Abstract
RNA possesses the ability to bind a wide repertoire of small molecules. Some of these binding interactions have been shown to be of primary importance in molecular biology. For example, several classes of mRNA domains, collectively referred to as riboswitches, have been shown to serve as RNA genetic control elements that sense the concentrations of specific metabolites (i.e. acting as direct sensors of chemical compounds). However, to date no RNA species binding a hormone has been reported. Here, we report that the use of an appropriate SELEX (systematic evolution of ligands by exponential enrichment) strategy results in the isolation of thyroxine-specific aptamers. Further biochemical characterization of these aptamers, including mutational studies, the use of transcripts with site-specific modified nucleotides, nuclease and chemical probing, binding-shift assays and CD, demonstrated that these RNA structures included a G-rich motif, reminiscent of a guanine quadruplex structure, adjacent to a helical region. The presence of the thyroxine appeared to be essential for the formation of the structural motif's scaffold. Moreover, the binding is shown to be specific to thyroxine (T4) and tri-iodothyronine (T3), the active forms of the hormone, whereas other inactive derivatives, including thyronine (T0), do not support complex formation. These results suggest that this aptamer specifically binds to the iodine moieties of the thyroxine, a previously unreported ability for an RNA molecule.
Keywords: aptamer, riboswitch, RNA ligand, thyroid hormone, thyroxine (T4), tri-iodothyronine (T3)
Abbreviations: DMS, dimethyl sulfate; G-quartet, guanine quadruplexes; ITP, inosine triphosphate; SELEX, systematic evolution of ligands by exponential enrichment; T0, L-thyronine; T2, 3,5-di-iodo-L-thyronine; T3, 3,3′,5-tri-iodo-L-thyronine; T4, L-thyroxine
INTRODUCTION
Several of the original discoveries of biological molecules binding RNA species occurred during the characterization of the self-splicing mechanism of the group I intron (reviewed in [1]). For example, the first step of this mechanism, which is composed of two trans-esterification reactions, requires the binding of a GTP that subsequently acts as a nucleophilic group. The amino acid arginine binds to the same site in the intron, leading to inhibition of the splicing reaction [2]. In addition, various antibiotics have been shown to be potential inhibitors of group I intron self-splicing [3]. These initial results revealed the interesting ability of RNA species to bind small molecules [4].
More recently, it has been demonstrated that specific metabolites can directly bind mRNA molecules (reviewed in [5]). Several classes of mRNA domains, collectively referred to as riboswitches and serving as RNA genetic control elements that sense the presence of specific metabolites, act as direct sensors of chemical compounds. Upon interaction with the appropriate small ligand molecule, riboswitch mRNAs undergo a structural reorganization that results in the modulation of the genes that they encode. Riboswitches are known to be responsible for sensing metabolites that are critical for a number of fundamental biochemical processes, metabolites such as coenzyme B12, thiamine pyrophosphate, flavin mononucleotide, S-adenosylmethionine, lysine, guanine, adenine and glucosamine-6-phosphate among others [5]. Clearly the ability of RNA molecules to bind a wide repertoire of small ligands is not confined to the test tube, but rather is of primary importance in molecular biology.
In vitro selection, or SELEX (systematic evolution of ligands by exponential enrichment), makes use of a large population of random RNA or DNA sequences as the raw material for the selection of rare functional molecules (reviewed in [6]). This method, basically, consists of sequentially repeating a process that includes the selection of a specific activity (e.g. the binding sequence) coupled to an amplification of either the RNA or DNA molecules possessing this activity. These techniques have broadened our appreciation of the abilities that nucleic acids are capable of. In vitro selection has been used to identify aptamers binding a diverse array of small molecules including nucleotides, amino acids, peptides, cofactors, basic antibiotics and transition-state analogues, among others [6,7]. However, RNA species binding either a steroid or a hormone have yet to be reported. We under-took to investigate whether or not T4 (L-thyroxine), as a model hormone, can bind to RNA molecules (Figure 1A). Here we describe the development of a SELEX strategy for the isolation of thyroxine-specific aptamers. The selected aptamers were then characterized further in terms of their binding activities.
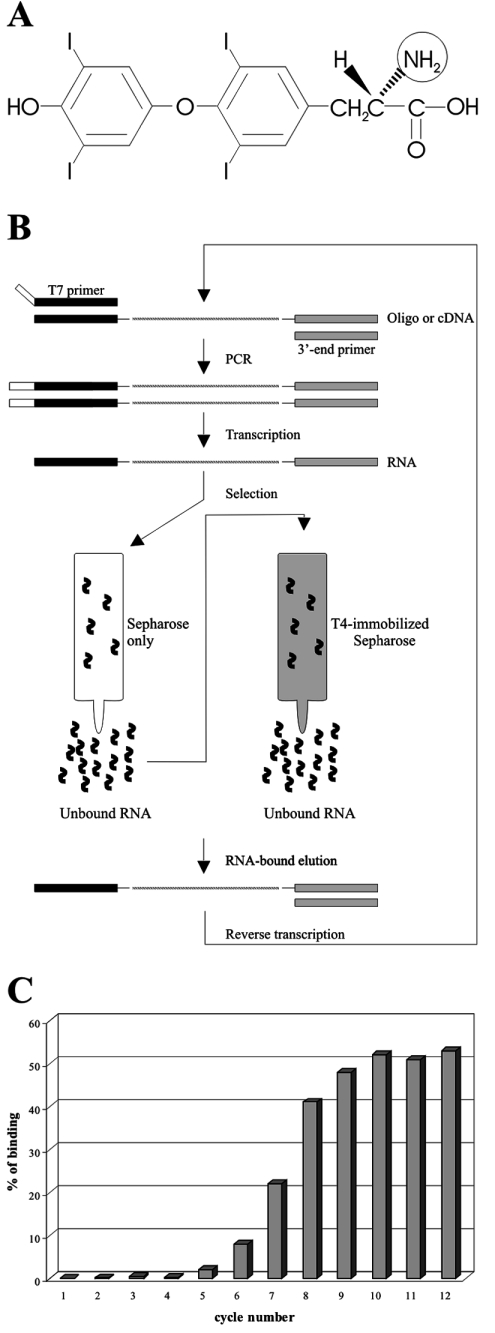
Figure 1. Atomic structure of T4 and selection cycle schematic.
(A) Atomic structure of T4. The circled amino group is involved in the reaction with the Sepharose. (B) Schematic representation of the selection cycle used to isolate T4–aptamer. (C) Enrichment profile of the T4–aptamers. The percentage of T4-bound aptamers after each cycle is illustrated in histogram form.
