents of recovery falling in all cases within 20% of that of the experiment shown (SI Table 1). In all experiments, the recovery data could be fit to a single exponential curve, from which a rate constant was derived.
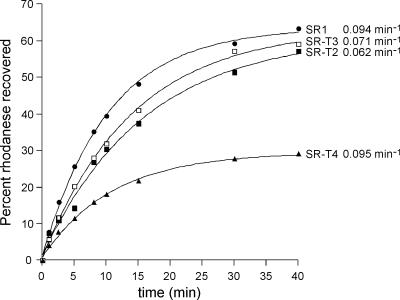
Fig. 1.
No acceleration of rhodanese refolding by tail-multiplied variants of SR1. Guanidine-HCl-denatured rhodanese was diluted into buffer containing the indicated chaperonin to form a binary complex. GroES and ATP were added, and at the indicated times, the recovery of native protein was measured by assay of rhodanese enzymatic activity. The amount of native rhodanese recovered is expressed as a percentage of the total input rhodanese.
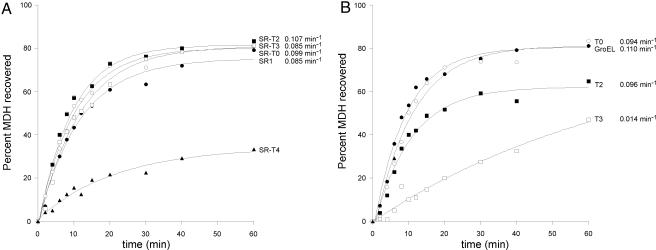
Fig. 2.
No acceleration of MDH refolding by tail-multiplied versions of either SR1 or GroEL. MDH denatured in guanidine-HCl was diluted into buffer containing the indicated single-ring (A) or double-ring (B) chaperonin to form binary complex. GroES and ATP were added, and at the indicated times, the recovery of native MDH was measured by assay of MDH enzyme activity. The yield of MDH activity recovered is expressed as a percentage of the initial MDH added to the reaction.
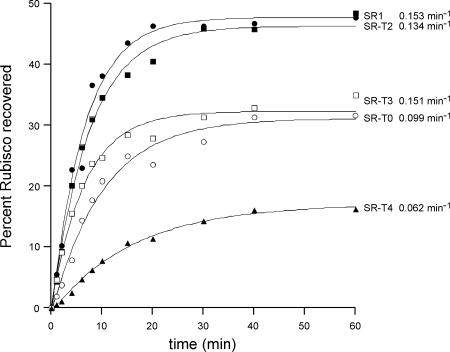
Fig. 3.
Rate of Rubisco refolding is not affected by tail-duplication or triplication in SR1. Guanidine-HCl-denatured Rubisco was diluted into buffer containing the indicated chaperonin. GroES and ATP were added, and the amount of native Rubisco recovered at the indicated times was determined by enzymatic assay. The recoveries are expressed as a percentage of the total input Rubisco.
As observed from the recovery data and derived rate constants for the three substrates, in no case was any significant acceleration of the rate of folding observed for tail-multiplied SR1 molecules when compared with the rate observed for SR1 itself (Figs. 1–3). In the case of rhodanese (Fig. 1), where a 50% acceleration had been reported for double-ring GroEL containing a duplicated tail (ref. 19; also observed here; SI Table 1), duplication and triplication of the tails of SR1 (SR-T2 and SR-T3) led to a modest decrease in the rate of folding. Quadruplication of the tails (SR-T4) produced a rate identical to SR1, although the extent of recovery was substantially reduced (see below). Likewise, in the case of MDH, another small stringent substrate whose subunit has the same mass as rhodanese, 33 kDa, no significant acceleration of folding was observed with duplication or triplication of the tails in SR1 (Fig. 2A); the rate constants for folding by SR-T2 and SR-T3 were all within 20% of the SR1-mediated reaction, approximating the range of experimental error. Deletion of the tails (T0) was without effect on the rate of recovery. Finally, as with rhodanese, the quadruplication of the tails in SR1 led to a strong reduction in the extent of MDH recovery (see below).
Rate of MDH Refolding Is Also Not Accelerated by Tail-Multiplied Double-Ring GroELs.
