Abstract
Molecular cloning of the gene encoding sterol Δ7 reductase from the filamentous fungus Mortierella alpina 1S-4, which accumulates cholesta-5,24-dienol (desmosterol) as the main sterol, revealed that the open reading frame of this gene, designated MoΔ7SR, consists of 1,404 bp and codes for 468 amino acids with a molecular weight of 53,965. The predicted amino acid sequence of MoΔ7SR showed highest homology of 51% with that of sterol Δ7 reductase (EC 1.3.1.21) from Xenopus laevis (African clawed frog). Heterologous expression of the MoΔ7SR gene in yeast Saccharomyces cerevisiae revealed that MoΔ7SR converts ergosta-5,7-dienol to ergosta-5-enol (campesterol) by the activity of Δ7 reductase. In addition, with gene silencing of MoΔ7SR gene by RNA interference, the transformant accumulated cholesta-5,7,24-trienol up to 10% of the total sterols with a decrease in desmosterol. Cholesta-5,7,24-trienol is not detected in the control strain. This indicates that MoΔ7SR is involved in desmosterol biosynthesis in M. alpina 1S-4. This study is the first report on characterization of sterol Δ7 reductase from a microorganism.
Sterols are major components of eukaryotic cell membranes. The end products and the sterol biosynthetic pathway differ among species. Cholesterol (cholesta-5-enol) is typical in animals, ergosterol (ergosta-5,7,22-trienol) is common in fungi, and sitosterol (24-ethyl cholesta-5-enol), campesterol (ergosta-5-enol), and stigmasterol (24-ethyl cholesta-5,22-dienol) are the main end sterols in plants (7). These end sterols are composed of a similar four-ringed structures, including an unsaturation at the Δ5 position on ring B and a specific branched side chain. Ergosterol, unlike other end sterols, contains an additional unsaturation at the Δ7 position on ring B. On the other hand, desmosterol (cholesta-5,24-dienol) was found to be an end sterol in the zygomycetes fungus Mortierella alpina (20). Recently, the sterol biosynthetic pathway in M. alpina was demonstrated to produce 13 sterols but no ergosterol (12).
The biosynthetic reactions that allow conversion of lanosterol (4,4-dimethyl cholesta-8,24-dienol) resulting from the oxidative cyclization of squalene to cholesterol have been well studied (14, 15, 18). As a representative of microorganisms, yeast, the biosynthetic steps of ergosterol and the related enzymes were characterized in Saccharomyces cerevisiae (5, 13). From zymosterol (cholesta-8,24-dienol) to cholesterol in mammals, ergosterol in fungi, and phytosterols in plants, there are multiple alternative pathways leading to terminal sterols. In the sequential steps of cholesterol biosynthesis in mammals, sterol Δ7 reductase (Δ7SR; EC 1.3.1.21) is a terminal enzyme, which is absent in yeast, catalyzes the reduction of the Δ7 double bond in sterol intermediates with presence of NADPH under anaerobic conditions (6, 10). A deficiency of this enzyme activity due to genetic mutation in humans has been found to cause Smith-Lemli-Opitz syndrome (SLOS) (4, 28). SLOS is an autosomal-recessive multiple congenital anomaly and/or mental retardation disorder caused by inborn error of post-squalene cholesterol biosynthesis that leads to the elevation of 7-dehydrocholesterol in serum body fluids and tissues (19, 21, 27).
Despite their importance, the isolation and functional analysis of the Δ7SR genes were just reported from the limited species. A gene encoding NADPH-dependent Δ7SR was first isolated from a higher plant, Arabidopsis thaliana (9). Functional analysis in a yeast expression system revealed that this plant enzyme efficiently reduced Δ5,7-ergosta- and cholesta-sterols in vivo, regardless of the structural variations on the side chain (9). Plant Δ7SR was used for steroid biosynthesis such as pregnenolone, progesterone, and hydrocortisone in yeast (3, 22). On the other hand, Δ7SR genes from humans (11) and rats (1) were isolated and characterized. At present, more than 100 different mutations have been identified in SLOS patients who represent a continuum of clinical severity (30).