EXPERIMENTAL
Preparation of T4-Sepharose
T4-Sepharose was prepared as previously described [8]. Briefly, 6 g of cyanogen bromide-activated Sepharose (Amersham Biosciences) was rinsed with 1 litre of 1 mM HCl and dried. Half of the Sepharose was then mixed with 20 ml of solution containing 50% (w/v) ethylene glycol, 50 mM NaHCO3 (pH 9.6) and 3 mM of T4 (Sigma). The other half of the Sepharose was prepared in the same solution without T4 (i.e. negative selection). Both solutions were stirred overnight at 4 °C, centrifuged at 5000 g for 15 min, washed twice with 50 ml of the solution lacking T4 for 2 h at 4 °C, twice with 50 ml of buffer containing 10 mM Tris/HCl (pH 7.5), 500 mM NaCl and 1 mM MgCl2 for 1 h, and then resuspended in 10 ml of buffer containing 20% (v/v) ethanol, 10 mM Tris/HCl (pH 7.5), 500 mM NaCl and 1 mM MgCl2 and stored at 4 °C. The same procedure was used for the preparation of T3 (3,3′,5-triiodo-L-thyronine)-, T2 (3,5-diiodo-L-thyronine)-, 3-iodo-L-tyrosine- and T0 (L-thyronine)-Sepharose (Sigma).
In vitro selection cycle
Synthetic 98-mer oligonucleotides manually synthesized for the random sequence of 58 nt (5′-GAATTCGTCGACGGATCC-N58-CTGCAGGTCGACGCATGCGCCG-3′; Biosource) were amplified by 30 PCR cycles using the T7 primer (5′-TAATACGACTCACTATAGGGAATTCGTCGACGGATCC-3′) and a 3′-end primer (5′-CGGCGCATGCGTCGACCTGCAG-3′). The PCR products were purified using the QIAquick PCR purification kit (Qiagen).
Purified PCR products were transcribed in 100 μl reactions containing 27 units of RNA guard (Amersham Biosciences), 80 mM Hepes/KOH (pH 7.5), 24 mM MgCl2, 2 mM spermidine, 40 mM dithiothreitol, 4mM of each of ATP, UTP, CTP and GTP, 30 μCi of [α32P]UTP (3000 Ci/mmol; New England Nuclear), 0.01 units of yeast pyrophosphatase (Roche Diagnostic) and 10 μg of purified T7 RNA polymerase at 37 °C for 4 h. The reactions were stopped by the addition of 5 units of RNase free DNase I (Promega) and incubation at 37 °C for 30 min. The resulting products were extracted twice with phenol/chloroform, the nucleic acids precipitated, and subsequently dissolved in 50 μl of water and 25 μl of formamide dye buffer [97.5% (v/v) formamide, 0.025% Xylene Cyanole and 0.025% Bromophenol Blue] prior to being fractionated by 8% (w/v) denaturing (7 M urea) PAGE (acrylamide/bisacrylamide, 19:1, w/v) in buffer containing 45 mM Tris/borate (pH 7.5) and 1 mM EDTA. Products were visualized by UV-shadowing, and the bands corresponding to the full-length RNAs were excised. The transcripts were eluted by a solution of 0.5 M ammonium acetate and 0.1% SDS from the gel slices overnight at 4 °C. The resulting products were passed through Sephadex G-50 spun columns (Amersham Biosciences) and the RNA precipitated. The quantity of RNA was determined by spectrophotometry at 260 nm.
Radiolabelled transcripts (300 pmol) were dissolved in 100 μl of loading buffer [10 mM Tris/HCl (pH 7.5), 500 mM NaCl and 1 mM MgCl2], heated at 65 °C for 2 min and then cooled to room temperature (22 °C). The mixtures were then loaded on to columns containing a 0.2 ml bed column of Sepharose without T4 and incubated for 20 min at room temperature. The columns were rinsed with 300 μl of loading buffer, and the eluates (unbound RNAs) were then loaded on to a second column containing a 0.2 ml bed column of T4-Sepharose and incubated for 20 min at room temperature. The columns were then rinsed with 1.2 ml of loading buffer. Bound transcripts were eluted by either 600 μl of ultrapure water (for cycles 1–6) or 300 μl of 5 M urea (cycles 7–12). Eluted RNAs were ethanol precipitated in the presence of 20 μg of glycogen (Roche Diagnostic). The resulting pellets were conserved for the subsequent reverse transcription reactions. The percentages of RNA bound to the column were calculated as follows: 5 μl of the solutions were recovered both before the loading onto the T4-Sepharose and after the elution, and the radioactivity measured using a scintillation counter.
The pellets from the column eluates were dissolved in 9 μl of ultrapure water and reversed transcribed using Superscript II reverse transcriptase (Invitrogen) as recommended by the manufacturer. The reactions were stopped by the addition of RNase A (10 μg) and incubation at room temperature for 5 min. Aliquots (6 μl) of the resulting products were used as cDNA sources for the PCRs of the subsequent selection cycles.
Cloning and sequencing
PCR products of various cycles were cloned into pGEM-T vector as recommended by the manufacturer (Promega). Several clones were sequenced using the T7 sequencing kit (USB), and the sequences were aligned using the Clustal multiple alignment program [9], followed by minor manual readjustments.
In vitro transcription and 32P labelling of T4-aptamers
A large collection of aptamer variants was synthesized. Briefly, pairs of complementary and overlapping DNA oligonucleotides corresponding to the T7 RNA promoter followed by the full-length aptamer were synthesized and annealed. The second strands were then synthesized by adding 2.5 units of Pwo DNA polymerase (Roche Diagnostic) in a final volume of 100 μl containing 200 μM dNTPs, 10 mM Tris/HCl (pH 8.9), 25 mM KCl, 5 mM (NH4)2SO4 and 2 mM MgSO4, followed by five cycles of amplification. The resulting products were purified by extraction with phenol/chloroform twice, then the nucleic acids were precipitated and the DNA templates were in vitro transcribed as described above, except that non-radioactive NTP was used. In the case of the aptamers, including either inosine or 7-deaza-GTP, the GTP was replaced by 5 mM GMP and 5 mM of either ITP (inosine triphosphate) or 7-deaza-GTP respectively. After the gel purification and extraction procedures, the RNA aptamers (20 pmol) were dephosphorylated in a final volume of 20 μl containing 200 mM Tris/HCl (pH 8.0), 10 units of RNA Guard and 0.2 units of calf intestinal alkaline phosphatase (Roche Diagnostic) at 37 °C for 30 min. The reactions were purified by extraction with phenol/chloroform twice, and the RNA was ethanol precipitated. Dephosphorylated RNA aptamers (5 pmol) were 5′-end-labelled in a final volume of 10 μl containing 3.2 pmol of [γ-32P]ATP (6000 Ci/mmol), 50 mM Tris/HCl (pH 7.5), 10 mM MgCl2, 50 mM KCl and 3 units of T4 polynucleotide kinase (Amersham Biosciences) at 37 °C for 30 min. The reactions were stopped by the addition of 5 μl of formamide dye buffer, and the mixtures fractionated through denaturing 8 or 12% (w/v) polyacrylamide gels. The bands containing the appropriate 5′-end-labelled RNAs were excised, and the nucleic acids recovered as described above. For the preparation of 3′-end-labelled aptamers, the latter were incubated in the presence of [32P]Cp (3000 Ci/mmol) and T4 RNA ligase as described by the manufacturer (New England Biolabs), and then purified as described above.