Because there were no effects of duplication, triplication, or deletion of the GroEL tails upon the rate of recovery of MDH by single-ring molecules (Fig. 2A), we tested whether any effects could be observed with tail-modified double-ring GroELs. As shown in Fig. 2B, here also no acceleration was observed. For example, tail duplication (GroEL-T2), in contrast with the 50% acceleration of recovery observed for rhodanese, produced virtually the same rate of folding of MDH as wild-type. By contrast, triplication (GroEL-T3) led to reduction of the rate of recovery, and quadruplication led to essentially no recovery (data not shown). Meanwhile, similar to the single-ring context, deletion of the tails altogether was without effect on MDH refolding. We conclude that, whatever the basis to the effects of tail duplication in double-ring GroEL on modestly increasing the rate of rhodanese recovery, it does not obtain for MDH. That is, such acceleration in the double-ring context is not a general effect.
No Inhibition of cis Folding Rate of the Large Substrate Protein Rubisco by Tail-Multiplication in SR1; No Evidence for “Tight” Volume Restriction.
The third stringent substrate examined was the large protein Rubisco, whose subunit mass, 51 kDa, suggests that it occupies a substantial volume of the cis cavity. The reported studies with double-ring GroEL indicated that multiplication of the GroEL tails led to a strong decrease in the rate of folding, with simple duplication reducing the rate of folding to ≈20% of wild-type (19). Here in SR1, in contrast, no such “critical” behavior of the tails was observed. When they were either duplicated or triplicated, the rate constant for folding was virtually identical, with a moderate reduction in extent of recovery for the triplicated SR-T3 (to ≈70% that of SR1). Quadruplication once again had a strong effect on the extent of recovery, reducing it to ≈30% that of SR1, with a similar effect observed on rate.
In the direction of increasing the cavity volume, deletion of the tails of double-ring GroEL was reported to cause a moderate reduction (50%) in the rate of Rubisco folding (19), and the same was observed here for SR1. Whether this is a volume effect or, instead, the result of a loss of an interacting “barrier” (i.e., the hydrophobic tails) cannot at present be resolved. In summary, however, duplication and triplication of the tails in SR1 was essentially without effect on Rubisco folding rate, whereas deletion had a moderate effect. These results do not seem consistent with the proposed critical volume requirement for cis folding of large proteins, at least within the limits of contracting the volume for folding by ≈8–10%, as is achieved with tail triplication.
Finally, we attempted to reproduce the reported effects on Rubisco refolding in the context of tail-multiplied double-ring GroEL, but did not observe such behavior (SI Table 1). Duplication of the tails produced no change of rate constant, whereas triplication produced a moderate reduction (by ≈40%). Once again, as for all of the substrates, quadruplication (GroEL-T4) drastically reduced extent of recovery (see next section).
Quadruplication of the Tails of Either GroEL or SR1 Leads to Strongly Reduced Extent of Binding Nonnative Substrate Proteins.
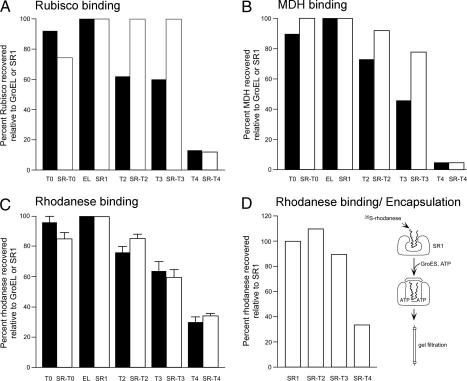
A reduced extent of recovery was observed for all three of the foregoing substrates, rhodanese, MDH, and Rubisco, when tested with quadruplicated tail molecules in either the double-ring (GroEL-T4) or single-ring (SR-T4) contexts (Figs. 1–3). This result could be consistent with “stuffing” the central cavity to a degree that a fraction of each of the substrate proteins was arrested in folding. However, considering the significantly different sizes of the various substrate proteins, it seemed unlikely that all three proteins would be similarly affected in the cis folding phase of the reaction. It seemed possible instead that the defect could lie at the level of initial substrate binding to the open rings of T4 chaperonins, for example, if the T4 tails interacted with the GroEL apical domains and competed for substrate binding. Failure of such initial binding would lead to irreversible misfolding and aggregation of the unbound fraction of molecules, and this result would be mirrored in the refolding reaction as reduced extent of recovery. To evaluate this result, we directly measured the association of the three substrates, Rubisco, MDH, and rhodanese, with the entire series of GroEL and SR1 tail-multiplied constructs (Fig. 4 A–C). 35S-substrate diluted from denaturant was allowed to bind to the various chaperonin molecules, and the mixture was separated by gel filtration. We compared the amount of substrate protein bound by the tail-multiplied mutants with that bound by SR1 or GroEL (EL), respectively. Reproducibly, the T4 molecules exhibited strong reduction of binding of all three substrates, achieving a level 5–30% of that of the other constructs. This result correlated with an in vivo rescue experiment in which all of the tail-multiplied (and tail-deleted) GroEL-encoding plasmids except GroEL-T4 could efficiently rescue growth of a GroEL-deficient Escherichia coli strain (LG6; see ref 22). No colonies were produced by the GroEL-T4 plasmid.
