Abstract
Many strains of Helicobacter pylori are naturally competent for transformation and able to transfer chromosomal DNA among different isolates using a conjugation-like mechanism. In this study, we sought to determine whether H. pylori can transfer DNA into Campylobacter jejuni, a closely related species of the Campylobacterales group. To monitor the transfer, a chromosomally encoded streptomycin resistance cassette prearranged by a specific mutation in the rpsL gene of H. pylori was used. Mating of the bacteria on plates or in liquid broth medium produced C. jejuni progeny containing the streptomycin marker. DNA transfer was unidirectional, from H. pylori to C. jejuni, and the progeny were genetically identical to C. jejuni recipient strains. DNase I treatment reduced but did not eliminate transfer, and DNase I-treated cell supernatants did not transform, ruling out phage transduction. Recombinants also did not occur when the mating bacteria were separated by a membrane, suggesting that DNA transfer requires cell-to-cell contact. Transfer of the streptomycin marker was independent of the H. pylori comB transformation system, the cag pathogenicity island, and another type IV secretion system called tfs3. These findings indicated that a DNase I-resistant, conjugation-like mechanism may contribute to horizontal DNA transfer between different members of the Campylobacteriales group. The significance of this DNA uptake by C. jejuni in the context of acquiring antibiotic resistance is discussed.
Within the Epsilonproteobacteria, the order Campylobacteriales comprises two families, the Helicobacteraceae and the Campylobacteraceae. Helicobacter pylori and the enterohepatic species Helicobacter hepaticus are classified as cancer-causing microorganisms because the infections they produce can lead to gastric cancer in humans and liver cancer in rodents, respectively. However, not every infection leads to disease development, and H. pylori can persist in the human stomach asymptomatically. Campylobacter jejuni is one of the main causes of bacterial food-borne illness worldwide. It is also the most common microbial antecedent to Guillain-Barré syndrome and persists as a commensal in avian hosts. The genus Wolinella is represented by Wolinella succinogenes, a bacterium that persists as a commensal in the gastrointestinal tracts of cattle by utilizing metabolites present in the rumen.
There are presently 11 complete genome sequences available from the Campylobacteriales group: six from the Helicobacteraceae—three distinct strains of H. pylori, 26695, J99, and HPAG1 (2, 3, 44, 53); Helicobacter acinonychis (17); W. succinogenes DSM 1740 (5); and H. hepaticus ATCC 51449 (51)—and five from the Campylobacteraceae—C. jejuni NCTC 11168 and RM1221, Campylobacter lari RM2100, Campylobacter upsaliensis RM3195, and Campylobacter coli RM2228 (22, 46). The availability of these genomes has allowed the study of genome rearrangements that have taken place since these bacteria diverged from their last common ancestor. These ongoing dynamic processes and the level of genome plasticity have been investigated on an intraspecies level, as well as on an interspecies level (2, 18). It is known that horizontal DNA transfer among bacteria is a major factor contributing to their genetic variability (13, 19, 20, 26, 42). Therefore, elucidating the mechanisms involved in DNA transfer will help us understand the adaptation of H. pylori and C. jejuni to changing environmental conditions and may have clinical relevance in the area of antibiotic resistance.
The various mechanisms of bacterial DNA transfer include (i) uptake of DNA by genetic transformation (16), (ii) bacteriophage transduction (35), and (iii) conjugative DNA transfer mediated by the so-called type IV secretion system (T4SS) (45). The H. pylori chromosome has been shown to encode at least three T4SSs (6, 7), although there is no evidence for either bacteriophage transduction or type IV pilin-like proteins associated with bacterial natural competence in H. pylori. The first identified T4SS of H. pylori is encoded by the 40-kb cytotoxin-associated gene (cag) pathogenicity island (PAI), consisting of up to 31 genes (14). This T4SS represents a major disease-associated determinant for the delivery of virulence factors, such as the CagA protein, into host target cells. The second T4SS of H. pylori is the comB system, consisting of the comB2 to comB4 and comB6 to comB10 genes. This system has been shown to mediate the uptake of naked DNA (29). The third T4SS gene cluster found in certain strains of H. pylori is a segment named type IV secretion system 3 (tfs3), located in one of the two plasticity zones (31). This cluster corresponds to a 16.3-kb DNA segment including up to 16 open reading frames, some of which are homologous to the genes virB4, virB7 to virB11, and virD4 in Agrobacterium tumefaciens. No specific function has been ascribed to this putative T4SS.
Although many H. pylori strains are naturally competent for transformation (29) and conjugation-like chromosomal-DNA transfer in vitro (33), the mechanisms for genetic exchange among H. pylori strains in nature remain enigmatic. The aim of this study was to further characterize the process of conjugative chromosomal DNA transfer in H. pylori and to examine whether H. pylori is able to transfer DNA into other related bacteria, such as C. jejuni. We show here that H. pylori is capable of transferring chromosomally encoded streptomycin resistance into C. jejuni by a conjugation-like mechanism in a manner independent of the three previously identified T4SSs.
MATERIALS AND METHODS
Bacterial strains.
