Abstract
Peroxiredoxins (Prx) are widely distributed peroxidases that can be divided into 1-Cys and 2-Cys Prx groups based on the number of conserved cysteine residues that participate in their catalytical cycle. Prx have been described to be strictly dependent on thiols, but here, we show that ascorbate (vitamin C) also reduces 1-Cys Prx, but not 2-Cys Prx, from several taxonomic groups. Reduction by ascorbate is partly related to the fact that the oxidized form of 1-Cys Prx is a stable sulfenic acid (Cys-SOH) instead of a disulfide. In addition, a histidine residue in the active site is required. In fact, we engineered a 2-Cys Prx with these two features, and it displayed ascorbate peroxidase activity. These data represent a breakthrough in the thiol-specific antioxidant paradigm. Ascorbate may be the long-sought-after biological reductant of 1-Cys Prx. Because ascorbate is present in high amounts in cells, the ascorbate/protein sulfenic acid pair represents an aspect of redox biochemistry that has yet to be explored in vivo.
Keywords: peroxidase, sulfenic acid, cellular redox processes, oxidative stress, ascorbate-dependent activity
Oxygen toxicity is related to the deleterious properties of the so-called reactive oxygen species (ROS) and reactive nitrogen species (RNS). Cellular defenses are comprised of a wide range of components that interact with each other to control the levels of ROS and RNS (1). One of them is vitamin C (ascorbic acid), which is not synthesized by human cells and, therefore, must be consumed in the diet. Some human diseases, such as scurvy, are related to low concentrations of vitamin C (reviewed in ref. 2). In addition to low-molecular-weight compounds, cells also rely on enzymes to cope with oxidative injury.
Peroxiredoxins (Prx) are ubiquitous enzymes that use their highly reactive cysteine residues to decompose peroxides (3). The importance of Prx enzymes is underlined by their high abundance and involvement in multiple cellular processes ranging from antioxidant defense (4, 5), parasite drug resistance (6), and cancer (5, 7, 8) to H2O2-mediated cellular signaling (9, 10). In addition to their well known peroxidase activity, some of these enzymes are also efficient molecular chaperones (11). The switch from chaperone to peroxidase activity is regulated by enzymes that reduce sulfinic acids only when they are generated in Prx cysteine residues (12, 13). This redox biochemistry adds a level of versatility to the well established fact that Prx recycle between disulfide/sulfenic acid to the sulfhydryl form (reviewed in ref. 14). These properties may be related to the fact that Prx protect against several stresses (15–17).
Prx can be divided into two main groups, 1-Cys and 2-Cys Prx, based on the number of conserved cysteine residues that participate in the catalytic cycle. Both 1-Cys and 2-Cys Prx contain a reactive cysteine residue, the so-called peroxidatic cysteine (Cysp-S−), that is oxidized to sulfenic acid (Cysp-SOH) when exposed to peroxides. In the 1-Cys mechanism, Cysp-SOH is directly reduced, whereas in the 2-Cys catalytic cycle, a second Prx cysteine residue, the resolving cysteine (Cysr-SH), condenses with the sulfenic acid to form a disulfide. Finally, the 2-Cys disulfide is reduced by another biothiol, particularly thioredoxin, a low-molecular-weight protein with two vicinal cysteine residues (3). So far, Prx have been described as strictly depending on thiols (RSH) as reducing agents. Indeed, these enzymes have been characterized as peroxidases undergoing cycles of peroxide-dependent oxidation and thiol-dependent reduction during their catalytic activity (14), as shown in Reaction 1:
Although all Prx are assumed to be thiol-specific antioxidant enzymes, the 1-Cys Prx mechanism is not fully understood (reviewed in refs. 3 and 14). An important point to be clarified is the nature of the biological reductant of 1-Cys Prx, which remains unknown in most cases. Here, we present results showing unequivocally that 1-Cys Prx are reduced by ascorbate, and thus advance a change in the thiol-specific antioxidant paradigm. Because ascorbate is present at high-millimolar level in cells, it may be the long-sought-after biological reductant of 1-Cys Prx. Finally, our characterization of the peroxiredoxin (Prx) sulfenic acid (Cys-SOH)/ascorbate pair may open perspectives in the understanding of cellular redox processes.
Results
Ascorbate Peroxidase Activity of Yeast 1-Cys Prx.