Binding assays
The binding assays of individual aptamers on T4-Sepharose were performed as described above for the selection. Briefly, both radioactive (200000 c.p.m.) and non-radioactive (300 pmol) aptamers were pooled, and then loaded on to the column. Several washes were performed, and the quantity of unbound aptamers was determined by 32P counting. At least two independent experiments were performed for the determination of the percentages of bound T4-aptamers.
Chemical probing
Either 5′- or 3′-32P-end-labelled (50 000 c.p.m.) ApT4-A' RNA and 30 μM non-radioactive RNA were added to a 9 μl final volume solution containing 1 mM MgCl2, 30 mM sodium cacodylate (pH 7.0) and 150 mM of LiCl, NaCl or KCl. The solution was kept at room temperature for 20 min before the addition of 1 μl of DMS (dimethyl sulfate; diluted 1:8 in 100% ethanol) and then incubated at room temperature for a further 20 min. The RNA samples were ethanol precipitated, and the pellets washed twice with ethanol in order to remove all traces of DMS. The resulting pellets were dissolved in 20 μl of 500 mM Tris/HCl (pH 7.5). Sodium borohydride (200 mM; 10 μl) was added to the samples, which were then kept on ice for 5 min in the dark. Next, 10 μl of aniline solution (aniline/glacial acetic acid/water, 10:6:93, by vol.) was added to the samples and the tubes incubated at 60 °C for 10 min in the dark. The aptamers were then ethanol precipitated, fractionated on denaturing 10% (w/v) PAGE and analysed.
Binding shift assay
A mixture of non-radioactive (3 μM) and radioactive (10000 c.p.m.) aptamers was incubated at room temperature in a final volume of 10 μl containing 10 mM Tris/HCl (pH 7.5) and 1 mM MgCl2 in the absence or the presence of 150 mM NaCl, LiCl or KCl, and with or without 100 μM of T4. After 1 h of incubation, 2 μl of native gel loading buffer was added and the mixtures, which were then analysed on native 15% (w/v) PAGE gels (acrylamide/bisacrylamide, 29:1, w/v) in buffer containing 45 mM Tris/borate (pH 7.5) and 1 mM EDTA, with or without 150 mM of the salt used for the incubation. When the incubations were performed in the presence of T4, the gel also included 100 μM of T4. The results were visualized with a PhosphorImager (Molecular Dynamics). In order to determine the equilibrium constant (Kd), the experiments were repeated in the presence of various concentrations (1 to 100 μM) of T4 in both the sample and the gel.
CD
CD measurements were performed with a Jasco J-810 spectropolarimeter. The samples were analysed in quartz cells with path-lengths of 1 cm at 22 °C. Far- and near-UV wavelength scans were recorded from 200–250 nm and from 250–340 nm respectively. All experiments were performed using 5 μM ApT4-A' dissolved in 50 mM Tris/HCl (pH 7.5) either in the absence of monovalent salt or in the presence of 50 mM of NaCl or KCl. When required, 100 μM of T4 was added to the samples. The means of at least three wavelength scans are presented. Substraction of the buffer was not required since control experiments in the absence of RNA sample showed negligible curves.
RESULTS
Selection of T4-specific aptamers
Initially an appropriate SELEX strategy was devised in order to isolate T4-specific aptamers (Figure 1). The 98 nt oligodeoxyribonucleotides used as templates for the first PCR amplification included a randomized domain of 58 positions (i.e. an equal likelihood of all four nucleotides at each of the 58 positions). The selection step included two columns. First, a column of Sepharose alone, permitting a negative selection, was used. The transcripts that passed through this column (i.e. the unbound flow-through), which therefore had no affinity for the support material, were selected. Next, a column of T4-Sepharose was used for positive selection from the above initial fraction. The T4 was immobilized through its amine group (Figure 1A). The concentration of T4-Sepharose was determined to be 7 mM by iodine reactivity [10]. The transcripts in the flow-through from this second column (i.e. those positively selected) were discarded, while those bound to the column were eluted and used in the subsequent cycles. Since the transcripts were 32P-radiolabelled, both the retention of the column and its elution were easily monitored. The proportion of transcripts retained on the T4-Sepharose column in each cycle is illustrated in Figure 1(C). This percentage was negligible for the four initial cycles, and then increased significantly at each cycle up to approx. 50% by cycle nine.
The aptamers resulting from cycle eight, prior to saturation, were cloned and sequenced in order to avoid any bias that might be created by using a limited set of sequences. Out of a total of 60 clones, 53 were found to include a characteristic UGGAGG sequence box and one possessed an UGGUGG box (Figure 2). Since the conserved UGGAGG box occupies various positions, it cannot be used as a common feature in attempting to produce an accurate alignment. In fact, even considering only the 54 clones possessing either the UGGAGG or the UGGUGG box did not yield a relevant sequence alignment. Moreover, the 54 sequences showed variable abundances. Four groups of closely related sequences accounted for 49 of the 54 sequences. Groups I, II, III and IV include 12, 10, 21 and 6 clones respectively. Ten aptamers from cycle ten, which corresponds to that after the saturation of enrichment, were also sequenced. Only representatives from groups I and II, which always include the UGGAGG box, were retrieved.
Figure 2. T4–aptamer enrichment.
Sequences of the majority of the clones analysed. The conserved UGGAGG sequence is boxed, and the substitution of a U in the ApT4-C is indicated in bold. The frequencies of each clone, after cycle eight, are indicated in parentheses. The groups of sequences (I–IV) are also indicated.
Smaller ApT4-A and structural characterization
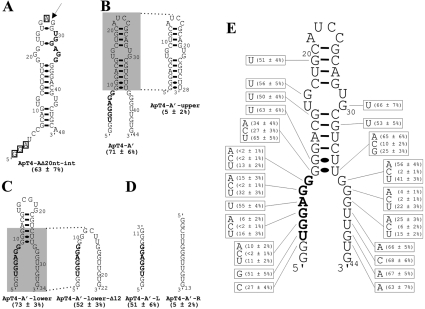
In order to identify a motif binding T4, we chose to study ApT4-A (Figure 2) for two reasons: it was abundant, and one variation of its UGGAGG box was observed (i.e. UGGUGG). Several deletion mutants were synthesized, and their binding on the T4-Sepharose column was assessed (Table 1). All individual mutants exhibited binding to the Sepharose column deprived of T4, although in very small amount (<5%). Therefore any binding activities smaller than 5% were considered negligible. In general, we observed that the estimated percentages were higher than those observed with the selected populations; for example, ApT4-A bound at 61±5%. Briefly, the sequences that were used as the primer-binding sites for PCR can be deleted from the 5′-end without any loss of binding capacity; however, any additional deletion results in a significant loss (i.e. compare ApT4-AΔ20nt-5′ with ApT4-AΔ32nt-5′; Table 1). Conversely, only 17 nt can be removed from the 3′-end without significant loss of binding capacity (compare ApT4-AΔ17nt-3′ with ApT4-AΔ22nt-3′; Table 1). The effects of the various modifications were confirmed with aptamers possessing deletions at both ends (Table 1). Subsequently, in order to further minimize the aptamer, internal deletions were performed (Table 1). When 20 nt from the middle of the aptamer were deleted, the binding ability of the resulting aptamer remained unchanged (ApT4-AΔ20nt-int, 63±7%). However, the further removal of an additional 10 nt significantly reduced the binding percentage (ApT4-AΔ30nt-5′int, 22±3%). Lastly, removal of a further 10 nt, including the UGGAGG box, was noticeably detrimental to binding (ApT4-AΔ30nt-3′int, 6±2%). Thus these results show that it is possible to reduce the aptamer to 48 nt (i.e. ApT4-AΔ20nt-int), and that the presence of the UGGAGG box was essential for efficient binding.