The wild-type and mutant H. pylori strains used in this study, as well as their origins, are listed in Table 1. These strains were selected because they do not contain any plasmids. This attribute was verified by plasmid preparations using the Wizard Plus SV Minipreps purification system (Promega) as described by Hofreuter and Hass (28). H. pylori was grown on solid or in liquid medium. The solid medium consisted of GC agar plates supplemented with 10% horse serum (Biochrom, Berlin, Germany), 10 μg/ml of vancomycin, 1 μg/ml of nystatin, and 5 μg/ml of trimethoprim (8). Antibiotics were purchased from Sigma-Aldrich (Deisenhofen, Germany). The liquid medium consisted of brain heart infusion (BHI) broth with 10% horse serum. Incubation was performed at 37°C for 2 days in an anaerobic jar under microaerophilic conditions (5% O2, 10% CO2, and 85% N2) generated by CampyGen (Oxoid, Wesel, Germany). C. jejuni strains (Table 2) were grown on Campylobacter Blood-Free Agar Base containing Campylobacter Growth Supplement (Oxoid) at 37°C under microaerophilic conditions for 48 h. C. jejuni 81-176 was isolated from an outbreak of campylobacteriosis and has been shown to cause disease in human volunteers (11, 32). Strains 1543 and ST3046 were isolated from feces of patients with diarrhea at the Institute of Medical Microbiology (Magdeburg, Germany).
TABLE 1.
Helicobacter pylori and Campylobacter jejuni strains used in this study
| Bacterial strain | Genotypea
|
Phenotypea
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Plasmid | cag PAI | vacA | tfs3 | comB | virB11 | flaA | ciaB | cdt | Strr | Sptr | Rifr | Kanr | Origin | Reference | |
| H. pylori | |||||||||||||||
| 26695 wtb | − | + | + | − | + | − | − | − | − | United Kingdom | 53 | ||||
| P1 wt | − | + | + | − | + | − | − | − | − | Germany | 43 | ||||
| 1061 wt | − | − | + | − | + | − | − | − | − | Canada | 25 | ||||
| 26695 | − | + | + | − | + | − | − | − | − | United Kingdom | This study | ||||
| P1 | − | + | + | − | + | + | − | − | − | Germany | This study | ||||
| 1061 | − | − | + | − | + | + | − | − | − | Canada | This study | ||||
| 26695ΔureB | − | + | + | − | + | + | − | − | + | United Kingdom | This study | ||||
| 1061ΔureB | − | − | + | − | + | + | − | − | + | Canada | This study | ||||
| P1ΔureB | − | + | + | − | + | + | − | − | + | Germany | This study | ||||
| P1ΔcomB7-10 | − | + | + | − | − | + | − | − | + | Germany | This study | ||||
| P1ΔcagPAI | − | − | + | − | + | + | − | − | + | Germany | This study | ||||
| C. jejuni | |||||||||||||||
| 1543 | − | − | + | + | + | − | + | + | − | Germany | This study | ||||
| ST3046 | − | − | + | + | + | − | + | + | − | Germany | This study | ||||
| 81-176 | +c | + | + | + | + | − | + | + | − | United States | 11, 32 | ||||
TABLE 2.
Effect of DNase I treatment on the frequency of recombination between H. pylori and C. jejuni strains
| Strain(s)a | Treatment with DNase I (200 μg/ml)b | Transconjugants
|
|
|---|---|---|---|
| Sptr/Strr (no. of CFU) | Relative %c | ||
| H. pylori alone | − | 0 | 0 |
| C. jejuni alone | − | 0 | 0 |
| H. pylori + C. jejuni | − | 3.4 × 10−7 | 100 |
| H. pylori + C. jejuni | + | 4.4 × 10−8 | 13d |
Plate matings were done in triplicate experiments. H. pylori P1 wild type (Strr); C. jejuni 1543 wild type (Sptr).
+, treated; −, untreated.
Relative to H. pylori plus C. jejuni without DNase I.
P < 0.01 in comparison with H. pylori plus C. jejuni without DNase I (Mann/Whitney test).
Mating experiments on GC agar plates.