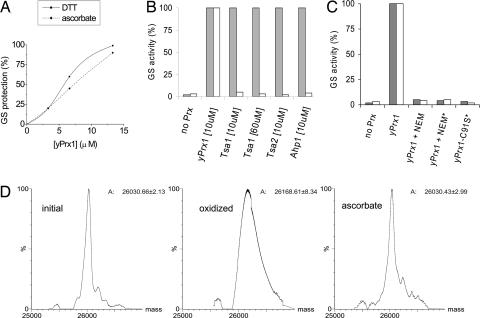
Although Prx are assumed to be peroxidases strictly dependent on thiols for regeneration of the reduced forms, we observed that a mitochondrial 1-Cys isoform from Saccharomyces cerevisiae, yPrx1, protected glutamine synthetase (GS) from inactivation in the presence of ascorbate. Protection was dose-dependent on yPrx1, and ascorbate and DTT were similarly effective as electron donors (Fig. 1A). The enzyme also prevented strand breaks in supercoiled plasmid DNA submitted to a similar oxidizing system (data not shown), indicating that yPrx1 is capable of protecting biomolecules by using ascorbate as reductant. The ability of yPrx1 to consume reducing equivalents from ascorbate was confirmed by the ascorbate oxidase assay, which is based on the ability of this enzyme to catalyze oxygen consumption in amounts proportional to ascorbate concentrations [see supporting information (SI) Fig. 4].
Fig. 1.
Characterization of yPrx1 ascorbate-dependent peroxidase activity. (A–C) Glutamine synthetase (GS) protection from inactivation by metal-catalyzed oxidation systems containing DTT (10 mM) (gray bars) or ascorbate (10 mM) (white bars) in 15-min incubations at 37°C. GS activity was considered 100% when it was not preincubated with metal-catalyzed oxidation systems. (A) GS protection by increasing yPrx1 concentrations. (B) Inability of other yeast Prx enzymes to use ascorbate. Tsa1, gi: 464970; Tsa2, gi: 2499475; or Ahp1, gi: 1709682. (C) Inhibition of yPrxI protective effects by inactivation of cysteine residues. yPrxI was modified by N-ethylmaleimide (NEM) and by site-specific mutagenesis as described in Experimental Procedures. Afterward, a GS protection assay was performed as described above by using yPrx1 at 3 μM or in excess of 70 μM, indicated by ∗. (D) Deconvoluted MS spectra of yPrxI in different oxidation states treated with dimedone (140 amu). Samples of reduced yPrx1 (initial), treated with H2O2 and dimedone (oxidized) or treated with H2O2, ascorbate, and dimedone (ascorbate) were prepared and analyzed as described in Experimental Procedures. Molecular masses were calculated by the Transform algorithm of the BioLynx Data Analysis package of MassLynx Software (Micromass). Data were acquired in an ESI-Q-TOF (Micromass) mass spectrometer under positive ionization mode. Calculated molecular masses are indicated at the right side of the corresponding spectra. These spectra were obtained by using a recombinant yPrx1 (100 μM) without the 40 first amino acids residues and containing only one cysteine residue, which is Cysp (see supporting information on the PNAS web site for more details). All excess reagents were washed away by ultrafiltration before activity analysis.
We tested other yeast Prx, all of which were expected to be oxidized to disulfide bonds during their catalytic cycles (2-Cys Prx), but none of them protected GS from inactivation in the presence of ascorbate (Fig. 1B). Therefore, this ascorbate-dependent property is a particular feature of yPrx1 that is not displayed by any of the tested yeast 2-Cys Prx.
Although the above assays showed that yPrx1 uses ascorbate as reductant, all of them are indirect. Therefore, dehydroascorbate, ascorbate, and peroxide concentrations were determined directly in the presence of DTPA to prevent metal-catalyzed processes. From the data of several experiments summarized in Table 1, it became clear that yPrx1 catalyzed the consumption of 1 mol of peroxide and 1 mol of ascorbate to produce 1 mol of dehydroascorbate. Thus, stoichiometric data (Table 1 and SI Fig. 5) indicated that yPrx1 catalyzes Reaction 2:
Table 1.