Table 1. Deletion mutants of the ApT4-A aptamer.
The sequences of the PCR primers located at both ends of the aptamer, and the conserved UGGAGG sequence, are boxed.
Transcripts complementary to ApT4-AΔ14nt-5′/Δ17nt-3′ were synthesized, and their binding to the column was tested (Table 1). The resulting ApT4-AΔ-Comp did not exhibit any binding activity, indicating that the nature of the sequence is important. Finally, when ApT4-AΔ14nt-5′/Δ17nt-3′ and ApT4-AΔ-Comp were heat-denatured and slowly cooled together in order to favour their annealing into a double-stranded structure, no binding activity was detected. These results indicate that the secondary structure of the aptamer is important for the binding activity.
The most stable secondary structure of ApT4-AΔ20nt-int was predicted using Mfold [11]. Only one putative structure with a ΔG of −18.1 kcal/mol (1 cal≈4.184 J) was obtained (Figure 3A). Overall, this structure consists of a hairpin that includes one internal loop and several bulges. In order to verify whether or not the positions of the ends were important for efficient binding, we used a circularly permuted RNA strategy [12]. A phosphodiester bond linked positions 4 and 48, while both the 5′- and 3′-ends were shifted to positions 28 and 26 respectively (i.e. in the upper loop, Figure 3A). During this process the nucleotides G1–A3 and A27 were deleted, generating the 44 nt ApT4-A' (Figure 3B). The binding capacity of ApT4-A' was found to be similar to that of the previous version (i.e. 71±6% compared with 63±7% for ApT4-AΔ20nt-int). The secondary structure of ApT4-A' received physical support from an analysis of RNase H (results not shown). The presence of oligonucleotides complementary to the single-stranded regions of both ends allow for the detection of cleavage product, while oligonucleotide complementary to the sequence of the middle stem did not. Thus ApT4-A' exhibited a single-stranded conserved UGGAGG box adjacent to a double-stranded region.
Figure 3. Determination of a minimal structure and important nucleotides for ApT4-A.
(A) Nucleotide sequence and proposed secondary structure of the deletion mutant ApT4-AΔ20nt-int. The binding activity is indicated in parentheses. The highly conserved sequence UGGAGG is shown in bold. The boxed nucleotides indicate bases that were deleted when designing ApT4-A' [see (B)]. The arrow indicates the position where the loop of ApT4-AΔ20nt-int was opened to produce ApT4-A'. (B) Nucleotide sequences and proposed secondary structures of ApT4-A' and ApT4-A'-upper. (C) Nucleotide sequences and proposed secondary structures of ApT4-A'-lower and ApT4-A'-lower-Δ12. (D) Nucleotide sequences and proposed secondary structures of ApT4-A'-L and ApT4-A'-R. (E) Nucleotide sequence and proposed secondary structure of ApT4-A'. All point mutations are indicated in the boxes along with the observed binding levels (averages from at least two independent experiments).
In order to more precisely define the region responsible for the binding of T4, various mutated aptamers were synthesized using ApT4-A' as a starting point. For example, when the lower portion containing the UGGAGG sequence was removed, the residual aptamer (ApT4-A'-upper) was deprived of any binding activity (i.e. 5±2%; Figure 3B). Conversely, when the upper hairpin region was deleted, the resulting aptamer (ApT4-A'-lower) retained full binding activity (i.e. 73±3%; Figure 3C). An additional deletion of 12 nt from the upper domain was also possible, without dramatic loss of binding activity (i.e. ApT4-A'-lower-Δ12, 52±3%; Figure 3C). This surprised us as the resulting aptamer appears likely to be unstructured. Moreover it indicated that the stem, adjacent to the UGGAGG box, had a contribution without being essential (i.e. reduction of 21%). Other mutated aptamers were produced and showed that the identity of the base pairs composing the stem did not significantly influence the binding activity (results not shown). When the last aptamer was split in two RNA strands, the one including the UGGAGG box exhibited a significant binding activity, while the other strand did not (i.e. ApT4-A'-L and ApT4-A'-R showed binding activity at 51±6% and 5±2% respectively; Figure 3D). Together, these results indicate that only the UGGAGG boxed is essential for binding to the T4-Sepharose.
Subsequently point mutations were introduced in many different positions of ApT4-A' in order to identify the nucleotides important for binding (Figure 3E). Only a few mutants resulted in a significant reduction of the binding activity. From the 5′-strand of the aptamer, only mutation of the guanosine residues of the conserved UGGAGG sequence resulted in RNA molecules with low affinities for T4 (i.e. positions 4, 5, 7 and 8). Even the uridine and adenine residues of the UGGAGG can be mutated without significantly reducing the binding percentage (51% and 55% respectively, compared with 71% for the original ApT4-A'). The latter mutation confirmed that the adenosine residue can be replaced by a uridine, as has been observed previously with ApT4-C. Moreover, substitution of the other guanosine residues also results in significant reduction of the binding (e.g. positions 2 and 9). The situation was similar for the point mutations performed on the 3′-strand: only replacement of the three consecutive guanosine residues led to aptamers with low T4 binding (positions 37, 38 and 39). If the aptamer ended with these three guanosine residues, the binding to the T4 was efficient, whereas the replacement of these residues by three adenosines was detrimental to the binding activity (results not shown). Together, these results show that the presence of guanosine residues is the basic building block of this RNA motif. Moreover, these guanosine residues appear to be located in single-stranded regions of the RNA that are adjacent to a stem.