Mating experiments were carried out using plasmid-free H. pylori strains. We introduced chromosomal antibiotic resistance markers into various strains using the mating experiments on solid medium described by Kuipers et al. (33). The markers were streptomycin resistance (Strr) and streptomycin resistance/kanamycin resistance (Strr/Kanr). To easily monitor transfer of the Strr marker, we selected C. jejuni strains 1543, ST3046, and 81-176 because they are naturally resistant to spectinomycin (Sptr) and rifampin (Rifr) (Table 1). The frequency of DNA transfer was assessed by quantitating the number of Strr/Sptr doubly resistant recombinants per parent. Each mating experiment involved one H. pylori and one C. jejuni strain, represented by A and B, respectively, with mutually exclusive antibiotic resistance markers. After 12 h of growth on GC agar, the bacterial growth was harvested and suspended in 3 ml of BHI broth. The number of bacteria was calculated from a standard curve of optical densities at 600 nm. Matings were assayed by mixing strains A and B containing known resistances (Table 1). Aliquots of 100 μl of the bacterial suspension (∼1 × 109 bacteria) from each strain were plated on four GC agar plates in the following order: plate 1, strain A; plate 2, strain B; plates 3 and 4, strain A and strain B together. Plates 3 were supplemented with DNase I (Roche, Mannheim, Germany) and MgCl2 at final concentrations of 200 μg/ml and 2 mM, respectively. In the control experiments, the bacteria were incubated in the presence of 0.1-μm-pore-size membranes (Millipore, Schwalbach, Germany) that blocked cell-to-cell contact between the donor and recipient strains. After incubation overnight, bacteria were harvested and suspended in 1 ml of BHI broth. The suspensions from plates 1 were serially diluted, and 100 μl of 10−5, 10−6, and 10−7 dilutions were inoculated on GC agar plates without antibiotics. To exclude the occurrence of spontaneous mutants, aliquots of 200 μl of the undiluted samples from plates 1 and 2 were plated on GC agar plates containing 10 μg per ml each of streptomycin and spectinomycin (GCSS plates). Undiluted suspensions of each individual sample of plates 3 and 4 were inoculated as follows: 200 μl on a GC agar plate without antibiotics and 250, 100, and 25 μl on three GCSS plates. All plates were incubated under microaerophilic conditions for 96 h, after which the colonies were counted. Single colonies from GCSS plates were subcultured onto GC agar plates without antibiotics. To determine the direction of DNA transfer, bacteria were cultured on GC agar plates containing either 10 μg of kanamycin per ml or 10 μg of rifampin per ml.
DNA exchange using cell extracts, purified total DNA, or heat-inactivated bacterial cells.
Mating experiments of the C. jejuni recipient strain with either cell extracts, purified DNA (2 μg), or heat-inactivated H. pylori cells were performed as described above. The total DNA of strain P1 was isolated using a genomic-DNA preparation kit (Roche, Mannheim, Germany). Cell extracts from 1 × 109 H. pylori cells were prepared by five freeze-thaw steps, followed by passage through a 0.2-μm sterile filter (Roth, Germany). Heat-inactivated H. pylori cells were prepared by incubation at 80°C for 10 min. These experiments were done in the presence and absence of 200 μg/ml DNase I and 2 mM MgCl2. After 12 h of incubation, bacteria from each of the suspensions were inoculated on nonselective and selective plates as described above.
Random amplified polymorphic DNA (RAPD) fingerprinting, PCR of bacterial marker genes, and sequencing of the transferred rpsL gene.
Chromosomal DNA was prepared from wild-type and recombinant bacteria using a genomic-DNA preparation kit (QIAGEN). RAPD PCRs were carried out in 25-μl mixtures that contained 20 ng genomic H. pylori or C. jejuni DNA, 3 mM MgCl2, 250 μM deoxynucleotide triphosphates, 1 unit of Taq polymerase in 1× buffer (QIAGEN), 30 pmol of the RAPD primer D9355 under cycling conditions described previously (1). The PCR products were resolved in 1.0% agarose gels and visualized by staining them with ethidium bromide.
Because the streptomycin resistance in H. pylori is mediated by a single point mutation (K43R) in the rpsL gene product (21), the rpsL gene of the recombinant progeny was sequenced using standard procedures. For this purpose, a 346-bp fragment specific for H. pylori rpsL was amplified using the following primers: RpsL-fwd (5′-GAA AAG AAA GGA AAA AGG TGG-3′) and RpsL-rev (5′-GCT TTA GTC TTT TTA GTC CCG-3′). Chromosomally encoded Strr clones of H. pylori were selected after transformation with the suicide plasmid pEG21, which carries the point mutation in the rpsL gene, and by growing clones on GC agar plates containing 10 μg/ml of streptomycin (21, 33).
Genotyping for the presence of cagA (H. pylori) and cadF (C. jejuni) genetic markers in the donors, recipients, and recombinants was done by PCR (24). The following primers were used: CagA-fwd (5′-AAA GGA TTG TCC CTA CAA GAA GC-3′) and CagA-rev (5′-GTA AGC GAT TGC TCT TGC ATC-3′) (a 330-bp fragment), and CadF-fwd (5′-TTG AAG GTA ATT TAG ATA TG-3′) and CadF-rev (5′-CTA ATA CCT AAA GTT GAA AC-3′) (a 377-bp fragment). The resulting PCR-amplified products were analyzed by standard agarose gel electrophoresis. We also examined the correlation between the conjugation phenotype and the presence of specific T4SS genes of H. pylori that may be involved in DNA transfer. Specific knockout mutants were generated by integration of aphA3 (Kanr) in the respective chromosomal genes (Table 1) as described previously (8, 33). The integration of the cassette into the genes of interest was confirmed by standard PCR analysis (7, 24, 33).
RESULTS
Chromosomal-DNA transfer between H. pylori and C. jejuni on GC agar plates.
