Abstract
A study was designed to describe the molecular epidemiology of carbapenem-resistant (CR) Pseudomonas aeruginosa in a large well-defined geographical region with a centralized laboratory system serving one pediatric and three large adult hospitals (acute care centers I, II, and III). Molecular characterization was done using PCR with sequencing of the integron-associated gene cassettes. Pulsed-field gel electrophoresis (PFGE) using a modified combined Stenotrophomas maltophilia and Streptococcus pneumoniae protocol with SpeI was performed on CR P. aeruginosa strains isolated in the Calgary Health Region during 2002-2006. The majority (96%) of metallo-β-lactamase (MBL)-producing isolates produced VIM-2 with gene cassettes aacC1 and aacA4, while 4% produced IMP-7 with gene cassettes aacC4 and aacC1. Eighty-six percent of VIM-2 producers belonged to a cluster (MBLV) that was responsible for nosocomial outbreaks during 2003 (intensive care unit) and 2004 (bone marrow transplant unit) at acute care center I. Environmental isolates from these units also belonged to MBLV. The majority of strains from cluster MBLVR (related to MBLV) were present in acute care center III. Isolates producing IMP-7 belonged to a different cluster (MBLI) and were related to strains described during the 1990s. PFGE of the MBL-negative CR strains showed that 37% belonged to a closely related cluster, NMBL, whose members were predominantly isolated from acute care center II. Our findings suggest that CR and dissemination of MBL clusters among P. aeruginosa populations in large geographic healthcare regions are dynamic processes that require continuous molecular surveillance.
Pseudomonas aeruginosa strains producing metallo-β-lactamases (MBLs) were first reported from Japan in 1991 and, since then, have been described from various parts of the world, including Asia, Europe, Australia, South America, and North America (25). Metallo-β-lactamases have the ability to hydrolyze a wide variety of β-lactam agents, such as penicillins, cephalosporins, and carbapenems. These enzymes require zinc for their catalytic activity and are inhibited by metal chelators, such as EDTA and thiol-based compounds. The genes responsible for the production of MBLs are typically part of an integron structure and are carried on large transferable plasmids (25). Therefore, P. aeruginosa strains producing MBLs are often resistant to different groups of antimicrobial agents, and resistance can be transferred to various types of bacteria. Acquired MBLs can be divided into five categories according to their molecular structures, namely, the IMP, VIM, GIM, SIM, and SPM types (10, 25). Metallo-β-lactamase-producing P. aeruginosa strains have been responsible for several nosocomial outbreaks in tertiary care centers from different parts of the world, illustrating the need for proper infection control practices (19). These isolates have also been responsible for serious infections, such as septicemia and pneumonia, and have been associated with failure of therapy with carbapenems.
In the absence of novel agents for the treatment of infections caused by multiresistant gram-negative bacteria, the uncontrolled spread of MBL producers may lead to treatment failures, with increased morbidity and mortality (22). Previous studies illustrated that MBL-producing P. aeruginosa strains are an important cause of carbapenem resistance (CR) among members of this species isolated from the Calgary Health Region (CHR) (9, 14). The CHR provides all publicly funded healthcare services to the population of >1 million residing in the cities of Calgary and Airdrie, Canada, and numerous adjacent surrounding communities covering an area of 37,000 km2 (1). Acute care is provided principally through one pediatric and three large adult hospitals (acute care centers I, II, and III). A centralized laboratory (Calgary Laboratory Services [CLS]) performs all routine clinical microbiology services for both the community and hospital sites within the CHR.
Very limited data are available regarding the epidemiology of MBL-producing P. aeruginosa strains in large geographical areas. We decided to investigate the molecular epidemiology of CR P. aeruginosa strains isolated in the CHR over a 4-year period, from April 2002 to March 2006.
MATERIALS AND METHODS
Bacterial isolates.