Stoichiometry of the yPrx1 ascorbate-dependent peroxidase activity
| Reaction time, min |
Initial peroxide, 1 mM |
Peroxide consumption, μM |
Ascorbate consumption, μM |
DHA produced, μM |
Peroxide/ ascorbate |
Ascorbate/DHA |
|---|---|---|---|---|---|---|
| 15 min | H2O2 | 156 ± 10 | 150 ± 20 | 150 ± 20 | 1.04 ± 0.14 | 1.0 ± 0.1 |
| 45 min | t-BOOH | 90 ± 15 | 98 ± 10 | 85 ± 25 | 0.92 ± 0.20 | 1.2 ± 0.3 |
The results are average of three independent experiments using HPLC analyses and ascorbate oxidase assay. Peroxides were measured by the FOX2 method. DHA, dehydroascorbate; initial ascorbate, 1 mM; yPrx1, 50 μM.
The amount of ascorbyl free radical produced in the incubations was monitored by electron spin resonance (ESR) and found to be in the nM range (SI Fig. 6). This is well below the amount of ascorbate consumed and of dehydroascorbate produced, both ≈100 μM. Because the ascorbyl free radical has a relatively long lifetime (18), our data indicated that yPrx1 ascorbate peroxidase activity is a two-electron process (SI Figs. 5 and 6) similar to the thiol-dependent activity.
More H2O2 than tert-butyl hydroperoxide (t-BOOH) was removed by the ascorbate-dependent catalysis (Table 1), similar to the yPrx1 thiol-dependent peroxidase activity described by Pedrajas et al. (19). Nevertheless, independent of the peroxide nature, yPrx1 was capable of reducing it by consuming a nonthiol reductant. These results led us to verify whether yPrx1 would also accept electrons from other reductants. We tested both thiolic (glutathione, β-mercaptoethanol, and DTT) and nonthiolic reductants (trolox, which is a hydrophilic analogue of vitamin E, and urate). Of all the low-molecular-weight compounds tested, only thiols and ascorbate supported the yPrx1 activity (SI Fig. 7).
Characterization of Amino Acid Residues Involved in 1-Cys Prx Ascorbate-Dependent Catalysis.
Because the thiol-dependent peroxidase activity of yPrx1 relies on a reactive cysteine residue, the importance of cysteine residues in ascorbate-dependent GS protection was investigated by alkylating the sulfhydryl groups with N-ethylmaleimide (NEM). Protection of GS was inhibited by NEM, implicating yPrx1 cysteine residues as the site of peroxide reduction during the ascorbate-dependent activity. Furthermore, mutation of the Cysp fully abolished DTT and ascorbate peroxidase activity, indicating that this residue is directly involved in peroxide reduction mediated by both reductants (Fig. 1C).
The results presented so far suggest that protection of biomolecules is due to yPrx1 peroxidase activity that depends on its reactive cysteine residue and ascorbate (Table 1 and Fig. 1 A–C). To characterize the mechanism of ascorbate-dependent activity, the redox state of the Cysp was analyzed by spectrophotometric and mass spectrometry methods in the presence of NBD-Cl (a thiol and sulfenic acid reagent) and dimedone (a specific sulfenic acid reagent), respectively (see also Experimental Procedures). The results showed that the catalytic cysteine residue was oxidized to sulfenic acid upon peroxide exposure and was rereduced upon ascorbate treatment (Fig. 1D and SI Fig. 8). To further test the hypothesis that active-site sulfenic acid is reduced by ascorbate, we examined the peroxidase activity of yPrx1 modified with dimedone. Abolishment of the yPrx1 capability to reduce H2O2 was observed only when the enzyme was previously oxidized and then incubated with dimedone. Treatment of reduced yPrx1 with dimedone in all other conditions (sample treated with DTT and sample oxidized then treated with DTT or ascorbate) resulted in no activity loss, suggesting that the Cysp sulfenic acid derivative was reduced by both reductants (SI Fig. 9).