Thus far, the microbial Δ7SR gene has never been isolated and characterized. In the present study, a Δ7SR cDNA, designated MoΔ7SR, was isolated by a PCR-based cloning method and characterized by a yeast expression system and RNA interference (RNAi) in M. alpina 1S-4. This is the first report on an Δ7SR gene from a microorganism.
MATERIALS AND METHODS
Strains and cultivation media.
For cloning a Δ7SR gene, M. alpina 1S-4, deposited in the AKU culture collection (Agriculture of Kyoto University) as AKU3998 (29), was used. The uracil auxotroph (ura5− strain) isolated previously from M. alpina 1S-4 was used for gene silencing of MoΔ7SR by RNAi. Yeast S. cerevisiae SX-1065 (MATα his3 leu2 erg5::TRP1 ura3), with its sterol Δ22 desaturase (ERG5) gene (8) disrupted, was kindly provided by Mitsubishi Chemical Co. (Tokyo, Japan) and used for functional analysis of the MoΔ7SR gene. A complete medium (GY medium) and a selection yeast minimal medium (uracil-free SC medium) previously reported (23) were used for cultivation of M. alpina. A selection yeast minimal medium (YNBD medium) and a complete medium (YPD medium) were used for cultivation of S. cerevisiae (16).
Chemicals.
Desmosterol and campesterol were purchased from Sigma-Aldrich Co. Cholesta-5,7,24-trienol was kindly provided by Mitsubishi Chemical Co. All other chemicals were of the highest commercial grades available. The polymerase used was Takara LA Taq (Takara Bio, Inc., Shiga, Japan).
PCR-based cloning.
Degenerate oligonucleotide primers were designed based on the highly conserved amino acid sequences of Δ7SR from human (GenBank accession no. AF034544) and African clawed frog (GenBank accession no. BC044995). Primer Δ7SR-DeF1 (5′-ATGGGIATHGARTTYAAYCC-3′) corresponding to amino acid sequence MGIEFNP and primer Δ7SR-DeR1 (5′-CATRTADATIAWRTARAARTA-3′) corresponding to YFY(F/I)IYM were used. PCR amplification was carried out with the genomic DNA extracted from M. alpina 1S-4 as described previously (17). The amplified 760-bp PCR fragment was cloned into the pT7Blue T-vector (Novagen/Merck KGaA, Darmstadt, Germany) and sequenced.
The complete MoΔ7SR cDNA was obtained by PCR using primers with primers designed from the sequence information mentioned above with the cDNA library as a template. The amplified 1.4-kb PCR fragment was cloned into the pT7Blue T-vector and sequenced. The resultant plasmid was designated pT7Mo7SR. Similarly, the genomic gene encoding this protein was obtained with the same primers and the genomic gene as a template. The amplified 1.9-kb PCR fragment was cloned into the pT7Blue T-vector and sequenced.
Functional analysis of MoΔ7SR cDNA.
For the expression of MoΔ7SR cDNA in yeast, an amplified product was obtained by PCR using the primer Δ7SR-ExF4 (5′-CTCCTCAAGCTTATGGCAGTGCAGCAGAGG-3′) containing an ATG start site (underlined) and a HindIII cloning site (italics) and the primer Δ7SR-ExR4 (5′-CATTCTAGATTAGTAAATGTAGGGAATGAGC-3′) containing a TAA stop site (underlined) and a XbaI cloning site (italics) and plasmid pT7Mo7SR as a template. The resultant PCR fragment was cloned into the pT7Blue T-vector and sequenced. The full-length gene was cloned into the HindIII/XbaI sites of yeast expression vector pYES2 (Invitrogen) to generate a MoΔ7SR gene expression vector, pYEMoΔ7SR, followed by transformation into S. cerevisiae SX-1065 by the electroporation method (2). Transformants were selected for uracil auxotrophy on YNBD medium without uracil. To characterize the function of MoΔ7SR, transformants were grown at 28°C for 24 h in YPD medium and then for 48 h after the addition of galactose (2% of final concentration) to induce gene expression, followed by whole-cell sterol analysis. The host strain transformed with pYES2 was used as a negative control in all experiments.
MoΔ7SR gene silencing.