Consecutive nonduplicate isolates of P. aeruginosa which were intermediate or resistant (nonsusceptible) to imipenem (IPM) (MIC, >8 μg/ml; referred to as CR), collected at CLS during April 2002 to March 2006, were included in this study. An additional 10 environmental isolates recovered from contaminated faucets at the intensive care unit (ICU) and the bone marrow transplant unit of acute care center I were also included in this study. Isolates were identified to the species level with a Vitek system (Vitek AMS; bioMérieux Vitek Systems Inc., Hazelwood, MO).
Antimicrobial susceptibility testing.
MICs of the following drugs were determined by Vitek (Vitek AMS; bioMérieux Vitek Systems Inc., Hazelwood, MO) and microdilution panels (Microscan gram-negative NMIC30; Dade Behring Canada Inc., Mississauga, Ontario, Canada): piperacillin (PIP), ceftazidime (CAZ), aztreonam (ATM), IPM, ciprofloxacin (CIP), gentamicin (GEN), amikacin (AMK), and tobramycin (TOB). Colistin (COL) susceptibility was determined using Etest (AB BioDisk Company, Solna, Sweden). The quality control strains used for this part of the study were Escherichia coli ATCC 25922, P. aeruginosa ATCC 27853, and Staphylococcus aureus ATCC 29213. Throughout this study, results were interpreted using CLSI criteria for broth dilution (12).
Identification of metallo-β-lactamases.
The presence of MBLs was evaluated in the clinical isolates of CR P. aeruginosa by using both an EDTA screen test (14) and the MBL Etest (AB BioDisk Company, Solna, Sweden) according to the manufacturer's instructions. DNA template preparation and duplex PCR amplification for the simultaneous detection of blaIMP and blaVIM β-lactamase genes were carried out on a Thermal Cycler 9700 instrument (Applied Biosystems, Norwalk, CT), using previously described primers and conditions (14). Sequencing of the VIM and IMP genes was performed using the class 1 integron primers 5CS (5′-GGC ATC CAA GCA GCA AG-3′) and 3CS (5′-AAG CAG ACT TGA CCT GA-3′) in combination with IMP and VIM primers, respectively. Two different amplicons per isolate were sequenced in an overlapping fashion to ensure that the entire MBL gene allele was obtained. Automated sequencing was performed on the PCR products with an ABI Prism 3100 genetic analyzer (Applied Biosystems, Norwalk, CT) as previously described, using Sequence Analysis software (9).
PFGE.
Pulsed-field gel electrophoresis (PFGE) was performed on the CR isolates by using a modification of two different protocols, previously described for Streptococcus pneumoniae (11) and Stenotrophomas maltophilia (2).
A single isolated colony of P. aeruginosa was inoculated into 2 ml brain heart infusion broth (PML Microbiologicals, Wilsonville, OR) and incubated for 16 to 18 h at 37°C. A 150- to 180-μl aliquot of the overnight culture was transferred to a 1.5-ml microcentrifuge tube, and cells were harvested by centrifugation. Following aspiration of the supernatant, the cell pellet was resuspended in 150 μl PIV buffer (10 mM Tris-HCl [pH 8.0], 1 mM NaCl). An equal volume of molten 1.6% low-melting-point agarose (Bio-Rad Laboratories, Hercules, CA) was added to the cell suspension, mixed thoroughly by pipetting, and then immediately cast into plug molds (Bio-Rad Laboratories, Hercules, CA), which were placed at 4°C for at least 10 min to allow plugs to completely solidify. Plugs were then transferred to 50-ml tubes containing 5 ml freshly prepared lysis buffer (6 mM Tris-HCl [pH 8.0], 1 M NaCl, 0.5% polyoxyethylene 20 cetyl ether, 0.2% deoxycholic acid, 0.5% N-lauroylsarcosine, 1 mg/ml lysozyme, 20 μg/ml RNaseA) (Sigma-Aldrich, St. Louis, MO). Plugs were incubated in lysis buffer in a 37°C water bath for 2 h, and the lysis buffer was then removed and replaced with 5 ml freshly prepared ESP buffer (0.5 M EDTA [pH 9.0], 1% N-lauroylsarcosine, 0.1 mg/ml proteinase K) (Roche Diagnostics, Laval, Quebec, Canada). Plugs were incubated in ESP buffer in a 56°C water bath for 2 to 20 h. Following incubation, ESP buffer was removed, and plugs were rinsed and washed in TE buffer (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA [pH 9.0]).