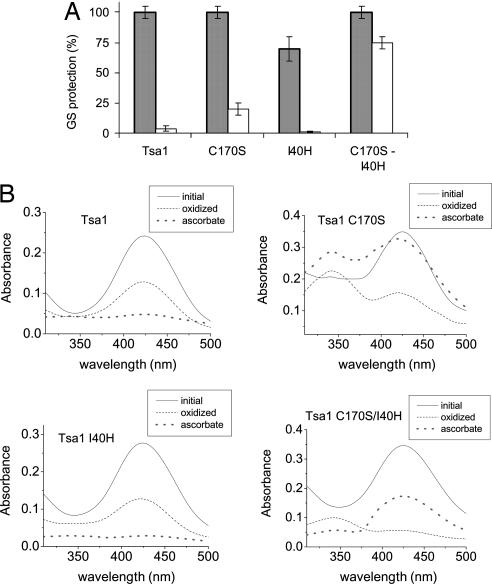
Next, we explored which specific features of 1-Cys Prx would enable them, and not 2-Cys Prx, to use reducing equivalents from ascorbate. One obvious difference between the two main mechanisms of Prx is the presence or absence of the Cysr (2-Cys and 1-Cys Prx, respectively). The simple absence of Cysr might predispose the sulfenic acid of 1-Cys Prx to ascorbate reduction, whereas the disulfide bond in 2-Cys Prx would be resistant. To test this hypothesis, we analyzed whether a recombinant yeast 2-Cys Prx (Tsa1-gi: 464970) whose Cysr was mutated to a serine residue (Tsa1-C170S) acquired the ascorbate-dependent activity. An absent Cysr provoked the appearance of a small ascorbate peroxidase activity (Fig. 2A), and the sulfenic acid derivative was detectable spectrophotometrically even after ascorbate addition (Fig. 2B). Therefore, the sulfenic acid form of Tsa1-C170S was only partially reduced by ascorbate (Fig. 2B).
Fig. 2.
The 2-Cys Prx engineered for acquiring the ascorbate-dependent peroxidase activity. (A) GS protection assay was used to measure DTT- (gray bars) and ascorbate-dependent (white bars) peroxidase activity of the Tsa1 mutants. Each enzyme (50 μM) was incubated as described in the Fig. 1 legend. (B) Analysis of the oxidation state of cysteine residues from Tsa1 and the mutants C170S, I40H, and C170S-I40H by the NBD-Cl assay. Treatments and assays were performed as described in Experimental Procedures. Sulfhydryl and sulfenic acid adducts peak at 420 nm and 347 nm, respectively, with similar extinction coefficients (46).
Several Prx showing 1-Cys mechanism belong to the PrxVI group, which comprises proteins sharing amino acid sequence similarity (3). PrxVI proteins have a highly conserved histidine residue (SI Fig. 10) in addition to an arginine (fully conserved in all Prx) in the active site (20), probably creating a positively charged environment. This histidine is absent in 2-Cys Prx. We hypothesized that these two features, i.e., the absence of Cysr and the presence of histidine, would render 1-Cys Prx capable of receiving electrons from ascorbate. The absence of Cysr should increase the half-life of the sulfenic acid form of the Cysp, whereas the presence of a positively charged histidine would further improve the sulfenic acid stabilization and favor the interaction with negatively charged molecules such as ascorbate. Therefore, we introduced a histidine residue in a position equivalent to the 1-Cys Prx group. The single addition of histidine in Tsa1 (Tsa1-I40H) did not transform it into an ascorbate peroxidase (Fig. 2A). This is probably related to the fast reaction of sulfenic acid with Cysr, because no Cysp-SOH was detected (Fig. 2B). Only when two mutations were introduced into Tsa1 (C170S and I40H) did this 2-Cys Prx acquire an ascorbate-dependent activity comparable with the DTT-dependent one (Fig. 2A). Furthermore, a sulfenic acid derivative was detectable in the double mutant, and it was rereduced by ascorbate (Fig. 2B). The ascorbate peroxidase activity of the Tsa1 double mutant was considerably lower than that of the natural 1-Cys Prx enzyme (yPrx1), indicating that additional structural features are required. Accordingly, the structures available so far in the Protein Data Bank suggest that 1-Cys Prx maintain the secondary structure near the active domain throughout the catalytic cycle, whereas the 2-Cys Prx active sites undergo an unfolding during the oxidation step (SI Fig. 11 and ref. 3). In principle, it is possible that any Prx might acquire ascorbate peroxidase activity if the appropriate amino acid changes are made.