Fig. 4.
Tail quadruplication greatly reduces substrate binding to both SR1 and GroEL. (A) Rubisco binding. (B) MDH binding. (C) Rhodanese binding. (D) Rhodanese binding and encapsulation. 35S-labeled Rubisco (A), MDH (B), or rhodanese (C and D) were denatured in guanidine-HCl and diluted into buffer containing the indicated chaperonin. In A–C, binary complexes were purified by gel-filtration chromatography, and the amount of substrate protein recovered was compared with that for GroEL (filled bars) or SR1 (open bars). In D, GroES and ATP were added before gel filtration to encapsulate bound 35S-rhodanese, and gel-filtration was carried out in the presence of 1 mM ADP. Error bars in C represent the standard deviation for three experiments.
The concern could be raised that the reduced binding by T4 molecules in part reflects that the gel filtration resin itself might compete for nonnative substrate protein that had initially been bound by the chaperonin. To address this concern, a further set of experiments was carried out with the SR1 series of molecules and 35S-rhodanese. After initial formation of the respective binary complexes, they were incubated with ATP and an excess of GroES, allowing the initially bound substrate to become encapsulated inside the SR1-GroES cis folding chamber before application of the mixture to the gel filtration column. Once encapsulated in this location, the substrate would be protected from interaction with the gel filtration resin, such that all of the input radioactive molecules that became initially bound in solution would be recovered at the elution position of SR1-GroES. The results obtained (Fig. 4D) were essentially identical to those in the absence of encapsulation, with SR1, SR-T2, and SR-T3 complexes all recovered with 90% or more of the input rhodanese molecules, whereas the SR-T4 complex was recovered with ≈30% that level, corresponding to the same fraction bound without a prior encapsulation step (compare Fig. 4C with Fig. 4D).
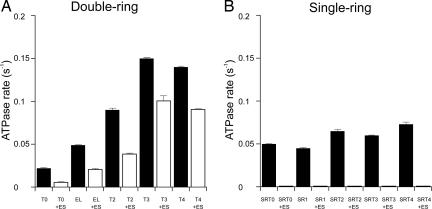
Tail Multiplication Incrementally Accelerates the ATPase Rate of GroEL and GroEL-GroES.
In earlier studies of a fusion protein joining the B subunit of the sweet protein monellin (51 aa) at the C termini of the GroEL subunits (total tail length, 68 aa), we had observed not only a substrate binding defect resembling that described above for T4 molecules (extended by 51 aa), but also an acceleration of the rate of ATP turnover of the fused GroEL–monellin complex (G.W.F., unpublished data). We thus investigated the steady-state rates of ATP turnover of the various tail-multiplied GroELs. As shown in Fig. 5A, the rate of steady-state ATP turnover was significantly affected by tail multiplication, approximately scaling with the multiplied length of the C-terminal tails up to triplication. For example, the rate increased by ≈50% for GroEL-T2 compared with GroEL and increased by more than two-fold over GroEL for T3. Concordantly, deletion of the tails reduced the rate of turnover by ≈50% as compared with GroEL. The addition of GroES, as in refolding reactions, reduced the rate of steady-state ATP turnover of wild-type GroEL by ≈50%, as observed reproducibly by many laboratories (e.g., ref. 23). Correspondingly, GroES addition reduced steady-state ATP turnover for each tail-multiplied complex by ≈25–50%. Overall, however, in the presence of GroES, the steady-state turnover maintained the scaled behavior with respect to tail length.