Mating experiments were carried out using plasmid-free H. pylori strains (see Materials and Methods) (Table 1). We introduced chromosomal antibiotic resistance markers, such as Strr or Strr/Kanr, into the various strains using procedures described previously (33). To easily monitor the transfer of Strr, we selected C. jejuni strains that are naturally Sptr/Rifr (Table 1). Matings were then performed on GC agar plates, and the frequency of DNA transfer was assessed by determining the number of Strr/Sptr doubly resistant recombinants per parent. Since no Strr/Sptr recombinant was obtained when each strain was incubated alone, the possibility of spontaneous mutation was eliminated. The results showed that H. pylori strain P1 and C. jejuni strain 1543 gave rise to Strr/Sptr doubly resistant recombinants at frequencies of approximately 3 × 10−7 to 4 × 10−7 per donor (Table 2). The addition of 200 μg DNase I per ml decreased this number to approximately 4 × 10−8 to 5 × 10−8 per donor. However, the presence of DNase I was unable to prevent the development of doubly resistant C. jejuni recombinants (Table 2). Similar results were obtained for other mating combinations (Table 3). The relative percentage of doubly resistant mutants observed after bacteria mated in the presence or absence of DNase I varied only slightly (10 to 13%) for all of the different combinations. Thus, both DNase I-sensitive and DNase I-resistant events occurred at stable frequencies under the experimental conditions.
TABLE 3.
Frequency of recombination between different H. pylori and C. jejuni strains
| H. pylori strain (Strr Kanr) | Treatment with DNase I (200 μg/ml)a | No. of C. jejuni transconjugants (Sptr/Strr) (CFU)
|
||
|---|---|---|---|---|
| 1543 | 81-176 | ST3046 | ||
| P1ΔureB | − | 3.6 × 10−7 | 6.8 × 10−8 | 2.8 × 10−7 |
| P1ΔureB | + | 4.2 × 10−8 | 1.2 × 10−8 | 3.3 × 10−8 |
| 26695ΔureB | − | 2.3 × 10−7 | 8.4 × 10−8 | 7.9 × 10−8 |
| 26695ΔureB | + | 2.1 × 10−8 | 2.2 × 10−8 | 6.4 × 10−9 |
| 1061ΔureB | − | 1.9 × 10−7 | 5.7 × 10−8 | 9.5 × 10−8 |
| 1061ΔureB | + | 3.1 × 10−8 | 7.9 × 10−9 | 1.2 × 10−8 |
+, treated; −, untreated.
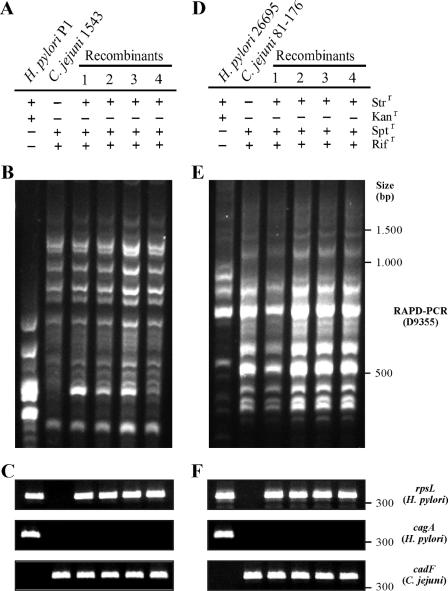
Unidirectional chromosomal-DNA transfer from H. pylori into C. jejuni.
Single recombinant colonies grew only on rifampin-supplemented GC agar plates, but not on kanamycin-containing plates, which suggested that DNA transfer occurred from H. pylori into C. jejuni and not vice versa (Fig. 1A to F). This unidirectional DNA transfer from H. pylori into C. jejuni was also confirmed by the growth of the recombinants on blood-free Campylobacter selective agar base and by the almost identical RAPD-PCR fingerprinting patterns observed between the recombinants and the parental C. jejuni strain (Fig. 1B and E). Moreover, PCR of a 346-bp rpsL gene product using H. pylori-specific primers confirmed that this gene was successfully transferred from H. pylori into the C. jejuni recombinants, because it is absent in the corresponding C. jejuni parental strains (Fig. 1C and F). Sequencing of these PCR products revealed the H. pylori rpsL gene sequence (data not shown). In addition, we performed control PCRs for species-specific genes, including well-known virulence/pathogenicity factors of H. pylori (cagA) and C. jejuni (cadF). The results demonstrate the presence of cadF in the C. jejuni parents and in all recombinants, while cagA was present in the H. pylori parents but absent in the recombinants (Fig. 1C and F). These data were also in agreement with the identities of strains as described above.
FIG. 1.
RAPD-PCR analysis of H. pylori and C. jejuni recombinants obtained from mating between the indicated parental strains (A and D) using primer D9355 (B and E). Antibiotic resistance phenotypes of selected Strr/Sptr doubly resistant recombinants are shown. Agar plates contained 10 μg/ml streptomycin, 10 μg/ml spectinomycin, 8 μg/ml kanamycin, or 10 μg/ml rifampin. A mixture of progeny showed profiles identical to that of the C. jejuni parental strain, which is consistent with unidirectional DNA transfer from H. pylori into C. jejuni. (C and F) Control PCRs for the indicated H. pylori and C. jejuni genes.
Characterization of the chromosomal-DNA transfer between H. pylori and C. jejuni.