Pseudomonas aeruginosa agarose plugs were digested with SpeI (Roche Diagnostics, Laval, Quebec, Canada); a reference strain, Salmonella enterica serovar Braenderup H9812, was digested with XbaI (New England Biolabs) and used as an electrophoresis molecular weight marker (7). Briefly, an agarose plug was sliced into thirds; one-third of the plug was transferred to a microcentrifuge tube and equilibrated in 150 μl of 1× restriction enzyme (RE) buffer at 37°C for at least 10 min. Following equilibration, RE buffer was removed and replaced with 100 μl of RE buffer containing either 30 U of SpeI enzyme (P. aeruginosa) or 40 U of XbaI (Salmonella serovar Braenderup H9812 reference strain). SpeI and XbaI digestion mixes were incubated at 37°C. Following SpeI digestion, the buffer was aspirated, and digested plugs were briefly melted at 65°C prior to being loaded into a 1% pulsed-field-certified agarose (Bio-Rad Laboratories, Hercules, CA) gel using ART 200G (Molecular BioProducts, San Diego, CA) wide-bore pipette tips; XbaI-digested reference strain Salmonella serovar Braenderup H9812 plugs were loaded directly into the gel. Electrophoresis was performed on a CHEF MAPPER XA apparatus (Bio-Rad Laboratories, Hercules, CA) at 14°C for 18 h under the following conditions: initial switch time, 5.3 s; final switch time, 34.9 s; included angle, 120°; voltage gradient, 6 V/cm; ramping factor, linear. Gels were stained with ethidium bromide and destained with distilled water prior to illumination under UV light. Images were captured with Quantity One software (Bio-Rad Laboratories, Hercules, CA).
Gel images in TIFF format were exported to BioNumerics software, version 3.0 (Applied Maths, Sint-Martens-Latem, Belgium), for analysis. Comparisons for P. aeruginosa were made using the band-based Dice coefficient, which is a binary coefficient measuring similarity based upon common and different bands. Dendrograms were generated using the unweighted-pair group method using average linkages, with a 1.5% position tolerance. DNA relatedness was calculated based on the Dice coefficient, and isolates were considered genetically related if the Dice coefficient correlation was 80% or greater, which corresponds to the “possibly related (4 to 6 bands difference)” criterion of Tenover et al. (23).
RESULTS
Bacterial isolates.
During the 4-year study period, a total of 3,668 P. aeruginosa strains were isolated at CLS, of which 518 (14.1%) were intermediate or resistant to imipenem: 227 (44%) were isolated from urines, 42 (8%) were isolated from blood, 111 (21%) were isolated from wounds, 106 (20%) were isolated from respiratory tract specimens, and the remaining 32 (6%) were isolated from various other specimens.
Antimicrobial susceptibility.
Of the 518 clinical and 10 environmental CR P. aeruginosa isolates included in this study, 47 (9%) were resistant to PIP, 181 (34%) were resistant to CAZ, 228 (43%) were resistant to TOB, 326 (62%) were resistant to GEN, and 258 (49%) were resistant to CIP (Table 1). The isolates producing MBLs were more resistant to PIP, CAZ, GEN, TOB, and CIP than were CR isolates not producing MBLs (Table 1). Additional antimicrobial susceptibility testing was performed on the MBL-positive isolates and showed resistance to ATM in 16% of isolates and to AMK in 80% of isolates. No COL resistance was detected among the MBL producers (Table 1). Paterson recently suggested definitions for multidrug-resistant and panresistant P. aeruginosa (13); all of the MBL-producing isolates and 52% of the CR isolates not producing MBLs were multidrug resistant, while 11% of the MBL-producing isolates and 8% of the CR isolates not producing MBLs were panresistant.