Fig. 5.
The steady-state ATPase activity of GroEL and GroEL-GroES scales with the number of C-terminal tails, whereas GroES completely suppresses the activity of tail-multiplied SR1. The rate of ATP hydrolysis for tail-multiplied versions of GroEL (A) and SR1 (B) was determined by the malachite green assay in the absence (filled bars) or presence (open bars) of GroES, using 1 mM ATP. Error bars represent the standard deviation in three experiments.
Discussion
Consequences of Accelerated ATP Turnover.
What are the effects of accelerating the rate of ATP turnover in the cycling tail-multiplied GroEL–GroES reactions by as much as three-fold? Earlier studies have shown that the rate-limiting step of the wild-type GroEL–GroES reaction, in the presence of nonnative substrate protein, is the step of ATP hydrolysis in a GroEL–GroES–ATP–substrate complex (24). This defines the “dwell time,” during which the substrate protein attempts to fold in the cis chamber. With an increased rate of ATP turnover in the tail-multiplied molecules, this dwell time must necessarily be decreased. Such speeding up of the GroEL cis “timer” would have significant effects on the lifetimes and population of various nonnative intermediate forms, and this in turn could have substantial effects on the rates of reaching the native state. The effects of speeding up the timer are not entirely predictable. A shortened dwell time could either disfavor formation of some unproductive intermediates, potentially accelerating the rate of folding, or disfavor formation of productive intermediates, slowing the rate of productive folding. More generally, effects of the multiplied tails on the rates of allosteric transitions and their cooperativity could lead to changes in folding rates (25).
By contrast with such perturbing effects of altering the ATPase in cycling GroEL–GroES reactions, tail multiplication of SR1 does not perturb the noncycling SR1-GroES reaction. In particular, tail-multiplied SR1 alone exhibited only modest increases of steady-state ATP turnover with tail multiplication, and adding GroES completely suppressed ATP turnover (Fig. 5B). This finding is consistent with the known single-turnover behavior of SR1-GroES, where a single event of ATP/GroES binding is followed within ≈10 sec by a single round of ATP hydrolysis in the seven equatorial ATP sites of SR1, producing a stable, long-lived, folding-active cis SR1-GroES-ADP7 chamber (9, 26). The lack of effects of tail elongation on rates of folding in the SR-GroES chamber makes clear that the volume decreases produced by multiplying the C-terminal tails, involving, in the case of triplication, an ≈10% decrease of volume, are generally insufficient to affect the folding process.
Further volume contraction of the cis cavity by tail multiplication beyond T4 was not pursued because quadruplication already largely blocked the step of polypeptide binding, presumably by association of the T4 tails with the apical binding surface of SR1 or GroEL, thus competing for binding of nonnative substrate proteins. It seems possible that at least some component of the increment of ATPase activity produced by tail multiplication in the context of double-ring GroEL or in SR1 alone, particularly for T4, involves allosteric effects of such putative interaction of the tails with the apical domains to stimulate the equatorial ATPase. This would mimic the effects seen with nonnative proteins such as casein (27) or RCMLA (28), which stimulate the steady-state ATPase activity of GroEL. As an alternative explanation for reduced binding, the quadruplicated tails may simply sterically block entry of nonnative substrate into the cavity. Importantly, we note that the volume of the open cavity of an unliganded GroEL ring is about half that of the encapsulated cis cavity, and, correspondingly, the same volume of quadruplicated tails that would occupy ≈12% of the cis cavity would occupy ≈25% of this space. Whatever the mechanism of blocked substrate binding, the point at which cis folding of any of the stringent substrates, including the largest substrate, Rubisco, becomes impaired by volume limitation of the cis cavity remains presently unknown. Likewise, whether any folding rate enhancement would be achieved before such volume restriction is also not known.
Influence of Cavity Wall vs. ATPase on Folding by GroEL-GroES.