According to the previous method (26), a vector for MoΔ7SR gene silencing was constructed. Four primers, Δ7SR-RiF5 (5′-CGGATCCTATGGCAGTGCAGCAGAGGAAAG-3′), Δ7SR-RiR5 (5′-GTCCATGGAACAATTCCTAGGCGACCGTTG-3′), Δ7SR-RiF6 (5′-GATCCCATGGCAGTGCAGCAGAGGAAAG-3′), and Δ7SR-RiR6 (5′-GAGCCCTAGGACTTGCGGTCGGCAGCATGC-3′) containing BamHI, NcoI and BlnI, NcoI, and BlnI sites, respectively, were designed for construction of a MoΔ7SR gene silencing vector. First, a PCR product (730 bp) amplified with a primer pair (Δ7SR-RiF5 and Δ7SR-RiR5) and pYEMoΔ7SR as a template was digested with NcoI and BamHI and then ligated to the NcoI/BamHI sites of pBlues-hpt, which was previously made for the construction of gene expression vector in M. alpina (25), resulting in the construction of pBluesΔ7-1. Next, the PCR product (623 bp) amplified with a primer pair (Δ7SR-RiF6 and Δ7SR-RiR6) and pYEMoΔ7SR as a template was digested with NcoI and BlnI and ligated to the NcoI/BlnI sites of pBluesΔ7-1, resulting in construction of pBluesΔ7-2. Finally, the DNA fragment containing histone H4 promoter and ura5 gene was prepared by digestion of pDura5 with EcoRI and BamHI, which had been previously constructed for the transformation of uracil auxotrophs of M. alpina (24) and then ligated to the EcoRV site of pBluesΔ7-2 with a DNA blunting kit, resulting in construction of pBluesΔ7U5. Plasmid pBluesΔ7U5, which brings about formation of double-strand RNA of the MoΔ7SR gene under the control of the homologous histone H4 promoter in M. alpina, was used for transformation of the M. alpina 1S-4 ura5− strain as described previously (24). Transformants were selected on uracil-free SC agar medium, and their sterol composition was analyzed.
Sterol analysis.
The cultivated yeast and mycelia were harvested by centrifugation and vacuum filtration, respectively, and their total sterols, extracted as described previously (20), were analyzed by gas-liquid chromatography (GLC) and gas chromatography-mass spectrometry (GC-MS). The analytical conditions for the GLC were as follows: apparatus, GC-17A (Shimadzu, Kyoto, Japan) equipped with a flame ionization detector; column, ULBON HR-52 (0.25 mm by 50 m; Shinwakakou, Kyoto, Japan); injection port temperature, 300°C; detector port temperature, 300°C; carrier gas, He; makeup gas, N2; and split ratio, 50:1. The initial column temperature of 220°C was raised at 5°C/min to 280°C and then maintained for 28 min at 280°C. A GC-MS QP5050 (Shimadzu) operating at an ionization voltage of 70 eV was used for mass spectral analyses.
DNA sequence.
The nucleotide sequences were determined with a CEQ dye terminator cycle sequencing kit (Beckman Coulter, Inc., Fullerton, CA) and an automated sequencer DNA analysis system CEQ 2000XL (Beckman Coulter, Inc.).
Nucleotide sequence accession number.
The genomic gene sequence coding for MoΔ7SR has been registered in the DDBJ database under nucleotide sequence accession number AB270696.
RESULTS
Isolation of a Δ7SR gene from M. alpina 1S-4.
A PCR-based cloning strategy was adopted to obtain a gene encoding Δ7SR from the genomic DNA of M. alpina 1S-4. The amino acid sequences of two Δ7SR derived from human and African clawed frog were aligned, and degenerate oligonucleotides were designed based on the conserved regions. The single band obtained on PCR amplification contained a putative Δ7SR gene fragment. This fragment was used to isolate the full-length genes from the genomic gene and the cDNA library. The open reading frame in the genomic gene was 1,886 bp in length, contained three introns (227, 142, and 113 bp) with 5′-GT and 3′-AG ends, and encoded a protein of 467 amino acids with a molecular weight of 53,965.