TABLE 1.
Resistance rates for clinical and environmental isolates of CR Pseudomonas aeruginosa isolated in the Calgary Health Region during 2002-2006
| Antibiotic | No. (%) of resistant isolates
|
||
|---|---|---|---|
| CR (n = 528)a | Non-MBL producer (n = 343)b | MBL producer (n = 185) | |
| PIP | 47 (9) | 27 (8) | 20 (11) |
| CAZ | 181 (34) | 20 (8) | 161 (88) |
| CIP | 258 (49) | 83 (24) | 175 (95) |
| GEN | 326 (62) | 159 (46) | 167 (90) |
| TOB | 228 (43) | 73 (21) | 155 (84) |
| AMKc | 148 (80) | ||
| ATMc | 30 (16) | ||
| COLc | 0 | ||
Intermediate or resistant to imipenem (MIC, >8 μg/ml).
IPM-intermediate or -resistant isolates not producing MBLs.
Additional antimicrobial testing was performed on the MBL-positive isolates.
Identification of metallo-β-lactamases and cassette arrays.
Of the 528 CR P. aeruginosa isolates included in this study, 185 (35%) were MBL positive using phenotypic methods: 178/185 (96%) were positive for blaVIM genes and 7/185 (4%) were positive for blaIMP genes. PCRs with primers 5C and 3C and with different combinations of IMP and VIM primers, respectively, amplified various amplicons, ranging from 900 bp to 1.2 kb. Sequence analysis of the products obtained with the VIM primers revealed 100% identity with the sequence of the blaVIM-2 allele (15). We detected the gene cassettes in eight of the VIM-2-producing P. aeruginosa isolates; two of eight profiles consisted of a sole blaVIM-2 cassette, three consisted of blaVIM-2 and aacC1, and the remaining three consisted of blaVIM-2, aacA4, and aacC1. Two isolates were randomly selected from 2002, two were selected from 2003, two were selected from 2004, and two were selected from 2005. Sequence analysis of IMP products revealed 100% identity with the sequence of the blaIMP-7 allele (6). We also detected the gene cassettes of all the IMP-7-producing P. aeruginosa isolates; three cassettes consisted of aacC4, blaIMP-7, and aacC1, and four consisted of blaIMP-7 and aacC1.
Pulsed-field gel electrophoresis.
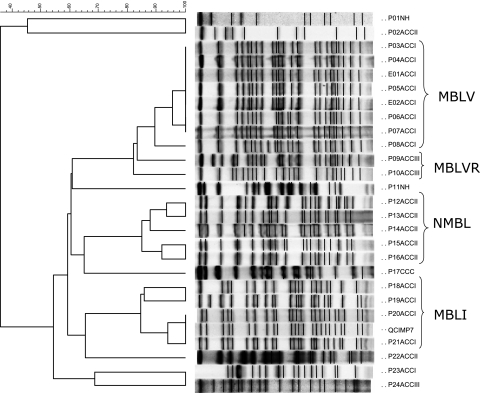
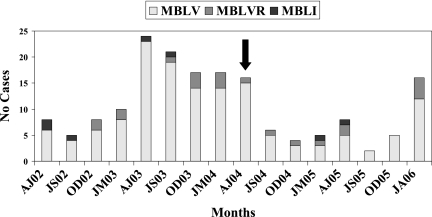
PFGE revealed two closely related restriction patterns (clusters MBLV [154 isolates] and MBLVR [related to MBLV] [21 isolates]) for the 178 VIM-2-producing isolates (Fig. 1). The remaining three VIM-2-producing strains were not related to these clusters or to each other. The majority (96/154 [62%]) of the strains belonging to cluster MBLV were isolated from patients admitted to acute care center I, 7/154 (4%) strains were isolated from acute care center II, 16/154 (10%) strains were isolated from acute care center III, 14/154 (9%) were isolated from nursing homes, and 11 (7%) were isolated from patients at community care centers. The remaining 10 isolates that belonged to cluster MBLV were recovered from contaminated faucets at the ICU and the bone marrow transplant unit of acute care center I. Of the 48 patients with cluster MBLV strains that received medical care outside acute care center I, 42 (88%) were transferred or discharged from acute care center I. An increased number of MBLV strains were isolated during April 2003 to June 2004 and again from January to April 2006 (Fig. 2).