A precedent for improved folding by GroEL-GroES has been reported, with a directed-evolution experiment in which GroEL-GroES were genetically altered by a recombinational shuffling procedure carried out to optimize the extent of GFP folding in vivo (the rate could not be measured in this context) (29). Interestingly, the characterized mutant pair with the best refolding action (mutant 3-1) combined an equatorial domain substitution of GroEL that increased ATPase activity by 50% with a GroES mutant affecting a cavity-facing residue, Y71H. The substitution affecting the ATPase, however, contributed the predominant effect, increasing GFP recovery by six-fold on its own in the company of wild-type GroES, whereas the Y71H produced alone only a two-fold effect when paired with wild-type GroEL. In the present study as well, the effects of tail multiplication on the ATPase in the double-ring system appear likely to account for most if not all of the effects on rates of folding, given that little or no effect was observed on folding rates in the single-ring system where ongoing ATP binding and turnover does not occur.
The issue thus of whether the GroEL cavity walls are active or passive in supporting cis folding appears to remain open. Evidence for an active role could come in the form of specific effects of amino acid substitution in GroEL upon folding in the cis cavity, as distinct from effects on polypeptide binding or encapsulation, necessary steps to forming a cis cavity in the first instance (see ref. 30 for further consideration). As mentioned, effects of neutralizing the predominantly negative charges that are present in the cis cavity wall were reported in in vitro studies (19), but only some substrates were affected, raising the question of whether these effects would be significant in an in vivo context. For example, in our own hands, a number of double substitutions and a triple one involving charged residues have not affected cell viability (SI Fig. 6). By contrast, two residues with hydrophobic character that face the cis cavity in GroEL–GroES complexes, F281 and L309, were shown in an earlier substitution analysis to be critical; expression plasmids carrying the substitutions F281D and L309K could not rescue GroEL-deficient E. coli (31). Interestingly, residual hydrophobicity has recently been proposed from a simulation experiment as potentially being able to facilitate production of the native state within the cis cavity (32). Further analysis of the 281 and 309 substitutions, however, does not resolve whether these residues have any significant role in the cis folding phase of the reaction cycle. For example, Yoshida and Taguchi and their coworkers have recently demonstrated (33) and we confirm (G.F., unpublished data) that L309 substitutions, including L309K, block the stable binding of GroES to substrate-GroEL binary complexes, thus preventing formation of a folding-active cis ternary complex in the first instance. This finding precludes any direct assessment of whether 309 has a role during the cis folding phase of the reaction. Our studies of F281D, by contrast, show that GroES binds to this mutant in its double ring form, but here the further turnover of ATP is blocked, simulating the behavior of SR1. Nevertheless, substrates such as rhodanese and trypsinogen are folded at normal rates. It thus remains unclear whether there is a direct and general role of the cis cavity wall beyond its overall hydrophilicity on the rate of cis folding.
Materials and Methods
Proteins.
The chaperonin variants used were produced from wild-type GroEL or SR1 by replacement of a fragment at the 3′-end of the coding sequence with appropriate synthetic double-stranded oligonucleotides encoding the desired changes. For T0, the coding sequence was terminated at amino acid 534; for the tail-multiplied species, one, two, or three iterations of (GGM)4M were added to the wild-type sequence to generate T2, T3, or T4, respectively. In addition, T2 and T3 had Ala-Ser substituted for Met-548, the terminal methionine in wild-type GroEL. Constructs were sequenced to confirm the substitutions. The chaperonins were overproduced and purified as described in ref. 26. The molecular mass of each of the purified proteins was determined by mass spectrometry and matched that calculated from the predicted amino acid sequence. In the case of the SR1 variants, the multiplied tails were determined to be localized inside the chaperonin cavity by their acquisition of resistance to digestion by proteinase K upon binding GroES in the presence of ATP. Rhodanese and Rubisco were overproduced and purified as before. Porcine MDH was obtained from Roche Applied Science (Indianapolis, IN). Substrate proteins labeled with 35S were produced by expression in the presence of 35S-TransLabel (GE Healthcare, Piscataway, NJ) as described in refs. 13 and 34.
Assays.