The deduced amino acid sequence of the cloned MoΔ7SR exhibited 52, 50, and 40% identity with those of Δ7SR from African clawed frog, human, and higher plant Arabidopsis thaliana (GenBank accession no. ATU49398), respectively. MoΔ7SR possesses an identical sequence from 431 to 454 amino acid residues with a sterol reductase family signature 2 (Prosite accession no. PS01018) and a similar sterol reductase family signature 1 from 205 to 220 residues with a sterol reductase family signature 1 (Prosite accession no. PS01017). The amino acid residue at position 212 in a similar sequence of the sterol reductase family signature 1 of MoΔ7SR is Leu, whereas the corresponding residue in signature 1 is Phe, Tyr, Trp, or Met. A hydropathy plot indicates that there is a large proportion (>55%) of hydrophobic amino acids throughout the polypeptide backbone (except for the region of amino acid residues 4 to 35 and 434 to 467) of the deduced amino acid sequence (data not shown). The existence of at least nine transmembrane α-helical domains is predicted by Kyte and Doolittle hydrophobicity analysis, which is identical with the profiles of animal and plant Δ7SR (data not shown).
Functional analysis of MoΔ7SR cDNA in yeast ERG5 disruptant.
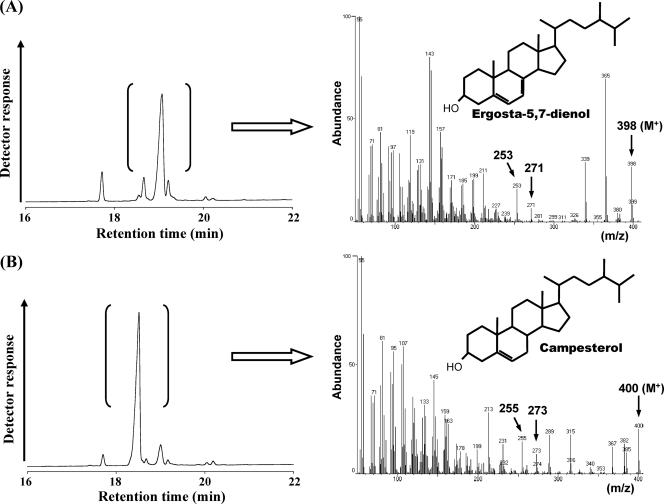
To identify the function of MoΔ7SR cDNA, an expression vector, pYEMoΔ7SR, constructed by ligation of MoΔ7SR cDNA under GAL1 promoter of yeast expression vector pYES2, was transformed into ERG5-disrupted yeast S. cerevisiae SX-1065. As shown in Fig. 1A, control strain transformed with pYES2 accumulated ergosta-5,7-dienol as the main sterol at 19.1 min of the GLC chromatogram, instead of ergosterol, because of its ERG5 disruption. GC-MS analysis of ergosta-5,7-dienol revealed a mass peak of m/z 398 and specific m/z 253 and 271 peaks. Ergosta-5,7-dienol is assumed to be derived from Δ24 reduction of ergosta-5,7,24(28)-trienol. On the other hand, GLC analysis of total sterols from the MoΔ7SR transformant revealed accumulation of another sterol (the main sterol of the transformant) at the retention time of 18.5 min as shown in Fig. 1B. Through further GC-MS analysis, this sterol showed specific ion peaks of m/z 255, 273, and 400, which were identical to the corresponding mass peaks of authentic campesterol. Moreover, it also showed identical retention time with the authentic campesterol in high-pressure liquid chromatography and GLC analysis (see the supplemental material). These results proved that ergosta-5,7-dienol was converted to campesterol as the reduction of a Δ7 double band by MoΔ7SR in the transgenic yeast.
FIG. 1.
GLC chromatogram of total sterols and GC-MS analysis of the main sterol from the S. cerevisiae ERG5 mutant. (A) Control strain (with the vector only); (B) MoΔ7SR transformant.
Effect of gene silencing of MoΔ7SR on sterol composition.