FIG. 1.
PFGE patterns and dendrogram of MBL-producing P. aeruginosa strains isolated from the Calgary Health Region during April 2002 to March 2006. The dendrogram was constructed by cluster analysis using Bionumerics software (version 3.0). Percentages of similarity are shown above the dendrogram. MBLV and MBLVR (related to V) strains produced VIM-2, and MBLI (producing IMP-7) and NMBL (MBL negative) indicate clusters with >80% homology. P01, patient isolate 01, etc.; E01, environmental isolate 01, etc.; ACCI, acute care center I; ACCII, acute care center II; ACCIII, acute care center III; NH, nursing home; CCC, community care center. These isolates were randomly selected to show the separation of DNA fragments.
FIG. 2.
Distribution of MBL-producing P. aeruginosa strains isolated in the Calgary Health Region during April 2002 to March 2006. MBLV, cluster producing VIM-2; MBLVR, cluster related to MBLV, producing VIM-2; MBLI, cluster producing IMP-7; AJ, April to June; JS, July to September; OD, October to December; JM, January to March; -02 to -06, 2002 to 2006, respectively. The arrow denotes the replacement of faucets at acute care center I.
The majority (8/21 [38%]) of strains belonging to cluster MBLVR were isolated from patients admitted to acute care center III, 5/21 (24%) were isolated from acute care center I (during 2006), 1/21 (5%) was isolated from acute care center II, 2/21 (10%) were isolated from nursing homes, and five (24%) were submitted from community care centers. These strains were consistently isolated over the 4-year period, without an increase in number like that seen with the MBLV cluster (Fig. 2).
The IMP-7-producing isolates (n = 7) belonged to a different cluster (MBLI) and were related to the strains described during an outbreak in the 1990s (6). Strains belonging to this cluster were isolated sporadically during the 4-year study period.
One hundred forty-four isolates that were CR but tested negative for MBLs were available for PFGE. Typing of these isolates showed the following patterns: 53/144 were closely related (cluster NMBL) (Fig. 1), 4/144 belonged to cluster MBLV, 1/144 belonged to cluster MBLVR, 3/144 belonged to cluster MBLI, and the remaining 83 were not related to any of the clusters or to each other. The majority (27/53 [51%]) of strains belonging to cluster NMBL were isolated from patients admitted to acute care center II, 13/53 (25%) were isolated from acute care center I, 5/53 (9%) were isolated from acute care center II, 5/53 (9%) were isolated from nursing homes, and 3 (6%) were submitted from community care centers. Of the 26 patients with cluster NMBL strains that received medical care outside acute care center II, 8 (31%) were transferred from or were in contact with acute care center II.