Matings were then carried out in 1 ml BHI broth, which routinely produced fewer recombinants than matings performed on GC plates (Table 4). This suggested that the exchange of chromosomal DNA requires close cell-to-cell contact, somehow reminiscent of bacterial conjugation (6, 33, 45). Indeed, no DNA transfer was detected when parents were separated by a 0.1-μm-pore-size membrane that excluded cell-to-cell contact or when C. jejuni was incubated with a cell-free lysate of the parental H. pylori. In addition, a relatively low number of C. jejuni recombinants resulted from the incubation of C. jejuni with purified total DNA from H. pylori strain P1, and no DNA transfer was detected when the H. pylori parent strain was heat inactivated prior to the mating experiment (Table 4).
TABLE 4.
Effect of DNase I treatment on the frequency of recombination in which either H. pylori cells or culture supernatants were used as an exogenous DNA sourcea
| H. pylori sample (Strr) | Medium | Treatment with DNase I (200 μg/ml)b | C. jejuni sample (Sptr) |
C. jejuni transconjugants
|
|
|---|---|---|---|---|---|
| Sptr/Strr (no. of CFU) | Relative %c | ||||
| Cells | GC agar | − | Cells | 3.6 × 10−7 | 100 |
| Cells | BHI broth (1 ml) | − | Cells | 1.2 × 10−9 | <1 |
| Cells | GC agar | + | Cells | 4.7 × 10−8 | 13d |
| Cells | GC agar + membrane | − | Cells | 0 | 0 |
| 2 μg total DNA | GC agar | − | Cells | 2.1 × 10−9 | <1 |
| 2 μg total DNA | GC agar | + | Cells | 0 | 0 |
| Cell extract | GC agar | − | Cells | 0 | 0 |
| Cell extract | GC agar | + | Cells | 0 | 0 |
| Heat-inactivated cells | GC agar | − | Cells | 0 | 0 |
Matings were done in triplicate experiments. H. pylori P1 wild type (Strr); C. jejuni 1543 wild type (Sptr).
+, treated; −, untreated.
Relative to H. pylori plus C. jejuniwithout DNase I.
P < 0.01 in comparison with H. pylori plus C. jejuni without DNase I (Mann/Whitney test).
Roles of specific T4SS genes in chromosomal-DNA transfer from H. pylori into C. jejuni.
Finally, we examined the correlation between the conjugation phenotype and the presence of specific T4SS genes that may be involved in the DNA transfer initiated by H. pylori. All H. pylori strains used in this study lack most of the reported tfs3 genes (7) (Table 1), which supports the view that tfs3 is not involved in the DNA transfer. Inactivation of individual H. pylori comB genes (ΔcomB7-10) in the mating donor led to a reduction in, but did not prevent, DNA transfer to the C. jejuni recipient (Table 5). Matings of isogenic H. pylori mutants lacking the entire cag PAI also yielded high numbers of recombinants (Table 5). Taken together, these data suggest that the cag PAI T4SS, tfs3, and the comB system do not play roles in the described conjugative DNA transfer of H. pylori into C. jejuni.
TABLE 5.
comB and cag PAI systems of H. pylori are not involved in DNA transfera
| H. pylori strain (Strr) | C. jejuni strain (Sptr/Rifr) |
C. jejuni transconjugants
|
|
|---|---|---|---|
| Sptr Strr (no. of CFU) | Relative %b | ||
| P1 wild type | 1543 wild type | 4.3 × 10−7 | 100 |
| P1ΔcomB7-10 | 1543 wild type | 3.5 × 10−7 | 81 |
| P1ΔcagPAI | 1543 wild type | 3.7 × 10−7 | 86 |
Matings on GC agar plates were done in triplicate experiments.
Relative to H. pylori plus C. jejuni without DNase.
DISCUSSION
One of the striking characteristics of H. pylori is the extensive genetic diversity found among strains (2, 30, 49, 52). The clinical significance of this genetic diversity is highlighted by the identification of unique genetic features in strains with enhanced virulence (39, 47). Interspecies and intraspecies DNA transfers by transformation contribute to the exchange of genetic material in H. pylori and play important roles in the adaptation of the pathogen to different environmental conditions. Evidence also suggests that horizontal DNA transfer among H. pylori strains can take place via a DNase-resistant, conjugation-like mechanism (7, 33). We continued these studies to determine if clinical H. pylori isolates were able to transfer DNA into C. jejuni.
The chromosomal DNA transfer detected in this study was clearly unidirectional, from H. pylori into C. jejuni. The way H. pylori transferred chromosomally encoded Strr to C. jejuni was independent of the three previously identified T4SSs, and because no Strr/Sptr recombinant was obtained when each strain was incubated alone, the possibility of spontaneous mutation was eliminated. The DNA exchange occurred at a frequency of approximately 10−7 to 10−8 and was largely independent of the extracellular presence of nucleases, such as DNase I. The uptake of naked DNA by natural transformation played a minor role in the DNA transfer, a finding that was corroborated by the fact that cell extracts or purified DNA from parental strains did not induce a high number of recombinant C. jejuni organisms. Analysis of Strr/Sptr doubly resistant progeny by resistance to secondary antibiotic markers, RAPD-PCR, and gene-specific PCR confirmed that the DNA transfer was unidirectional. The exact integration site of the H. pylori rpsL gene into the C. jejuni chromosome, however, needs to be identified and characterized in future studies.