Rhodanese and MDH were assayed spectrophotometrically and Rubisco was assayed radiochemically as described in refs. 11 and 13. ATPase activity of the various chaperonins was determined with 1 mM ATP by using malachite green.
Binary Complex Formation and Refolding.
Binary complexes between denatured substrate proteins and the various chaperonins were formed as described in ref. 11, except that Rubisco was denatured in 6 M guanidine-HCl. In all cases, a 200-fold dilution into buffer containing a two-fold molar excess of chaperonin was performed. Refolding was carried out at a binary complex concentration of 0.5–1.0 μM as described in refs. 11 and 13 by adding a two-fold molar excess (relative to GroEL or SR1) of GroES and 5 mM ATP. Refolding data were fit to a single-exponential equation, using KaleidaGraph software. All refolding experiments were carried out at least three times.
Assay of Substrate Binding by Chaperonins.
Binary complexes formed as described above between various chaperonins and radiolabeled substrates were subjected to chromatography on a 7.8 × 300 mm G4000SWxl HPLC size exclusion column (TOSOH Biosep, Montgomeryville, PA) as described in ref. 11. Fractions were collected and analyzed by scintillation counting. For the experiments involving rhodanese encapsulated by the single-ring variants, GroES and ATP were added to the binary complexes before loading on the column, and the column buffer contained 5 mM MgCl2 and 1 mM ADP.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health and the Howard Hughes Medical Institute.
Abbreviation
- MDH
malate dehydrogenase.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700820104/DC1.
References
- 1.Thirumalai D, Lorimer GH. Annu Rev Biophys Biomol Struct. 2001;30:245–269. doi: 10.1146/annurev.biophys.30.1.245. [DOI] [PubMed] [Google Scholar]
- 2.Hartl FU, Hayer-Hartl M. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 3.Horovitz A, Willison KR. Curr Op Struct Biol. 2005;15:646–651. doi: 10.1016/j.sbi.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Horwich AL, Farr GW, Fenton WA. Chem Rev. 2006;106:1917–1930. doi: 10.1021/cr040435v. [DOI] [PubMed] [Google Scholar]
- 5.Chapman E, Farr GW, Usaite R, Furtak K, Fenton WA, Chaudhuri TK, Hondorp ER, Matthews RG, Wolf SG, Yates JR, et al. Proc Natl Acad Sci USA. 2006;103:15800–15805. doi: 10.1073/pnas.0607534103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viitanen PV, Gatenby AA, Lorimer GH. Protein Sci. 1992;1:363–369. doi: 10.1002/pro.5560010308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gragerov A, Nudler E, Komissarova N, Gaitanaris GA, Gottesman ME, Nikiforov V. Proc Natl Acad Sci USA. 1992;89:10341–10344. doi: 10.1073/pnas.89.21.10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Feng H, Landry SJ, Maxwell J, Gierasch LM. Biochemistry. 1999;38:12537–12546. doi: 10.1021/bi991070p. [DOI] [PubMed] [Google Scholar]
- 9.Weissman JS, Rye HS, Fenton WA, Beechem JM, Horwich AL. Cell. 1996;84:481–490. doi: 10.1016/s0092-8674(00)81293-3. [DOI] [PubMed] [Google Scholar]
- 10.Mayhew M, da Silva ACR, Martin J, Erdjument-Bromage H, Tempst P, Hartl FU. Nature. 1996;379:420–426. doi: 10.1038/379420a0. [DOI] [PubMed] [Google Scholar]
- 11.Rye HS, Burston SG, Fenton WA, Beechem JM, Xu Z, Sigler PB, Horwich AL. Nature. 1997;388:792–798. doi: 10.1038/42047. [DOI] [PubMed] [Google Scholar]