Secondary structures of the different aptamers
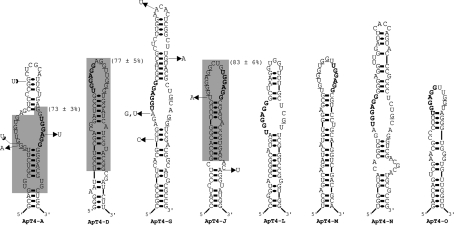
We next asked whether or not the structural features of ApT4-A' could be found in the other aptamers. All aptamers were folded using Mfold, and several of the most stable structures were analysed. After minimal manual adjustments, such as the removal of a G·U wobble base pair formed by one of the highly conserved guanosine residues, all isolated aptamers had the ability to fold into a structure reminiscent of ApT4-A' (Figure 4). In some aptamers, the UGGAGG sequence was located within an internal loop, whereas in others it was found in an external loop (compare ApT4-A, -G, -L and -N with ApT4-D, -J, -M and -O). The loops were always relatively large, ranging in size from 10 to 17 nt with an average of 14.4 nt. Furthermore, the location of the UGGAGG sequence in either the 5′- or 3′-strand of the aptamer loops appears to be unimportant. In ApT4-A it is located in the 3′-strand of the internal loop, whereas in ApT4-G, -L and -N it is in the 5′-strand. The same observation was made when analysing the location of the UGGAGG sequence in the external loop. In ApT4-D and -O it is located in the 5′ portion of the loop, while it is located in the 3′ portion in both ApT4-J and -M. Importantly, in all cases the UGGAGG sequence is juxtaposed to a double-stranded region that, for the most part, appears to be stable. Finally, it is noteworthy that all aptamers showed a high predominance of guanosine residues in the strands opposite to the UGGAGG boxes. For example, ApT4-A, -D, -J and -M had five guanosine residues in these positions, whereas only one or two would be expected if no bias existed. Clearly the presence of guanosine residues is the basic building block of this RNA motif.
Figure 4. Putative secondary structure of all isolated aptamers.
These proposed secondary structures include all requirements known to be important for T4 binding. The lines at both the 5′- and 3′-ends denote sequences that have not been represented in order to simplify the illustration. The natural variants are identified. The UGGAGG sequences are indicated in bold, while the guanosine residues located in front of them are underlined. The grey sections define the smaller versions synthesized and tested for binding and their binding percentages are in parentheses.
In order to verify if the predicted structures were logical, smaller versions of the aptamers ApT4-D and -J, which include the UGGAGG in either the 5′ or 3′ region of the external loop, were synthesized and found to efficiently bind to the T4-Sepharose column (Figure 4, grey sections). Moreover a second in vitro selection experiment was performed using the smaller version of the ApT4-J aptamer (results not shown). In this experiment the 16 positions of the loop, and the two corresponding to the adjacent base pair, were randomized and the selection performed as described above. Fifty clones were sequenced and a predominance of guanosine residues was found in the sequence (i.e. an average of 9.8 guanosine residues in the 18 positions), and the randomized regions appeared to form a single-stranded structure. A large proportion of the aptamers were observed to contain the UGGAGG box. Even when the aptamer did not contain this box, several GG-dimers or GGG-trimers were detected in the loop. Together these data revealed that the selection process isolated a single motif that can be located in many different RNA species.
Evaluation of the G-quartet (guanine quadruplexes) hypothesis
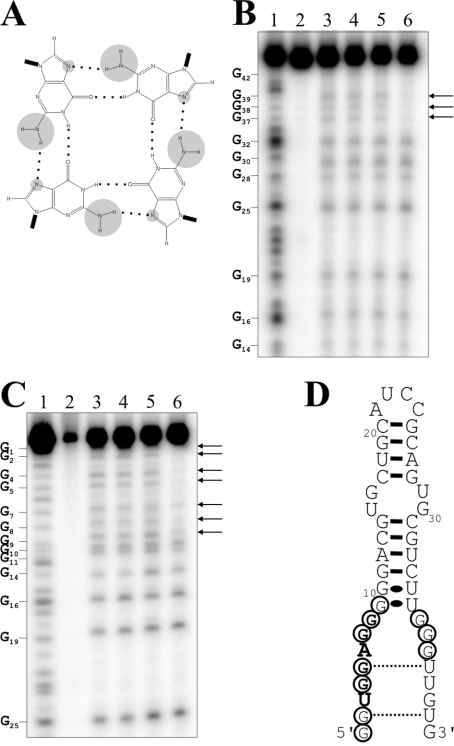
G-rich nucleic acid sequences are well known to adopt intermolecular or intramolecular quadruplex structures that are stabilized by the presence of G-quartets (Figure 5A; and reviewed in [13,14]). Since the G-rich single-stranded domain of the aptamers is reminiscent of a G-quartet structure, ApT4-A' aptamers with GTP replaced by either 7-deaza-GTP or ITP were synthesized. In the case of the 7-deaza-GTP, this substitution replaces the nitrogen and its lone pair at position 7 by a CH group, while in the inosine version, the modification removes the NH2 located at position 2 of the purine ring [15]. With the 7-deaza-GTP, position 7 is incapable of serving as a hydrogen bond acceptor, while the inosine version loses the potential to serve as a hydrogen bond donor from the secondary amine. Consequently, both resulting aptamers should lose four of the eight stabilizing hydrogen bonds per quartet and therefore become unstable. The deaza-GTP would retain the capacity to form Watson–Crick base pairs, but not the inosine version. The ability to bind the T4-Sepharose column was drastically reduced from 71±6% to 26±5% and 18±2% for the deaza- and inosine-aptamers respectively. This supports the hypothesis that the aptamers fold into a structure reminiscent of a G-quartet, rather than into a secondary structure motif.
Figure 5. Investigation of a G-quartet-like structure.
(A) Schematic representation of a G-quartet. The hydrogen bonds between the guanosine residues are indicated by dotted lines. The NH2 groups linked to the C2 and the N7 positions are circled in grey. (B) and (C) are autoradiograms of DMS probing of 5′- and 3′-32P-end-labelled ApT4-A' aptamers respectively. Lanes 1: alkaline hydrolyses; lanes 2: the experiments were performed in absence of salt and DMS. Lanes 3–6: show DMS treatments performed either in the absence of salt or in the presence of LiCl, NaCl or KCl respectively. The arrows indicate the resistant guanosine residues. The positions of the guanosine residues are indicated beside the gels. (D) Secondary structure and nucleotide sequence of ApT4-A'. The resistant guanosine residues are circled.
The hypothesis of a G-rich structure received additional physical support from DMS probing using either 5′- or 3′-32P-end-labelled ApT4-A' aptamers (Figures 5B–5D). The DMS treatment modifies the N7 group of all guanosine residues and was performed either in the absence of salt, or in the presence of LiCl, regardless of whether or not the guanosine residues were present in single- or double-stranded structures. LiCl is well known to suppress G-quartet formation [14,16]. The guanosine residues were also modified in the presence of NaCl, which supports G-quartet formation only under conditions different from those used here [14,16]. Conversely, the seven guanosine residues from the 5′-end and the first three residues from the 3′-end were not modified by DMS treatment when the aptamers were incubated in the presence of KCl (Figures 5B and 5C), which is well known to support G-quartet formation [14,16]. These residues are located in a single-stranded region, and, consequently, should be easily modified by the chemical reaction. Thus the G-quartet was formed only in the presence of KCl and absence of T4. When the experiments were repeated in the presence of T4, the results were similar, with the exception that some protection of the guanosine residues was also observed in the presence of NaCl, although at a considerably reduced level compared with that observed in the presence of KCl (results not shown). These observations support the idea that, in the presence of both NaCl and T4, a small proportion of the aptamers fold into a G-rich structure. Several versions of the experiment were repeated in the presence of T4 in order to permit observation of a higher level of protection of the guanosine residues in the presence of the NaCl. Unfortunately these experiments were unsuccessful, most probably due to the limited solubility of the hormone in water (150 μM; [17]), a property that also prevented several other experiments which required higher hormone concentrations. It is also possible that the T4 was modified during the DMS treatment, which would have the effect of limiting its reactivity with the guanosine residues. More importantly, these results are in agreement with the hypothesis of a G-rich structure. Considering the fact that the ApT4-A' aptamers do not include four G-rich segments, quadruplexes should be formed from either two or four RNA molecules.