Our experiments suggested that the DNA transfer from H. pylori to C. jejuni was likely to occur by a conjugative mechanism requiring close cell-to-cell contact between the donor and recipient strains. Since cell-free H. pylori extracts did not produce recombinants, we were able to exclude phage transduction, thereby providing more evidence for the existence of a conjugative DNA transfer apparatus in H. pylori. Similar processes of chromosomal-DNA transfer without the involvement of conjugative plasmids have been described for Neisseria gonorrhoeae (15) and Legionella pneumophila (38). We do not know if a classical mechanism of mating-pair formation and DNA transfer was involved in our experimental system (45). However, our current findings suggest that mobilization of chromosomal DNA from H. pylori occurs in a fashion similar to the high-frequency recombination described for some Escherichia coli strains (37, 50).
Although the natural habitats of H. pylori and C. jejuni in the gastrointestinal tract are distinct, it is conceivable that they may come into contact with each other. Such contact could occur when C. jejuni passes the stomach. H. pylori, which is continuously eliminated from the stomach by peristaltic movements and washout with the luminal fluid, could in turn lead to contact of H. pylori with C. jejuni in the intestine. Thus, we propose that the observed mechanism of DNA exchange may contribute to the heterogeneity of H. pylori strains and also other bacteria, such as Campylobacter, and may help the bacteria adapt to the hostile milieu of the gastrointestinal tract, where free chromosomal DNA is short-lived. Assuming that the transferred single-stranded DNA is unlikely to be recognized by restriction enzymes (4, 36, 41), this conjugative exchange of genetic material could be a means of overcoming interstrain restrictions. An understanding of how H. pylori uses its conjugative transfer system during the course of infection will provide important insights into the evolutionary strategies of this clinically important pathogen.
From the standpoint of the recipient, our findings confirm that C. jejuni may use the conjugation-like mechanism to acquire DNA from other species. We do not know if this DNA acquisition helps to enhance genetic diversity, to repair bacterial DNA, or other purposes (12). Apparently, C. jejuni can use T2SS- and T4SS-related complexes for DNA uptake (9). Orthologues of the H. pylori genes comB8 to comB10 have been identified in a plasmid from C. jejuni (9), and genes involved in competence in C. jejuni were shown to have similarities to the T2SS/T4SS-related proteins (54). However, mutation of comB7 to comB10 in this organism led to only a small reduction in transformation frequency. These findings may be important in the transmission of antibiotic resistance. For instance, resistance to tetracycline in Campylobacter spp. is common in many countries (40, 48), and although a high level of tetracycline resistance is usually associated with the tet(O) gene carried on transmissible plasmids, this gene has also been found in the chromosomes of C. jejuni and C. coli (23, 34). Recently, the sequencing of two plasmids carrying tetracycline resistance genes, pCC31 from C. coli and plasmid pTet from C. jejuni, revealed an identity of 94.3% between the plasmids—a remarkably high similarity for plasmids from strains isolated 20 years apart and on different continents. All of the genes present in these plasmids had amino acid similarities to the T4SS of different Brucella species. However, the highest homology was to a mating-pair formation gene cluster from Actinobacillus actinomycetemcomitans, a periodontal pathogen. Only three genes of unknown function were found on one of the plasmids, two of which have known homologues in H. pylori (10).
Besides mediating plasmid transfer, some specific T4SS can mediate DNA secretion in Neisseria gonorrhoeae (27) and DNA uptake in H. pylori (29). It has been postulated that the mechanism of DNA translocation by the comB system might be comparable to the transport of transfer DNA from the cytoplasm of Agrobacterium tumefaciens into plant host cells by the VirB/D T4SS or by bacterial plasmid conjugation systems (29). When the correlation between the conjugation phenotype and the presence of specific T4SS genes in H. pylori was addressed, we surprisingly found that the cag PAI T4SS, the comB system, and tfs3 did not play any role in conjugative DNA transfer. The identification of the H. pylori genes involved in the DNA transfer is currently under way in our laboratories.
Acknowledgments
We are grateful to Rainer Haas and Wolfgang Fischer for providing plasmids pEG21 and P1ΔcomB7-10.
The work of S.B. is supported through the NBL-3 project (Magdeburger Forschungsverbund PFG4) and Priority Program SPP1150 of the Deutsche Forschungsgemeinschaft (Ba1671/3-2).
Footnotes
Published ahead of print on 29 November 2006.