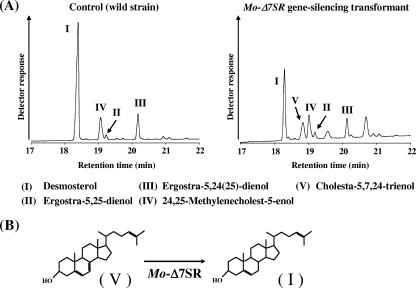
M. alpina 1S-4 ura5− strain was transformed with plasmid pBluesΔ7U5 constructed for MoΔ7SR gene silencing by biolistic transformation. The sterol composition of the obtained MoΔ7SR-RNAi transformant and the control strain with an empty vector, pDura5, were analyzed by GLC (Fig. 2A). When these transformants were cultivated in GY medium at 28°C for 5 days, desmosterol was the main sterol (75.7%) in the total sterols of the control strain, whereas the proportion of desmosterol decreased to 21.0% in the total sterols of the MoΔ7SR-RNAi transformant and some unidentified sterols were detected. One of the unidentified sterols, designated peak V in Fig. 2A, consisted of 9.9% of the total sterols. On GC-MS analysis, the mass spectra of peak V exhibited a mass peak of m/z 382, and the fragmentation pattern was identical with that of the authentic of cholesta-5,7,24-trienol (data not shown). Furthermore, the retention time of peak V on both GLC and HPLC analyses was identical to that of the authentic cholesta-5,7,24-trienol. These results indicate that peak V was identified as cholesta-5,7,24-trienol, which is converted to desmosterol by reduction at the Δ7 position of the sterol skeleton as shown in Fig. 2B. RNAi analysis revealed that MoΔ7SR was involved in desmosterol biosynthesis in M. alpina 1S-4.
FIG. 2.
(A) GLC chromatograms of Mortierella transformants. (B) Putative biosynthesis of desmosterol from cholesta-5,7,24-trienol by MoΔ7SR.
DISCUSSION
Δ7SR genes have been isolated from mammals (1, 11), frogs, amoebas, and higher plants (9). It catalyzes reduction of a double bond at the Δ7 position on the sterol skeleton and, in particular, exhibits a significant function in the biosynthesis from 7-dehydrocholesterol to cholesterol. SLOS in humans was characterized as an abnormal accumulation of 7-dehydrocholesterol, which was caused by congenital mutation of the Δ7SR gene (30). On the other hand, plant Δ7SR gene has been used for steroid production in recombinant yeast (3, 22). With respect to microorganisms, M. alpina and Thraustochytrium sp. were found to produce desmosterol and cholesterol, respectively, whereas the microbial Δ7SR gene has never been reported.
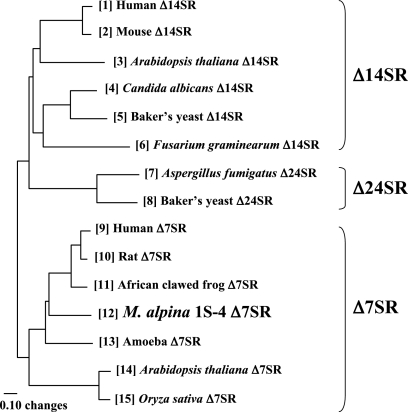
MoΔ7SR exhibits 39 to 51% homology with other Δ7SR proteins at the amino acid level and shows two typical sterol family signatures. However, the Leu residue at position 212 in MoΔ7SR is not consistent with the amino acid residue of the sterol family signature 1. This indicates that the sterol family signature 1 may be revised based on the Leu residue in MoΔ7SR. MoΔ7SR is inferred to be present in the membrane of endoplasmic reticulum because of the nine putative transmembrane domains. We propose a phylogenetic tree of representative Δ7SR, sterol Δ14 reductases (Δ14SR), and sterol Δ24 reductases (Δ24SR) as shown in Fig. 3. MoΔ7SR was found to be closer to mammal and frog Δ7SR than to A. thaliana and Oryza sativa Δ7SR. Δ7SR exhibits a relatively high similarity with Δ14SR and Δ24SR at the polypeptide level. In particular, MoΔ7SR exhibited high identities with Δ7SR (40 to 52%) from other organisms and some identities with Δ14SR (34 to 38%) and Δ24SR (28%). There is no information about fungal Δ7SR other than the present study, although some fungal Δ14SR and Δ24SR were reported. To elucidate sterol biosynthetic pathways in M. alpina 1S-4, isolation and characterization of Δ14SR and Δ24SR genes from M. alpina 1S-4 is also considered very important.
FIG. 3.