DISCUSSION
Pseudomonas aeruginosa isolates producing IMP-7 were responsible in the 1990s for an outbreak at acute care center I in the Calgary Health Region (6). We needed to establish a technique at Calgary Laboratory Services for the reliable typing of P. aeruginosa strains producing MBLs. Therefore, we tried several previously published PFGE methods for P. aeruginosa (18, 20, 21) and found that although the discriminatory powers of the individual techniques varied, the overall genetic clustering was independent of the PFGE technique, either by visual inspection or by computer-generated analysis of dendrograms. However, the results obtained with these methods were not optimal in our laboratory, e.g., we experienced poor band resolution and low DNA yields using these techniques (data not shown). Based on previous experience with typing of nonfermenting gram-negative bacteria, we decided to combine two previously described typing methods, for S. pneumoniae (11) and S. maltophilia (2). This new technique (described in detail in Materials and Methods) provided the best separation of DNA fragments and was easier to interpret than the previously published methods. The reproducibility of this method was demonstrated on at least three separate occasions, using five different isolates. We describe a modified PFGE technique for the typing of P. aeruginosa and recommend it as an alternative for molecular surveillance of this medically important organism.
The molecular epidemiology of MBL-producing P. aeruginosa strains on a regional or countrywide scale has been described previously for Poland (4), Greece (5), Italy (17, 24), and Japan (8). These studies consisted of typing of MBL-producing P. aeruginosa strains and the identification of integron-associated gene cassettes collected from different parts of the respective countries over specific periods of time ranging from 1 month to 3 years. The majority of these studies have shown that the emergence of MBL-producing P. aeruginosa strains (especially VIM-2-producing strains) occurs simultaneously and seemingly independently in different parts of the respective countries. The situation in Canada is very different in that MBL-producing bacteria have only been described for the Calgary Health Region (6, 9) and do not seem to be present in other parts of the country. Our study describes the molecular epidemiology of CR P. aeruginosa in a large, well-defined geographical region over a 4-year period. We studied all of the MBL-producing isolates as well as a portion of the non-MBL-producing isolates collected in the Calgary Health Region during that period. We identified the class 1 integron-associated gene cassettes in a small number of VIM-2-producing isolates, which had striking similarities to integron-associated structures previously described for VIM-2-producing P. aeruginosa strains from Portugal (16). This suggests that the emergence of VIM-2-producing P. aeruginosa occurs independently in different parts of the world.
Molecular typing of VIM-2-producing P. aeruginosa strains showed that clonally related strains (cluster MBLV) were responsible for a nosocomial outbreak in the ICU of acute care center I during April to December 2003 (Fig. 2). This cluster then spread to the bone marrow transplant unit and caused a similar outbreak during January to May 2004. Environmental strains isolated from faucets in both of these units also belonged to cluster MBLV (Fig. 1). This outbreak was contained with strict contact isolation practices and the replacement of faucets at both of the units (arrow in Fig. 2). An increase in the number of newly infected patients with clusters MBLV and MBLVR occurred during January and February 2006 at acute care center I (Fig. 2). This was the first appearance of MBLVR at acute care center I. These cases originated from different wards and units, and strict isolation and barrier precautions prevented a large-scale outbreak at this institution, underscoring the importance of infection control practices and continuous molecular surveillance of MBL-producing organisms.
Molecular typing further illustrated the ease with which VIM-2-producing strains from cluster MBLV accompanied patients transferred to other acute care centers, nursing homes, or the community. However, these strains did not cause an outbreak outside acute care center I, underlining yet again the importance of environmental reservoirs as a cause of nosocomial outbreaks due to P. aeruginosa (3). Typing also showed that three clusters were present at the different adult acute care centers within the Calgary Health Region. Of particular interest is the fact that MBL-producing P. aeruginosa strains have not been isolated from patients in the pediatric hospital. Cluster MBLI, which was responsible for a nosocomial outbreak at acute care center I during the late 1990s, practically disappeared during the study period, being replaced by cluster MBLV. Strains from clusters MBLV and MBLVR have nearly identical susceptibility profiles to that of cluster MBLI strains and share the aacC1 gene cassette, but they behave differently since infections caused by MBLV and MBLVR strains were associated with a higher case fatality rate and invasive disease (9). In our previous population-based study, we described the MBLA, MBLAR, and MBLB clusters. There is some overlap with the present study, as cluster MBLV is the same as MBLA, MBLVR is the same as MBLAR, and MBLI is the same as MBLB. Future investigations, including the study of microbiological and ecological factors that make MBLV such a successful pathogen, will hopefully provide some insight into these interesting issues. Our findings suggest that the spread and dissemination of MBL clusters among P. aeruginosa populations in large geographic healthcare regions are dynamic processes that require continuous molecular surveillance.