REFERENCES
- 1.Akopyanz, N., N. O. Bukanov, T. U. Westblom, S. Kresovich, and D. E. Berg. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 20:5137-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm, R. A., and T. J. Trust. 1999. Analysis of the genetic diversity of Helicobacter pylori: the tale of two genomes. J. Mol. Med. 77:834-846. [DOI] [PubMed] [Google Scholar]
- 3.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 4.Aras, R. A., A. J. Small, T. Ando, and M. J. Blaser. 2002. Helicobacter pylori interstrain restriction-modification diversity prevents genome subversion by chromosomal DNA from competing strains. Nucleic Acids Res. 30:5391-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baar, C., M. Eppinger, G. Raddatz, J. Simon, C. Lanz, O. Klimmek, R. Nandakumar, R. Gross, A. Rosinus, H. Keller, P. Jagtap, B. Linke, F. Meyer, H. Lederer, and S. C. Schuster. 2003. Complete genome sequence and analysis of Wolinella succinogenes. Proc. Natl. Acad. Sci. USA 20:11690-11695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Backert, S., and T. F. Meyer. 2006. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr. Opin. Microbiol. 9:207-217. [DOI] [PubMed] [Google Scholar]
- 7.Backert, S., T. Kwok, and W. König. 2005. Conjugative plasmid DNA transfer in Helicobacter pylori mediated by chromosomally encoded relaxase and TraG-like proteins. Microbiology 151:3493-3503. [DOI] [PubMed] [Google Scholar]
- 8.Backert, S., S. Moese, S. Selbach, V. Brinkmann, and T. F. Meyer. 2001. Phosphorylation of tyrosine 972 of the Helicobacter pylori CagA protein is essential for induction of a scattering phenotype in gastric epithelial cells. Mol. Microbiol. 42:631-644. [DOI] [PubMed] [Google Scholar]
- 9.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batchelor, R. A., B. M. Pearson, L. M. Friis, P. Guerry, and J. M. Wells. 2004. Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology 150:3507-3517. [DOI] [PubMed] [Google Scholar]
- 11.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472-479. [DOI] [PubMed] [Google Scholar]
- 12.Chen, I., and D. Dubnau. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 241:241-249. [DOI] [PubMed] [Google Scholar]
- 13.Cornelis, G. R., A. Boland, A. P. Boyd, C. Geuijen, M. Iriarte, C. Neyt, M. P. Sory, and I. Stainier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Covacci, A., J. L. Telford, G. Del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 15.Dillard, J. P., and H. S. Seifert. 2001. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol. Microbiol. 41:263-277. [DOI] [PubMed] [Google Scholar]
- 16.Dubnau, D. 1998. DNA uptake in bacteria. Annu. Rev. Microbiol. 53:217-244. [DOI] [PubMed] [Google Scholar]
- 17.Eppinger, M., C. Baar, B. Linz, G. Raddatz, C. Lanz, H. Keller, G. Morelli, H. Gressmann, M. Achtman, and S. C. Schuster. 2006. Who ate whom? Adaptive Helicobacter genomic changes that accompanied a host jump from early humans to large felines. PLOS Genet. 2:1097-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eppinger, M., C. Baar, G. Raddatz, D. H. Huson, and S. C. Schuster. 2004. Comparative analysis of four Campylobacterales. Nat. Rev. Microbiol. 2:872-885. [DOI] [PubMed] [Google Scholar]
- 19.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer, W., D. Schwan, E. Gerland, G. E. Erlenfeld, S. Odenbreit, and R. Haas. 1999. A plasmid-based vector system for the cloning and expression of Helicobacter pylori genes encoding outer membrane proteins. Mol. Gen. Genet. 262:501-507. [DOI] [PubMed] [Google Scholar]
- 22.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLOS Biol. 3:72-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibreel, A., D. Tracz, L. Nonaka, T. M. Ngo, S. R. Connell, and D. E. Taylor. 2004. Incidence of antibiotic resistance in Campylobacter jejuni isolated in Alberta, Canada, from 1999 to 2002, with special reference to tet(O)-mediated tetracycline resistance. Antimicrob. Agents Chemother. 48:3442-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gieseler, S., B. Konig, W. Konig, and S. Backert. 2005. Strain-specific expression profiles of virulence genes in Helicobacter pylori during infection of gastric epithelial cells and granulocytes. Microbes Infect. 7:437-447. [DOI] [PubMed] [Google Scholar]
- 25.Goodwin, A., D. Kersulyte, G. Sisson, S. J. O. Veldhuyzen van Zanten, D. E. Berg, and P. S. Hoffman. 1998. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 28:383-393. [DOI] [PubMed] [Google Scholar]
- 26.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 27.Hamilton, H. L., N. M. Dominguez, K. J. Schwartz, K. T. Hackett, and J. P. Dillard. 2005. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol. Microbiol. 55:1704-1721. [DOI] [PubMed] [Google Scholar]
- 28.Hofreuter, D., and R. Haas. 2002. Characterization of two cryptic Helicobacter pylori plasmids: a putative source for horizontal gene transfer and gene shuffling. J. Bacteriol. 184:2755-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hofreuter, D., S. Odenbreit, and R. Haas. 2001. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol. 41:379-391. [DOI] [PubMed] [Google Scholar]