Phylogenetic analysis of Δ7SR, sterol Δ24 reductase (Δ24SR), and sterol Δ14 reductase. The phylogenetic tree was prepared by using the GENETYX version 7.0.3 (Genetyx Co.). For each numbered sequence, the EMBL (em) or TrEMBL (tr) accession no. is as follows: sequence 1, em, AF048704; sequence 2, em, AF480070; sequence 3, em, AF256535; sequence 4, tr, Q59LS4; sequence 5, em, M99419; sequence 6, tr, Q41852; sequence 7, tr, Q4WW43; sequence 8, em, S58126; sequence 10, em, AB016800; sequence 13, em, AY653733; and sequence 15, em, AP005514. The gene accession numbers of sequences 9, 11, 12, and 14 are noted in the text.
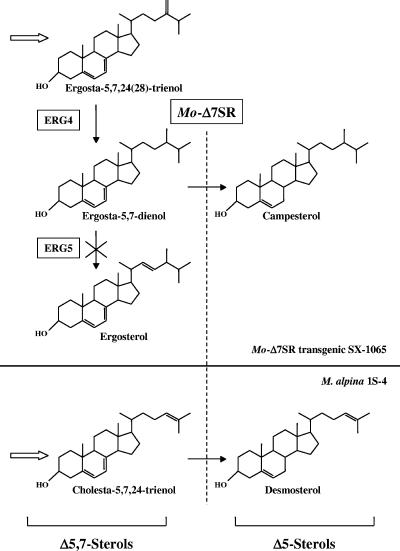
The ERG5 disruptant SX-1065 from S. cerevisiae accumulates ergosta-5,7-dienol instead of ergosterol. Campesterol, which is the reduction product of ergosta-5,7-dienol, is produced in the presence of MoΔ7SR in transformed SX-1065. Both ergosta-5,7-dienol and campesterol are not detected in M. alpina 1S-4. Furthermore, almost all ergosta-5,7-dienol was converted to campesterol in the transformed SX-1065. These points suggest that MoΔ7SR has wide substrate specificity and high activity for Δ7 reductase. M. alpina 1S-4 accumulates more than 70% of desmosterol in the total sterols. RNAi analysis revealed that MoΔ7SR plays a significant function in vivo, converting cholesta-5,7,24-trienol to desmosterol. The GLC chromatogram of the MoΔ7SR-RNAi transformant was very different from that of the control strain. Some unidentified peaks other than cholesta-5,7,24-trienol were detected in the MoΔ7SR-RNAi transformant. We hypothesize that these compounds have a Δ5,7 structure and are converted to ergosta-5,24(25)-dienol, ergosta-5,25-dienol, 24,25-methylenecholest-5-enol, and so on by Δ7 double bond reduction. MoΔ7SR can tolerate various sterol side chains while being specific for the Δ7 double bond reduction in a Δ5,7 structure, as shown in Fig. 4.
FIG. 4.
Partial biosynthetic pathways of the sterols in MoΔ7SR transgenic yeast and M. alpina 1S-4.
In conclusion, our results on the molecular characterization of MoΔ7SR gene may provide some useful opportunities with regard to (i) the study of steroid production by microbial fermentation, (ii) investigation of structure-function relationships in Δ7SR, and (iii) the elucidation of evolution of sterol reductases in various organisms.
Supplementary Material
Acknowledgments
We thank Makoto Ueda, Masahiro Yamagishi, and Takeshi Sakamoto, Mitsubishi Chemical Co., for GC-MS analysis of the sterols and kind suggestions regarding identification of the sterols.
This study was supported in part by a grant-in-aid for scientific research (grant 15658024) to S.S. from the Ministry of Education, Science, Sports, and Culture of Japan; a COE award for Microbial-Process Development Pioneering Future Production Systems (COE Program of the Ministry of Education, Science, Sports, and Culture of Japan) to S.S.; and the Project of the New Energy and Industrial Technology Development Organization of Japan (grant 05A07003d to E.S.).