Our results suggest that ascorbate probably supports the peroxidase activity of 1-Cys Prx because it can fully reduce sulfenic acids, but not disulfides, in a positively charged environment. Little is known about the reaction of ascorbate with oxidized forms of thiols to prove or disprove this view. In fact, we found no reported second-order rate constants for ascorbate reacting with disulfides or sulfenic acids. Nevertheless, ascorbate does not reduce the intermolecular disulfide bond of 2-Cys Prx (21) and reduces the low molecular disulfide DTNB only slowly. In our hands, reaction of ascorbate (100 mM) with DTNB (500 μM) for 24 h at room temperature produced only 15 μM of reduced DTNB. In contrast, DTNB reacts very fast with thiols (22). Furthermore, reduction of known proteinic and nonproteinic disulfide bonds by ascorbate is thermodynamically unfavorable (see Thermodynamic Considerations on the Reaction of Ascorbate with Disulfides in SI Text), but the sulfenic acid formed in glyceraldehyde 3-phosphate dehydrogenase has been reported to be reduced by ascorbate (23).
Ascorbate Peroxidase Is a Conserved Activity in 1-Cys Prx.
The identity of most 1-Cys Prx electron donors are unknown, and the yPrx1 studied here is an exception because its peroxidase activity has been shown to be supported by a mitochondrial thioredoxin system (19). yPrxI belongs to the PrxVI group, whose members share structural similarities, with most of them acting by the 1-Cys Prx catalytical mechanism (3). Ascorbate should also support the peroxidase activity of other PrxVI proteins because they are likely to produce stable sulfenic acid intermediates, because of the absence of a Cysr and the presence of a histidine in the active site. Supporting this scenario, all tested PrxVI from different taxonomic groups displayed ascorbate peroxidase activity (Table 2). Therefore, the ability of ascorbate to support the peroxidase activity of 1-Cys Prx is a highly conserved property. In contrast, tested 2-Cys enzymes from diverse organisms (Ohr from Xylella fastidiosa, PDB code 1ZB8; 2-Cys Prx from Crinipellis perniciosa, ID DQ813344; and OsmC from Escherichia coli, PDB ID 1QWI) did not reduce peroxides in the presence of ascorbate.
Table 2.
Comparison of 1-Cys Prx enzymes ascorbate- and DTT-dependent activities
| 1-Cys Prx protein | Species | Relative peroxidase activity (DTT/ascorbate) |
|---|---|---|
| yPrx1 | S. cerevisiae | 1.9 ± 0.1 |
| Prdx6 | R. norvegicus | 0.8 ± 0.1 |
| rPf1-Cys-Prx | P. falciparum | 1.05 ± 0.15 |
| AtPER1 | A. thaliana | 0.93 ± 0.21 |
| Dpx-2540 | D. melanogaster | 0.8 ± 0.14 |
| Dpx-6005 | D. melanogaster | 1.1 ± 0.17 |
| AhpE | M. tuberculosis | 0.73 ± 0.2 |
The FOX2 assay was used to monitor the peroxidase activity (enzyme concentration, 50 μM; peroxide, 400 μM; DTPA, 0.5 mM; Hepes, 50 mM; DTT or Ascorbate, 500 μM). The results are the average of three independent experiments.
We also tested the ascorbate peroxidase activity of AhpE from Mycobacterium tuberculosis, which shares moderate sequence identity (≈25%) to PrxVI proteins and presents the 1-Cys Prx mechanism (24). AhpE does not possess the PrxVI conserved histidine (SI Fig. 10) but is capable of decomposing peroxides at the expense of ascorbate (Table 2). Interestingly, its active site environment contains several positively charged residues (Arg-53, His-77, Lys-78, Arg-116, Lys-133, and Arg-139) that may play the role attributed to the conserved histidine.
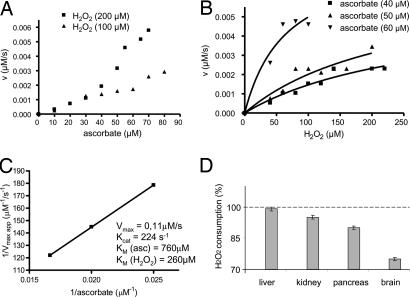
Next, we analyzed the enzymatic activity of Prdx6, a 1-Cys Prx from rat, in more detail by monitoring ascorbate oxidation at 265 nm (Fig. 3). Substrate consumption was continuously monitored, providing more reliable values of reaction rates than the FOX assay. Applying the bisubstrate kinetic approach and assuming a ping-pong mechanism (25), we found no evidence of enzyme saturation by ascorbate in the used range of concentrations (Fig. 3A). Because ascorbate possesses a very high absorption coefficient (ε265 = 14.500 M−1 cm−1), it could not be added at concentrations >80 μM. H2O2 concentrations were then varied under fixed ascorbate concentrations. In this case, a trend of enzyme saturation was observed (Fig. 3B) permitting a nonlinear fit through the Michaelis–Menten equation (Prism 4 for Windows, GraphPad Software, San Diego, CA). Thus, different Vmax app values were estimated and plotted against the reciprocal of the ascorbate concentration to provide the kinetic parameters shown in Fig. 3C.