- 30.Israel, D. A., N. Salama, U. Krishna, U. M. Rieger, J. C. Atherton, S. Falkow, and R. M. Peek, Jr. 2001. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc. Natl. Acad. Sci. USA 98:14625-14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kersulyte, D., B. Velapatino, A. K. Mukhopadhyay, L. Cahuayme, A. Bussalleu, J. Combe, R. H. Gilman, and D. E. Berg. 2003. Cluster of type IV secretion genes in Helicobacter pylori's plasticity zone. J. Bacteriol. 185:3764-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korlath, J. A., M. T. Osterholm, L. A. Judy, J. C. Forfang, and R. A. Robinson. 1985. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152:592-596. [DOI] [PubMed] [Google Scholar]
- 33.Kuipers, E. J., D. A. Israel, J. G. Kusters, and M. J. Blaser. 1998. Evidence for a conjugation-like mechanism of DNA transfer in Helicobacter pylori. J. Bacteriol. 180:2901-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, C.-Y., C.-L. Tai, S.-C. Lin, and Y.-T. Chen. 1994. Occurrence of plasmids and tetracycline resistance among Campylobacter jejuni and Campylobacter coli isolated from whole market chickens and clinical samples. Int. J. Food Microbiol. 24:161-170. [DOI] [PubMed] [Google Scholar]
- 35.Letellier, L., L. Plancon, M. Bonhivers, and P. Boulanger. 1999. Phage DNA transport across membranes. Res. Microbiol. 150:499-505. [DOI] [PubMed] [Google Scholar]
- 36.Lin, L. F., J. Posfai, R. J. Roberts, and H. Kong. 2001. Comparative genomics of the restriction-modification systems in Helicobacter pylori. Proc. Natl. Acad. Sci. USA 98:2740-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Low, K. B. 1987. Hfr strains of Escherichia coli K-12, p. 1134-1337. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger, (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol 2. ASM Press, Washington, DC. [Google Scholar]
- 38.Miyamoto, H., S. Yoshida, H. Taniguchi, and H. A. Shuman. 2003. Virulence conversion of Legionella pneumophila by conjugal transfer of chromosomal DNA. J. Bacteriol. 185:6712-6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montecucco, C., and R. Rappuoli. 2001. Living dangerously: how Helicobacter pylori survives in the human stomach. Nat. Rev. Mol. Cell. Biol. 2:457-466. [DOI] [PubMed] [Google Scholar]
- 40.Nachamkin, I., J. Engberg, and F. Aarestrup. 2000. Diagnosis and antimicrobial susceptibility of Campylobacter species, p. 45-66. In I. Nachamkin, and M. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, DC.
- 41.Nobusato, A., I. Uchiyama, and I. Kobayashi. 2000. Diversity of restriction-modification gene homologues in Helicobacter pylori. Gene 259:89-98. [DOI] [PubMed] [Google Scholar]
- 42.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 43.Odenbreit, S., B. Wieland, and R. Haas. 1996. Cloning and genetic characterisation of Helicobacter pylori catalase and construction of a catalase deficient mutant. J. Bacteriol. 178:6960-6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oh, J. D., H. Kling-Backhed, M. Giannakis, J. Xu, R. S. Fulton, L. A. Fulton, H. S. Cordum, C. Wang, G. Elliott, J. Edwards, E. R. Mardis, L. G. Engstrand, and J. I. Gordon. 2006. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc. Natl. Acad. Sci. USA 103:9999-10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pansegrau, W., and E. Lanka. 1996. Enzymology of DNA transfer by conjugative mechanisms. Prog. Nucleic Acid Res. Mol. Biol. 54:197-251. [DOI] [PubMed] [Google Scholar]
- 46.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. M. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 47.Peek, R. M., Jr., and M. J. Blaser. 2002. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer 2:28-37. [DOI] [PubMed] [Google Scholar]
- 48.Pratt, A., and V. Korolik. 2005. Tetracycline resistance of Australian Campylobacter jejuni and Campylobacter coli isolates. J. Antimicrob. Chemother. 55:452-460. [DOI] [PubMed] [Google Scholar]
- 49.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, G. R. 1991. Conjugational recombination in E. coli: myths and mechanisms. Cell 64:19-27. [DOI] [PubMed] [Google Scholar]
- 51.Suerbaum, S., C. Josenhans, T. Sterzenbach, B. Drescher, P. Brandt, M. Bell, M. Droge, B. Fartmann, H. P. Fischer, Z. Ge, A. Horster, R. Holland, K. Klein, J. Konig, L. Macko, G. L. Mendz, G. Nyakatura, D. B. Schauer, Z. Shen, J. Weber, M. Frosch, and J. G. Fox. 2003. The complete genome sequence of the carcinogenic bacterium Helicobacter hepaticus. Proc. Natl. Acad. Sci. USA 100:7901-7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suerbaum, S., and M. Achtman. 2004. Helicobacter pylori: recombination, population structure and human migrations. Int. J. Med. Microbiol. 294:133-139. [DOI] [PubMed] [Google Scholar]
- 53.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, and B. A. Dougherty. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 54.Wiesner, R. S., D. R. Hendrixson, and V. J. DiRita. 2000. Natural transformation of Campylobacter jejuni requires components of a type II secretion system. J. Bacteriol. 185:5408-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]