Acknowledgments
We thank Jo-Anne McClure and Philip Le for their technical support of this study and Terry Ross for database management.
This study was funded by the Antibiotic Resistant Organism (ARO) Research fund, which is sponsored by a partnership between Calgary Laboratory Services, the Calgary Health Trust, the University of Calgary, and the Calgary Health Region.
Footnotes
Published ahead of print on 22 November 2006.
REFERENCES
- 1.Church, D. L., C. Don-Joe, and B. Unger. 2000. Effects of restructuring on the performance of microbiology laboratories in Alberta. Arch. Pathol. Lab. Med. 124:357-361. [DOI] [PubMed] [Google Scholar]
- 2.Denton, M., N. J. Todd, K. G. Kerr, P. M. Hawkey, and J. M. Littlewood. 1998. Molecular epidemiology of Stenotrophomonas maltophilia isolated from clinical specimens from patients with cystic fibrosis and associated environmental samples. J. Clin. Microbiol. 36:1953-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deplano, A., O. Denis, L. Poirel, D. Hocquet, C. Nonhoff, B. Byl, P. Nordmann, J. L. Vincent, and M. J. Struelens. 2005. Molecular characterization of an epidemic clone of panantibiotic-resistant Pseudomonas aeruginosa. J. Clin. Microbiol. 43:1198-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiett, J., A. Baraniak, A. Mrowka, M. Fleischer, Z. Drulis-Kawa, L. Naumiuk, A. Samet, W. Hryniewicz, and M. Gniadkowski. 2006. Molecular epidemiology of acquired-metallo-β-lactamase-producing bacteria in Poland. Antimicrob. Agents Chemother. 50:880-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giakkoupi, P., G. Petrikkos, L. S. Tzouvelekis, S. Tsonas, N. J. Legakis, and A. C. Vatopoulos. 2003. Spread of integron-associated VIM-type metallo-β-lactamase genes among imipenem-nonsusceptible Pseudomonas aeruginosa strains in Greek hospitals. J. Clin. Microbiol. 41:822-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibb, A. P., C. Tribuddharat, R. A. Moore, T. J. Louie, W. Krulicki, D. M. Livermore, M. F. Palepou, and N. Woodford. 2002. Nosocomial outbreak of carbapenem-resistant Pseudomonas aeruginosa with a new blaIMP allele, blaIMP-7. Antimicrob. Agents Chemother. 46:255-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunter, S. B., P. Vauterin, M. A. Lambert-Fair, M. S. Van Duyne, K. Kubota, L. Graves, D. Wrigley, T. Barrett, and E. Ribot. 2005. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43:1045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura, S., J. Alba, K. Shiroto, R. Sano, Y. Niki, S. Maesaki, K. Akizawa, M. Kaku, Y. Watanuki, Y. Ishii, and K. Yamaguchi. 2005. Clonal diversity of metallo-β-lactamase-possessing Pseudomonas aeruginosa in geographically diverse regions of Japan. J. Clin. Microbiol. 43:458-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laupland, K. B., M. D. Parkins, D. L. Church, D. B. Gregson, T. J. Louie, J. M. Conly, S. Elsayed, and J. D. Pitout. 2005. Population-based epidemiological study of infections caused by carbapenem-resistant Pseudomonas aeruginosa in the Calgary Health Region: importance of metallo-β-lactamase (MBL)-producing strains. J. Infect. Dis. 192:1606-1612. [DOI] [PubMed] [Google Scholar]