Fig. 3.
Enzymatic and biological investigations of the ascorbate-dependent peroxidase activity of rat Prdx6. (A–C) Enzymatic activity was monitored by ascorbate consumption at 265 nm in incubations containing Prdx6 (0.5 nM), H2O2 and ascorbate at the concentrations specified in the figures. (D) The ability of rat homogenates to support the peroxidase activity of Prdx6 was investigated before and after treatment with ascorbate oxidase as described in Experimental Procedures. The 100% value means the amount of H2O2 consumed during 15 min at 37°C by each homogenate in the absence of ascorbate oxidase pretreatment. The bars represent the percentage of H2O2 consumed when homogenates were pretreated with ascorbate oxidase.
Because the used ascorbate concentrations were well below the KM(ascorbate) (760 μM) due to experimental constraints, the performed kinetic analysis presents a high degree of uncertainty. Nevertheless, the estimated KM(ascorbate) value suggests that ascorbate can support Prxd6 activity in vivo because its intracellular concentration is in the millimolar range (2). Remarkably, the catalytic efficiency of the ascorbate-dependent activity estimated here [kcat/KM(ascorbate) 3 × 105 M−1 s−1; kcat/KM(H2O2) 0.9 × 106 M−1 s−1] is comparable with those previously reported for thiol-dependent processes (26). Thus, the nonthiol peroxidase activity is likely to be a major process. The ascorbate-dependent activity of yPrx1 from yeast was also monitored by the A265nm decay. However, because yPrx1 is less efficient than its mammalian counterpart, it was not possible to determine the individual kinetic parameters with confidence. Nevertheless, it was possible to estimate that the catalytic efficiencies of yPrx1 [kcat/KM(ascorbate) and kcat/KM(H2O2)] are on the order of 104 and 105 M−1 s−1, respectively (data not shown). These values are also comparable with those of thiol-dependent processes (19). The detailed characterization of the ascorbate peroxidase activity of 1-Cys Prx will require novel kinetic approaches.
The biological relevance of the 1-Cys Prx activity was also indicated by experiments performed with homogenates of rat organs. As anticipated from the high content of proteinic and nonproteinic reductants in these extracts, they sustained H2O2 consumption that depended on Prdx6. Remarkably, however, H2O2 consumption consistently decreased when ascorbate was depleted by pretreatment of the homogenates with ascorbate oxidase (Fig. 3D), suggesting a major role for ascorbate in Prxd6 reduction.
Discussion
The 2-Cys Prx are involved in several physiological processes, probably because they modulate the intracellular level of H2O2, an oxidant with signaling properties (14). In comparison with 2-Cys Prx, 1-Cys Prx have been little studied, partly because their biological reductant remains unknown, except for a few described exceptions (19, 26). Indeed, most 1-Cys Prx are not efficiently reduced by thioredoxin, glutaredoxin, or glutathione. Therefore, the data presented here indicated that ascorbate is a strong candidate to support 1-Cys Prx peroxidase activity in vivo. Furthermore, our work provides some mechanistic details about 1-Cys Prx catalysis.
The physiological relevance of the ascorbate-dependent peroxidase activity in the yeast S. cerevisiae may be questionable. As mentioned before, this microorganism possesses a mitochondrial thioredoxin system capable of reducing yPrx1 (19). Furthermore, S. cerevisiae does not synthesize ascorbate; it synthesizes an analog, erythroascorbate, whose antioxidant role remains controversial (27). At this point, it is difficult to compare the relevance of erythroascorbate with the thioredoxin system because their relative abundance in yeast has yet to be determined. In any case, our results show that ascorbate and thioredoxin have similar catalytic efficiencies [kcat/KM(ascorbate) ≈104 M−1 s−1 and kcat/KM(H2O2) ≈105 M−1 s−1].