T4 is essential for the formation of the G-rich structure
Subsequently, we asked whether or not T4 was important for the formation of this G-rich structure. In order to answer this question, binding shift assays on native gels were performed. The ApT4-A' aptamer was 5′-end labelled, pre-incubated for 1 h in binding buffer, either with or without T4, and the mixtures then fractionated by native PAGE (15% gels) either without or with T4 in the gel. In the absence of T4, the transcripts did not shift when the experiments were performed in the presence of 150 mM NaCl, regardless of the aptamer tested (Figure 6A). Similar results were obtained using either a buffer that did not include a monovalent salt, or one containing LiCl, two conditions incompatible with G-quartet formation (results not shown). In fact, in all of these cases, a faint band corresponding to a product with slower electrophoretic migration was observed under all conditions tested. This product most probably corresponds to a misfolded structure adopted by the ApT4-A' and the several mutated versions tested, or to intermolecular products formed by two molecules due to their complementary sequences in the double-stranded region (i.e. as opposed to intramolecular base-pairing). When the experiments were repeated in the presence of both NaCl and 100 μM T4 in the buffer, a shift of the ApT4-A' aptamers was observed (Figure 6A). In fact, almost all of the aptamers showed a slower electrophoretic migration, a property that is characteristic of the formation of an intermolecular G-quartet like structure [18]. Using a range of concentrations in the sample and the gel preparations, the Kd for T4 was estimated to be 50±10 μM. When the experiment was repeated using a mutated version of ApT4-A' in which the guanosine residue at position 2 was substituted by a cytosine residue (ApT4-A'/G2C), no shift was observed (Figure 6A). This confirmed that the slower mobility observed previously was the result of the formation of the G-rich structure. Several other mutated versions were tested, and the results always correlated with the results of binding to the T4-Sepharose: an aptamer that bound the T4-Sepharose shifted on the native gel (results not shown). Finally, the experiment was repeated using the 44nt ApT4-A' and the 22nt ApT4-A'-lower-Δ12nt together. If the formation of the G-like structure is intramolecular, one shifted band for each aptamer should be detected. Experimentally, we observed three predominant shifted bands of different intensities. Using only one radioactive aptamer at a time permitted the detection of intermolecular complexes including only either ApT4-A' or ApT4-A'-lower-Δ12nt, or the two aptamers together, indicating that at least two RNA molecules were involved in the G-quartet-like structure. However, we also consistently detected, in smaller amounts, two other shifted bands that most probably correspond to other complexes formed under these conditions.
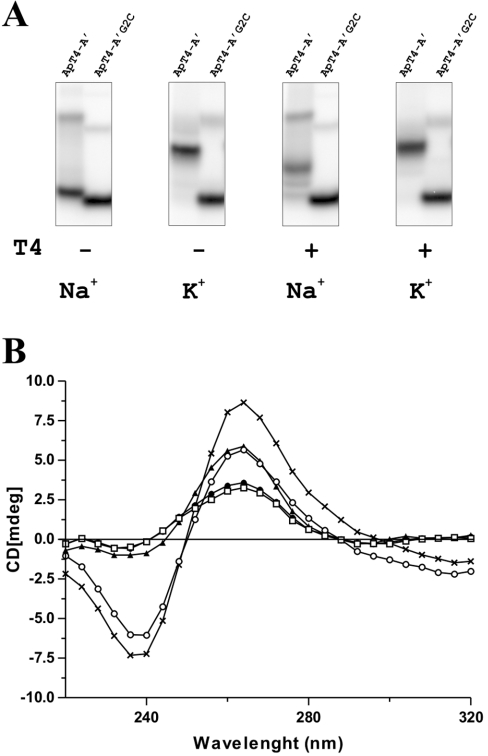
Figure 6. Characterization of the contribution of T4 to the G-quartet-like structure.
(A) Binding shift assays performed with both the ApT4-A' and ApT4-A'G2C aptamers in the presence of 150 mM of either NaCl or KCl, and with (+) or without (−) 100 μM T4. (B) CD spectra performed for the ApT4-A' aptamer under various conditions: no monovalent salt (□), 50 mM NaCl (●), 50 mM KCl (▲) and 100 μM T4 either alone (○) or with 50 mM NaCl (×).
When the experiment was repeated in the presence of 150 mM KCl in the buffers, a condition known to favour G-quartet structure, a shift was observed with the ApT4-A' aptamers, regardless of the presence or absence of the T4 (Figure 6A). However, it is important to note that the position of the latter shift was slightly higher than that observed in the presence of NaCl. This might be an indication that different structures were formed depending on whether NaCl or KCl was present. In the presence of KCl, we also accumulated evidence supporting the notion that the G-quartet structures were intermolecular complexes including at least two aptamers (e.g. it was aptamer concentration dependent). More importantly, together, these results suggest that the T4 is essential for the G-quartet-like structure formation when the buffer contains NaCl.
In light of the results described above, we investigated the conformation of the G-quartet structure adopted by the ApT4-A' aptamer using CD. A quadruplex formed by parallel strands is characterized by a long-wavelength positive maximum peak near 265 nm (and a negative peak at 240 nm), whereas a structure including antiparallel DNA strands is associated with a peak near 293 nm [16]. No specific peaks were detected at these wavelengths when the aptamer was incubated either in the absence of monovalent ions or in the presence of NaCl (Figure 6B). Conversely the addition of either KCl or T4 alone in the samples was sufficient to cause detection of a peak at 265 nm, suggesting that these two conditions were sufficient for a proportion of the aptamers to adopt a G-quartet structure. Interestingly, the addition of both the T4 and NaCl to the ApT4-A' yielded a significantly larger peak at 265 nm, indicating that the G-quartet structure is formed by parallel RNA strands. More importantly, it confirmed the essential role of T4 in the formation of the G-quartet-like structure.