- 12.Xu Z, Horwich AL, Sigler PB. Nature. 1997;388:741–751. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 13.Weissman JS, Kashi Y, Fenton WA, Horwich AL. Cell. 1994;78:693–702. doi: 10.1016/0092-8674(94)90533-9. [DOI] [PubMed] [Google Scholar]
- 14.Ranson NA, Burston SG, Clarke AR. J Mol Biol. 1997;266:656–664. doi: 10.1006/jmbi.1996.0815. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt M, Buchner J, Todd MJ, Lorimer GH, Viitanen PV. J Biol Chem. 1993;269:10304–10311. [PubMed] [Google Scholar]
- 16.Brinker A, Pfeifer G, Kerner MJ, Naylor DJ, Hartl FU, Hayer-Hartl M. Cell. 2001;107:223–233. doi: 10.1016/s0092-8674(01)00517-7. [DOI] [PubMed] [Google Scholar]
- 17.Houry WA, Frishman D, Eckerskorn C, Lottspeich F, Hartl FU. Nature. 1999;402:147–154. doi: 10.1038/45977. [DOI] [PubMed] [Google Scholar]
- 18.Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang H-C, Sines AP, Georgopoulos CP, Frishman D, Hayer-Hartl M, Mann M, Hartl FU. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 19.Tang YC, Chang HC, Roeben A, Wischnewski D, Wischneweski N, Kerner MJ, Hartl FU, Hayer-Hartl M. Cell. 2006;125:903–914. doi: 10.1016/j.cell.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 20.Saibil HR, Zheng D, Roseman AM, Hunter AS, Watson GMF, Chen S, auf der Mauer A, O'Hara BP, Wood SP, Mann NH, et al. Curr Biol. 1993;3:265–273. doi: 10.1016/0960-9822(93)90176-o. [DOI] [PubMed] [Google Scholar]
- 21.Todd MJ, Viitanen PV, Lorimer GH. Science. 1994;265:659–666. doi: 10.1126/science.7913555. [DOI] [PubMed] [Google Scholar]
- 22.Horwich AL, Low KB, Fenton WA, Hirshfield IN, Furtak K. Cell. 1993;74:909–917. doi: 10.1016/0092-8674(93)90470-b. [DOI] [PubMed] [Google Scholar]
- 23.Chandrasekhar GN, Tilly K, Woolford C, Hendrix R, Georgopoulos C. J Biol Chem. 1986;261:12414–12419. [PubMed] [Google Scholar]
- 24.Rye HS, Roseman AM, Chen S, Furtak K, Fenton WA, Saibil HR, Horwich AL. Cell. 1999;97:325–338. doi: 10.1016/s0092-8674(00)80742-4. [DOI] [PubMed] [Google Scholar]
- 25.Yifrach O, Horovitz A. Proc Natl Acad Sci USA. 2000;97:1521–1524. doi: 10.1073/pnas.040449997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weissman JS, Hohl CM, Kovalenko O, Chen S, Braig K, Saibil HR, Fenton WA, Horwich AL. Cell. 1995;83:577–587. doi: 10.1016/0092-8674(95)90098-5. [DOI] [PubMed] [Google Scholar]
- 27.Martin J, Langer T, Boteva R, Schramel A, Horwich AL, Hartl FU. Nature. 1991;352:36–42. doi: 10.1038/352036a0. [DOI] [PubMed] [Google Scholar]
- 28.Yifrach O, Horovitz A. J Mol Biol. 1996;255:356–361. doi: 10.1006/jmbi.1996.0028. [DOI] [PubMed] [Google Scholar]
- 29.Wang JD, Herman C, Tipton KA, Gross CA, Weissman JS. Cell. 2002;111:1027–1039. doi: 10.1016/s0092-8674(02)01198-4. [DOI] [PubMed] [Google Scholar]
- 30.Kawe M, Plückthun A. J Mol Biol. 2006;357:411–426. doi: 10.1016/j.jmb.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Fenton WA, Kashi Y, Furtak K, Horwich AL. Nature. 1994;371:614–619. doi: 10.1038/371614a0. [DOI] [PubMed] [Google Scholar]
- 32.Jewett AI, Baumketner A, Shea J-E. Proc Natl Acad Sci USA. 2004;101:13192–13197. doi: 10.1073/pnas.0400720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koike-Takeshita A, Shimamura T, Yokoyama K, Yoshida M, Taguchi H. J Biol Chem. 2006;281:962–967. doi: 10.1074/jbc.M506298200. [DOI] [PubMed] [Google Scholar]
- 34.Farr GW, Fenton WA, Chaudhuri TK, Clare DK, Saibil HR, Horwich AL. EMBO J. 2003;22:3220–3230. doi: 10.1093/emboj/cdg313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.