- 10.Lee, K., J. H. Yum, D. Yong, H. M. Lee, H. D. Kim, J. D. Docquier, G. M. Rossolini, and Y. Chong. 2005. Novel acquired metallo-β-lactamase gene, blaSIM-1, in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob. Agents Chemother. 49:4485-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louie, M., L. Louie, G. Papia, J. Talbot, M. Lovgren, and A. E. Simor. 1999. Molecular analysis of the genetic variation among penicillin-susceptible and penicillin-resistant Streptococcus pneumoniae serotypes in Canada. J. Infect. Dis. 179:892-900. [DOI] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. 2004. Performance standards for antimicrobial susceptibility testing; 14th informational supplement M100-S14. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 13.Paterson, D. L. 2006. The epidemiological profile of infections with multidrug-resistant Pseudomonas aeruginosa and Acinetobacter species. Clin. Infect. Dis. 43(Suppl. 2):S43-S48. [DOI] [PubMed] [Google Scholar]
- 14.Pitout, J. D., D. B. Gregson, L. Poirel, J. A. McClure, P. Le, and D. L. Church. 2005. Detection of Pseudomonas aeruginosa producing metallo-β-lactamases in a large centralized laboratory. J. Clin. Microbiol. 43:3129-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J. D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase, and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quinteira, S., J. C. Sousa, and L. Peixe. 2005. Characterization of In100, a new integron carrying a metallo-β-lactamase and a carbenicillinase, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:451-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riccio, M. L., L. Pallecchi, J. D. Docquier, S. Cresti, M. R. Catania, L. Pagani, C. Lagatolla, G. Cornaglia, R. Fontana, and G. M. Rossolini. 2005. Clonal relatedness and conserved integron structures in epidemiologically unrelated Pseudomonas aeruginosa strains producing the VIM-1 metallo-β-lactamase from different Italian hospitals. Antimicrob. Agents Chemother. 49:104-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romao, C. M., Y. N. Faria, L. R. Pereira, and M. D. Asensi. 2005. Susceptibility of clinical isolates of multiresistant Pseudomonas aeruginosa to a hospital disinfectant and molecular typing. Mem. Inst. Oswaldo Cruz 100:541-548. [DOI] [PubMed] [Google Scholar]
- 19.Rossolini, G. M. 2005. Acquired metallo-β-lactamases: an increasing clinical threat. Clin. Infect. Dis. 41:1557-1558. [DOI] [PubMed] [Google Scholar]
- 20.Speijer, H., P. H. Savelkoul, M. J. Bonten, E. E. Stobberingh, and J. H. Tjhie. 1999. Application of different genotyping methods for Pseudomonas aeruginosa in a setting of endemicity in an intensive care unit. J. Clin. Microbiol. 37:3654-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spencker, F. B., S. Haupt, M. C. Claros, S. Walter, T. Lietz, R. Schille, and A. C. Rodloff. 2000. Epidemiologic characterization of Pseudomonas aeruginosa in patients with cystic fibrosis. Clin. Microbiol. Infect. 6:600-607. [DOI] [PubMed] [Google Scholar]
- 22.Talbot, G. H., J. Bradley, J. E. Edwards, Jr., D. Gilbert, M. Scheld, and J. G. Bartlett. 2006. Bad bugs need drugs: an update on the development pipeline from the Antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin. Infect. Dis. 42:657-668. [DOI] [PubMed] [Google Scholar]
- 23.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toleman, M. A., D. Biedenbach, D. M. Bennett, R. N. Jones, and T. R. Walsh. 2005. Italian metallo-β-lactamases: a national problem? Report from the SENTRY Antimicrobial Surveillance Programme. J. Antimicrob. Chemother. 55:61-70. [DOI] [PubMed] [Google Scholar]
- 25.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]