Specificity of T4 binding
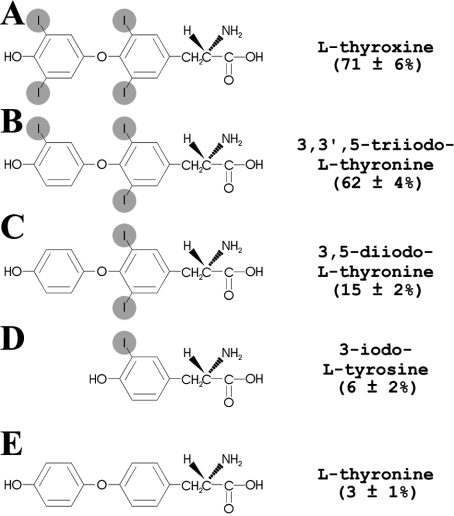
Initially we investigated whether or not the binding of the aptamer to the column was specific to T4. Columns with immobilized T0, iodotyrosine, T2, T3 and T4 were produced, and the binding of ApT4-A' aptamers to these columns was compared (Figure 7). ApT4-A' bound to the T0, iodotyrosine, T2, T3 and T4 columns at 3±1%, 6±2%, 15±2%, 62±4% and 71±6% respectively. In other words, only the T3 and T4 hormones allowed efficient binding, indicating that at least three atoms of iodine must be present for binding to occur. Both the outer and inner rings of T4 appeared to be essential for the binding, and both rings must possess an iodine atom.
Figure 7. Atomic structure of T4-derivatives and binding activity of the ApT4-A'.
(A) T4, (B) T3, (C) T2, (D) 3-iodo-L-tyrosine and (E) T0.
DISCUSSION
The designed in vitro selection protocol identified several RNA species with the capacity to bind the hormone T4. The mutational analysis and RNase H probing of the ApT4-A' in combination with a comparison of the predicted secondary structures of the various aptamers, led to the identification of several common structural features (Figures 3 and 4). Together, these results show that the presence of guanosine residues is the basic building block of this RNA motif. These guanosine residues (and more specifically the UGGAGG box) were located in single-stranded regions that are adjacent to a double-stranded stem. Mutational analysis demonstrated that RNA species possessing this sequence were able to bind to T4-Sepharose better; therefore it is expected that these aptamers will be more abundant. Similar results were obtained in two other independent selection experiments using different preparations of randomized oligonucleotides (results not shown).
The high guanosine residue content, as well as the fact that the randomized region appears to be unable to fold into a secondary structure, led us to postulate that the RNA species may fold into a G-quartet or a G-quartet-like structure. This hypothesis was confirmed by DMS probing, by T4-Sepharose binding using aptamers synthesized with site-specific modified nucleotides and by electrophoresis binding shift assay experiments (Figures 5 and 6). Together, these experiments provide a strong case in favour of the formation of a G-quartet or G-quartet-like structure that most probably involves either two or four parallel aptamer molecules. Interestingly the ApT4-A' DNA aptamer was also observed to bind to the T4-Sepharose, although at a reduced level (results not shown). Considering that DNA molecules can also fold into G-quartet structures, as observed with the telomeric sequence [19], this provides evidence in favour of such a structure. Some RNA and DNA sequences known to form a G-quartet were tested for their ability to bind to T4-Sepharose; however, at best, only weak binding was observed (e.g. the thrombin-binding DNA aptamer; results not shown). This indicates that G-quartet structures do not have the natural ability to bind T4. One way to reconcile these data is to propose that the sequence of the aptamer is important for the G-quartet formation and that the adjacent base-paired region also contributes to the structure involved in the binding to T4. Preliminary in-line probing experiments support this hypothesis. Specifically, when the probing was performed in the presence of T4 the adjacent helical region appears not to be hydrolysed, whereas in the absence of T4 it is (results not shown). However, the latter observation might also result from the fact that once the G-rich structure is formed between several copies of the aptamer, the stems of each one became parallel and were thus protected from hydrolysis.
The mechanism of the binding of T4 to the RNA aptamer is another intriguing issue. One interesting possibility is the formation of halogen bonds. Halogen bonds in biomolecules can be defined as a short C–X···O–Y interaction in which the X is a carbon-bonded chlorine, bromine or iodine, and the O–Y is a carbonyl, hydroxyl, charged carboxylate or phosphate group [20]. Study of the geometry of halogen bonds in small molecules showed that the interaction is primarily electrostatic, with additional contributions from polarization, dispersion and charge transfer [21]. Although halogen bonds are generally referred to as weak interactions, they have been reported to be exploited in the design of very specific and efficient recognition systems involving proteins [22–24]. For example, the binding between T4 and its transport protein transthyretin has been shown to involve the formation of several I···O halogen bonds [22]. It has been suggested that this large number of short halogen bonds plays an essential role in the recognition of this hormone by its cognate proteins [20]. The demonstration of halogen bonds in nucleic acid structures is limited to only two cases [25,26]. In both of these cases the presence of halogen bonds was shown, by crystallographic study, to take place in complex structures adopted by DNA molecules. More specifically, a Br···O–P halogen bond was shown to be formed in a complex four-stranded junction formed by the oligonucleotide d(CCAGTACbr5UGG) (br5U, 5-bromouridine) [25], whereas an I···O–P short link was detected in a six-stranded complex adopted by the oligonucleotide d(Gi5CGAAAGCT) (i5C, 5-iodocytosine) [26]. To our knowledge such halogens bonds have not yet to be observed in RNA structures either in solution or in crystal form. Therefore if they contribute to the specific binding of T4 to the RNA aptamer characterized here, it would be an original observation. Additional high-resolution structural studies using both NMR and X-ray diffraction should provide definitive proof of G-quartet formation, as well as of the involvement of any halogen bonds between the RNA aptamer and T4.
An interesting question is whether or not aptamers were in fact selected for their ability to bind to T4-Sepharose, or for their ability to fold into a G-quartet-like structure. In this regard, the binding shift assays performed in the presence of NaCl are the most relevant since the initial selection was performed in the presence of this salt. In the absence of T4, only a negligible fraction of ApT4-A' shifted, whereas in the presence of T4 most of the aptamers formed complexes with a Kd of approx. 50 μM. Since T4 has a limited solubility in water of 150 μM [17], this Kd value is impressive, because it is impossible to fully saturate the complex with T4. Moreover this situation renders the accurate determination of the aptamer–T4 stoichiometry within the complex almost impossible. However, the binding shift experiments suggest that T4 has a role in the formation of the G-quartet-like motif, explaining why all of the isolated aptamers possessed this feature. Moreover, it eliminates the possibility that the G-rich structures were formed in the solution of transcripts before their application on to the column. More likely, the T4 molecules that bound to the column served as scaffolds for the formation of this structure, in a manner reminiscent of the switch role of sodium-potassium in the formation of the DNA G-quartet [19]. Such structural motifs are not only restricted to the telomeric sequence. For example, DNA G-quartets have been proposed to be formed by the human c-myc oncogene promoter [27], while RNA equivalents have been found in mRNA and proposed to be important for ribonucleoprotein particle formation and mRNA localization [28,29]. As a result, we were not necessarily surprised to find one more example, although in the present case it has no demonstrated biological relevance.
This work provides an original demonstration that RNA species can specifically bind a hormone. Previously a DNA aptamer has been reported to bind to the T4 hormone [30]; however, the structure of this DNA aptamer bears no relation whatsoever to those of the RNA aptamers isolated here. This difference most probably reflects the different conditions used in both experiments. We do not know whether or not such RNA aptamers occur in natural RNA species found in living cells. If so, it might have a biological importance such as the riboswitch reported to regulate mRNA expression in bacteria [5,31]. Clearly, thyroid hormones are a suitable metabolite with which to search for a potential human riboswitch. Finding equivalent structures within natural mRNAs would most likely lead to a breakthrough in the molecular biology of the thyroid hormones.