Although it is premature to reach a conclusion about the biological relevance of the ascorbate-dependent peroxidase activity in yeast, the results obtained with mammalian enzymes favor such relevance. Despite the limitations of the assays used to monitor enzyme activity, it was possible to conclude that the ascorbate-dependent peroxidase activity of Prdx6 is comparable with the thiol-dependent process (Fig. 3 A–C). Furthermore, ascorbate depletion considerably decreased the ability of rat homogenates to support Prdx6 peroxidase activity (Fig. 3D). This is consistent with the fact that ascorbate is present at millimolar levels in most mammalian tissues (reviewed in ref. 2). It has been recently shown that 1-Cys Prx from mammals can heterodimerize with π-glutathione transferase and accept electrons from glutathione (reviewed in ref. 26). Our results indicate that ascorbate is also a significant source of reducing power for these enzymes (Fig. 3). Further studies are necessary to establish the relative importance of ascorbate and glutathione as physiological electron donors for mammalian 1-Cys Prx. Interestingly, ascorbate depletion in the rat homogenates resulted in different levels of a drop in Prdx6 activity (Fig. 3D). Among other reasons, these differences may be due to the endogenous ascorbate content of each organ, either intrinsic or that depend on the level of redox-active transition metal ions capable of promoting ascorbate oxidation (2).
It is also significant that ascorbate in mammalian tissues, especially in blood plasma, may serve as reductant for 1-Cys Prx from pathogens. Several pathogens have to cope with oxidative stress generated by phagocytic cells in blood. Diverse microorganisms such as Plasmodium falciparum and Trypanosoma cruzi possess no catalase or selenocysteine-dependent glutathione peroxidase and, therefore, rely on Prx to decompose peroxides (28).
An ascorbate-dependent peroxidase activity of 1-Cys Prx should be also relevant for plants (Table 2) because these organisms present very high levels of vitamin C in all subcellular compartments, particularly in chloroplasts, where 50 mM concentration can be attained (29). Consistent with these findings, intracellular redox homeostasis in plants is governed not only by GSH/GSSG ratios but also by ascorbate levels (30).
The product of the ascorbate-supported peroxidase activity of 1-Cys Prx, dehydroascorbate, is immediately reduced to ascorbate in the intracellular media, where it accumulates at millimolar levels (reviewed in ref. 2). Several mechanisms have been proposed to account for this fast reduction, such as chemical reaction with biothiols (31, 32) and enzymatic processes that depend on NADPH and glutathione (2). Glutaredoxin has often been considered an important dehydroascorbate reductase whose catalytic efficiency (kcat/KM) ranges from 104 to 107 M−1 s−1 (33, 34). However, other enzymes have also been considered to be relevant (35–39). Most likely, intracellular reduction of dehydroascorbate is a very fast reaction because many of these pathways operate together.
In conclusion, our studies extended previous observations about the redox versatility of Prx. All previously reported Prx reductants are thiol groups of low-molecular-weight substances or of proteins (thioredoxins and sulfiredoxins). Thus, the ascorbate-mediated reduction of protein sulfenic acids reported here represents a modification of the peroxiredoxin-thiol-specific antioxidant paradigm. In addition, our results reveal a previously uncharacterized antioxidant function for ascorbate. Of note, both sulfenic acids and ascorbate have been proposed in cell signaling (29, 40). The ascorbate/sulfenic acid pair may operate in the case of other proteins, representing an aspect of redox biochemistry that has yet to be explored in vivo. Thus, our results open perspectives in the understanding of cellular redox processes.
Experimental Procedures
Standard Reaction Buffer and Sample Preparation.
The standard mixture for most experiments was Hepes (50 mM), pH 7.5, containing azide (1 mM) and DTPA (0.5 mM). All enzymes were previously reduced with DTT (100 mM) overnight at 4°C. Proteins were subsequently gel filtered (PD10; Amersham Pharmacia, Piscataway, NJ) to eliminate excess DTT. This procedure was performed immediately before the experiments to obtain homogeneously reduced samples.
GS Protection Assay.