Footnotes
Published ahead of print on 12 January 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bae, S. H., J. N. Lee, B. U. Fitzky, J. Seong, and Y. K. Paik. 1999. Cholesterol biosynthesis from lanosterol: molecular cloning, tissue distribution, expression, chromosomal localization, and regulation of rat 7-dehydrocholesterol reductase, a Smith-Lemli-Opitz syndrome-related protein. J. Biol. Chem. 274:14624-14631. [DOI] [PubMed] [Google Scholar]
- 2.Delorme, E. 1989. Transformation of Saccharomyces cerevisiae by electroporation. Appl. Environ. Microbiol. 55:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duport, C., R. Spagnoli, E. Degryse, and D. Pompon. 1998. Self-sufficient biosynthesis of pregnenolone and progesterone in engineered yeast. Nat. Biotechnol. 16:186-189. [DOI] [PubMed] [Google Scholar]
- 4.Fitzky, B. U., M. Witsch-Baumgartner, M. Erdel, J. N. Lee, Y. K. Paik, H. Glossmann, G. Utermann, and F. F. Moebius. 1998. Mutations in the Δ7-sterol reductase gene in patients with the Smith-Lemli-Opitz syndrome. Proc. Natl. Acad. Sci. USA 95:8181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fryberg, M., A. C. Oehlschlager, and M. Unrau. 1973. Biosynthesis of ergosterol in yeast: evidence for multiple pathways. J. Am. Chem. Sci. 95:5747-5757. [DOI] [PubMed] [Google Scholar]
- 6.Gaylor, J. L. 1981. Formation of sterols in animals, p. 481-543. In J. W. Porter and S. L. Springer (ed.), Biosynthesis of isoprenoid compounds. Wiley, New York, NY.
- 7.Hartmann, M. A., and P. Benveniste. 1987. Plant membrane sterols: isolation, identification, and biosynthesis. Methods Enzymol. 148:632-650. [Google Scholar]
- 8.Hayashi, Y., and M. Nakajima. May 2004. Manufacturing process for fungi improved by metabolic engineering and sterols produced by them. Japan patent 2004-141125.
- 9.Lecain, E., X. Chenivesse, R. Spagnoli, and D. Pompon. 1996. Cloning by metabolic interference in yeast and enzymatic characterization of Arabidopsis thaliana sterol Δ7-reductase. J. Biol. Chem. 271:10866-10873. [DOI] [PubMed] [Google Scholar]
- 10.Lee, J., and Y. K. Paik. 1997. Cholesterol biosynthesis from lanosterol: development of a novel assay method, characterization, and solubilization of rat hepatic microsomal sterol Δ7-reductase. J. Biochem. Mol. Biol. 30:370-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moebius, F. F., B. U. Fitzky, J. N. Lee, Y. K. Paik, and H. Glossmann. 1998. Molecular cloning and expression of the human Δ7-sterol reductase. Proc. Natl. Acad. Sci. USA 95:1899-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nes, W. D., and S. D. Nichols. 2006. Phytosterol biosynthesis pathway in Mortierella alpina. Phytochemistry 67:1716-1721. [DOI] [PubMed] [Google Scholar]
- 13.Osumi, T., S. Taketani, H. Katsuki, T. Kuhara, and I. Matsumoto. 1978. Ergosterol biosynthesis in yeast. Pathways in the late stages and their variation under various conditions. J. Biochem. 83:681-691. [DOI] [PubMed] [Google Scholar]
- 14.Popjak, G., A. Meenan, E. J. Parish, and W. D. Nes. 1989. Inhibition of cholesterol synthesis and cell growth by 24(R,S),25-iminolanosterol and triparanol in cultured rat hepatoma cells. J. Biol. Chem. 264:6230-6238. [PubMed] [Google Scholar]
- 15.Reinhart, M. P., J. T. Billheimer, J. R. Faust, and J. L. Gaylor. 1987. Subcellular localization of the enzymes of cholesterol biosynthesis and metabolism in rat liver. J. Biol. Chem. 262:9649-9655. [PubMed] [Google Scholar]
- 16.Sakuradani, E., T. Abe, K. Iguchi, and S. Shimizu. 2005. A novel fungal ω3-desaturase with wide substrate specificity from arachidonic acid-producing Mortierella alpina 1S-4. Appl. Microbiol. Biotechnol. 66:648-654. [DOI] [PubMed] [Google Scholar]