Acknowledgments
We thank Dr M. Bisaillon (Département de Biochimie, Faculté de Médecine, Université de Sherbrooke, Québec, Canada) for access to the CD. This work was supported by grants from the Canadian Institute of Health Research (CIHR) and the Natural Sciences and Engineering Research Council (NSERC) of Canada to J.-P.P. The RNA group is supported by grants from Génome Québec and Université de Sherbrooke. J.-P.P. holds the Canada Research Chair in Genomics and Catalytic RNA.
References
- 1.Cech T. R. Self-splicing of group I introns. Annu. Rev. Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 2.Yarus M. A specific amino acid binding site composed of RNA. Science. 1988;240:1751–1758. doi: 10.1126/science.3381099. [DOI] [PubMed] [Google Scholar]
- 3.Schroeder R., Waldsich C., Wank H. Modulation of RNA function by aminoglycoside antibiotics. EMBO J. 2000;19:1–9. doi: 10.1093/emboj/19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chow C. S., Bogdan F. M. A structural basis for RNA–ligand interactions. Chem. Rev. 1997;97:1489–1514. doi: 10.1021/cr960415w. [DOI] [PubMed] [Google Scholar]
- 5.Tucker B. J., Breaker R. R. Riboswitches as versatile gene control elements. Curr. Opin. Struct. Biol. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Wilson D. S., Szostak J. W. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 7.Brody E. N., Gold L. Aptamers as therapeutic and diagnostic agents. J. Biotechnol. 2000;74:5–13. doi: 10.1016/s1389-0352(99)00004-5. [DOI] [PubMed] [Google Scholar]
- 8.Cuatrecasas P., Anfinsen C. B. Affinity chromatography. Methods Enzymol. 1971;22:345–378. [Google Scholar]
- 9.Thomson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Kennedy R., Bator J. M., Reading C. A microassay for the determination of iodide and its application to the measurement of the iodination of proteins and the catalytic activities of iodo compounds. Anal. Biochem. 1989;179:138–144. doi: 10.1016/0003-2697(89)90214-5. [DOI] [PubMed] [Google Scholar]
- 11.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nolan J. M., Burke D. H., Pace N. R. Circularly permuted tRNAs as specific photoaffinity probes of ribonuclease P RNA structure. Science. 1993;261:762–765. doi: 10.1126/science.7688143. [DOI] [PubMed] [Google Scholar]
- 13.Davis J. T. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 14.Keniry M. A. Quadruplex structures in nucleic acids. Biopolymers. 2001;56:123–146. doi: 10.1002/1097-0282(2000/2001)56:3<123::AID-BIP10010>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 15.Kuzmine I., Gottlieb P. A., Martin C. T. Structure in nascent RNA leads to termination of slippage transcription by T7 RNA polymerase. Nucleic Acids Res. 2001;29:2601–2606. doi: 10.1093/nar/29.12.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dapic V., Abdomerovic V., Marrington R., Peberdy J., Rodger A., Trent J. O., Bates P. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boulton D. J., Fawcett J. P., Woods D. J. Stability of an extemporaneously compounded levothyroxine sodium oral liquid. Am. J. Health Syst. Pharm. 1996;53:1157–1161. doi: 10.1093/ajhp/53.10.1157. [DOI] [PubMed] [Google Scholar]
- 18.Tang C. F., Shafer R. H. Engineering the quadruplex fold: nucleoside conformation determines both folding topology and molecularity in guanine quadruplexes. J. Am. Chem. Soc. 2006;128:5966–5973. doi: 10.1021/ja0603958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sen D., Gilbert W. A sodium-potassium switch in the formation of four stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 20.Auffinger P., Hays F. A., Westhof E., Ho P. S. Halogen bonds in biological molecules. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16789–16794. doi: 10.1073/pnas.0407607101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corradi E., Meille S. V., Messina M. T., Metrangolo P., Resnati G. Halogen bonding versus hydrogen bonding in driving self-assembly processes perfluorocarbon-hydrocarbon self-assembly, part IX. Angew. Chem. Int. Ed. Engl. 2000;15:1782–1786. doi: 10.1002/(sici)1521-3773(20000515)39:10<1782::aid-anie1782>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 22.Wojtczak A., Cody V., Luft J. R., Pangborn W. Structure of rat transthyretin (rTTR) complex with thyroxine at 2.5 Å resolution: first non-biased insight into thyroxine binding reveals different hormone orientation in two binding sites. Acta Crystallogr. Sect. D. 2001;57:1061–1070. doi: 10.1107/s0907444901007235. [DOI] [PubMed] [Google Scholar]
- 23.Howard E. I., Sanishvili R., Cachau R. E., Mitschler A., Chevrier B., Barth P., Lamour V., Van Zandt M., Sibley E., Bon C., et al. Ultrahigh resolution drug design I: details of interactions in human aldose reductase-inhibitor complex at 0.66 A. Proteins. 2004;55:792–804. doi: 10.1002/prot.20015. [DOI] [PubMed] [Google Scholar]
- 24.De Moliner E., Brown N. R., Johnson L. N. Alternative binding modes of an inhibitor to two different kinases. Eur. J. Biochem. 2003;270:3174–3181. doi: 10.1046/j.1432-1033.2003.03697.x. [DOI] [PubMed] [Google Scholar]
- 25.Hays F. A., Vargason J. M., Ho P. S. Effect of sequence on the conformation of DNA holiday junctions. Biochemistry. 2003;42:9586–9597. doi: 10.1021/bi0346603. [DOI] [PubMed] [Google Scholar]
- 26.Sunami T., Kondo J., Hirao I., Watanabe K., Miura K. I., Takenaka A. Structure of d(GCGAAAGC) (hexagonal form): a base-intercalated duplex as a stable structure. Acta Crystallogr. Sect. D. 2004;60:90–96. doi: 10.1107/s0907444903024703. [DOI] [PubMed] [Google Scholar]
- 27.Phan A. T., Kuryavyi V., Gaw H. Y., Patel D. J. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darnell J. C., Jensen K. B., Jin P., Brown V., Warren S. T., Darnell R. B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 29.Kostadinov R., Malhotra N., Viotti M., Shine R., D'Antonio L., Bagga P. GRSDB: a database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res. 2006;34:D119–D124. doi: 10.1093/nar/gkj073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito Y., Kawazoe N., Imanishi Y. In vitro selected oligonucleotides as receptors in binding assays. Methods. 2000;22:107–114. doi: 10.1006/meth.2000.1041. [DOI] [PubMed] [Google Scholar]
- 31.Winkler W. C. Riboswitches and the role of noncoding RNAs in bacterial metabolic control. Curr. Opin. Chem. Biol. 2005;9:594–602. doi: 10.1016/j.cbpa.2005.09.016. [DOI] [PubMed] [Google Scholar]