- 17.Sakuradani, E., M. Kobayashi, and S. Shimizu. 1999. Δ9-Fatty acid desaturase from arachidonic acid-producing fungus: unique gene sequence and its heterologous expression in a fungus, Aspergillus. Eur. J. Biochem. 260:208-216. [DOI] [PubMed] [Google Scholar]
- 18.Schroepfer, G. J., Jr. 1982. Sterol biosynthesis. Annu. Rev. Biochem. 51:555-585. [DOI] [PubMed] [Google Scholar]
- 19.Shefer, S., G. Salen, A. Honda, A. Batta, S. Hauser, G. S. Tint, M. Honda, T. Chen, M. F. Holick, and L. B. Nguyen. 1997. Rapid identification of Smith-Lemli-Opitz syndrome homozygotes and heterozygotes (carriers) by measurement of deficient 7-dehydrocholesterol-Δ7-reductase activity in fibroblasts. Metabolism 46:844-850. [DOI] [PubMed] [Google Scholar]
- 20.Shimizu, S., H. Kawashima, M. Wada, and H. Yamada. 1992. Occurrence of a novel sterol, 24,25-methylenecholest-5-en-3β-ol, in Mortierella alpina 1S-4. Lipids 27:481-483. [DOI] [PubMed] [Google Scholar]
- 21.Smith, D. W., L. Lemli, and J. M. Opitz. 1964. A newly recognized syndrome of multiple congenital anomalies. J. Pediatr. 64:210-217. [DOI] [PubMed] [Google Scholar]
- 22.Szczebara, F. M., C. Chandelier, C. Villeret, A. Masurel, S. Bourot, C. Duport, S. Blanchard, A. Groisillier, E. Testet, P. Costaglioli, G. Cauet, E. Degryse, D. Balbuena, J. Winter, T. Achstetter, R. Spagnoli, D. Pompon, and B. Dumas. 2003. Total biosynthesis of hydrocortisone from a simple carbon source in yeast. Nat. Biotechnol. 21:143-149. [DOI] [PubMed] [Google Scholar]
- 23.Takeno, S., E. Sakuradani, S. Murata, M. Inohara-Ochiai, H. Kawashima, T. Ashikari, and S. Shimizu. 2004. Cloning and sequencing of the ura3 and ura5 genes, and isolation and characterization of uracil auxotrophs of the fungus Mortierella alpina 1S-4. Biosci. Biotechnol. Biochem. 68:277-285. [DOI] [PubMed] [Google Scholar]
- 24.Takeno, S., E. Sakuradani, S. Murata, M. Inohara-Ochiai, H. Kawashima, T. Ashikari, and S. Shimizu. 2004. Establishment of an overall transformation system for an oil-producing filamentous fungus, Mortierella alpina 1S-4. Appl. Microbiol. Biotechnol. 65:419-425. [DOI] [PubMed] [Google Scholar]
- 25.Takeno, S., E. Sakuradani, S. Murata, M. Inohara-Ochiai, H. Kawashima, T. Ashikari, and S. Shimizu. 2005. Molecular evidence that the rate-limiting step for the biosynthesis of arachidonic acid in Mortierella alpina is at the level of an elongase. Lipids 40:25-30. [DOI] [PubMed] [Google Scholar]
- 26.Takeno, S., E. Sakuradani, A. Tomi, M. Inohara-Ochiai, H. Kawashima, T. Ashikari, and S. Shimizu. 2005. Improvement of the fatty acid composition of an oil-producing filamentous fungus, Mortierella alpina 1S-4, through RNA interference with Δ12-desaturase gene expression. Appl. Environ. Microbiol. 71:5124-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tint, G. S., M. Irons, E. R. Elias, A. K. Batta, R. Frieden, T. S. Chen, and G. Salen. 1994. Defective cholesterol biosynthesis associated with the Smith-Lemli-Opitz syndrome. N. Engl. J. Med. 13:107-113. [DOI] [PubMed] [Google Scholar]
- 28.Waterham, H. R., F. A. Wijburg, R. C. Hennekam, P. Vreken, B. T. Poll-The, L. Dorland, M. Duran, P. E. Jira, J. A. Smeitink, R. A. Wevers, and R. J. Wanders. 1998. Smith-Lemli-Opitz syndrome is caused by mutations in the 7-dehydrocholesterol reductase gene. Am. J. Hum. Genet. 63:329-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamada, H., S. Shimizu, and Y. Shinmen. 1987. Production of arachidonic acid by Mortierella elongata 1S-5. Agric. Biol. Chem. 51:785-790. [Google Scholar]
- 30.Yu, H., and S. B. Patel. 2005. Recent insights into the Smith-Lemli-Opitz syndrome. Clin. Genet. 68:383-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.