GS activity was measured by the γ-glutamyltransferase assay essentially as described (41). GS protection was evaluated by the amount of γ-glutamyltransferase activity remaining in solution after metal-catalyzed oxidation. GS oxidative inactivation was carried out as described (42, 43). The reagent concentrations used were DTT or ascorbate (10 mM), GS (0.2 mg/ml), and FeCl3 (3 μM) in Hepes (50 mM), pH 7.5, containing azide (1 mM). The incubation conditions were 15 min at 37°C. Remaining GS activity was measured by adding 1 ml of assay solution containing ADP (0.4 mM), glutamine (150 mM), Na2HAsO4 (10 mM), NH2OH (20 mM), MnCl2 (0.4 mM), and Hepes (0.1 M), pH 7.4. The reaction was incubated for 15 min at 37°C and then stopped by addition of 0.2 ml of FeCl3 (55 mg/ml) in HCl (6 M). Absorbance of the γ-glutamylhydroxamate–Fe3+ complex was spectrophotometrically measured at 540 nm. The concentrations of tested enzymes are described in the corresponding figure legends. GS activity was considered 100% when the enzyme was preincubated in the absence of metal-catalyzed oxidation systems. “No Prx” is the negative control establishing the extent to which the metal-catalyzed oxidation system inactivated the GS without the antioxidant enzymes.
Mass Spectrometry.
Accurate molecular mass determination was performed by mass spectrometry. Measurements were taken in positive ionization mode on a Q-TOF Ultima API fitted with an electrospray ion source (Micromass, Manchester, U.K.). The protein was solubilized in NH4HCOOH (50 mM) and directly injected into the instrument by using a Rheodyne 7010 sample loop coupled to a LC-10A VP Shimadzu pump at a constant flow rate (20 μl/min). Instrument control and data acquisition were conducted by a MassLynx 4.0 data system (Micromass), and the experiments were performed by scanning from a mass-to-charge ratio (m/z) of 50–1,800 by using a scan time of 2 seconds applied during the entire infusion. The mass spectra corresponding to the total ion current (TIC) chromatogram were averaged, allowing an accurate molecular mass determination. External calibration of the mass scale was performed with NaI.
Ferrous Oxidation Xylenol Orange (FOX2) Assay.
Peroxide concentration was determined by this method as described (44). Reaction mixtures containing standard reaction buffer with various enzyme concentrations (indicated in the legend or abscissa axis of the figures) and peroxide (500 μM) (H2O2 or tert-butyl hydroperoxide), ascorbate or DTT (1 mM) were incubated at 37°C for the indicated time. Reactions were stopped by addition of HCl at a final concentration of 0.2 M. Samples containing ascorbate were incubated for an additional 15 min with 1 unit of ascorbate oxidase (Sigma) at room temperature before ferrous xylenol orange solution addition. This was required because an excess of ascorbate reduces Fe+3, interfering with peroxide quantitation. Absorbance due to the nonenzymatic reaction, i.e., standard reaction buffer plus peroxide and reductant, was subtracted from the absorbance corresponding to the whole reaction mixture. Furthermore, appropriate controls were performed in each experiment (positive control, standard reaction buffer plus peroxide; negative Prx controls, standard reaction buffer plus peroxide and Prx without addition of any reductant).
NBD-Cl Assay.
The 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole (NBD-Cl) reacts with both sulfhydryl (-SH) and sulfenic acid (-SOH) to generate products that can be distinguished by the UV-visible absorption spectra. Sulfhydryl adducts of NBD-Cl peak at 420 nm (ε = 13,000 M−1 cm−1) (45). Sulfenic acid of NBD-Cl peaks at 347 nm, with a similar extinction coefficient (46).
Modification of Proteins with Thiol or Sulfenic Acid Reagents.
The proteins were previously reduced with DTT (100 mM) overnight in standard reaction buffer. Excess DTT was removed by gel filtration through a PD-10 column (Amersham Pharmacia). An aliquot, named “initial,” was taken from the reduced protein and added to 10 equimolar NBD-Cl or 1,000 equimolar 5,5-dimethyl-1,3-cyclohexanedione (dimedone). Then, the reduced protein was subsequently treated with 1.5 equimolar of H2O2 for 30 min at room temperature under anaerobic conditions (in the standard reaction buffer). An aliquot of this sample was treated with 10 equimolar NBD-Cl or 1,000 equimolar dimedone and named “oxidized.” Finally, the residual oxidized protein was treated with ascorbate (10 mM) or DTT (10 mM) for 1 h at room temperature; the excess reductant was washed away by ultrafiltration, and the protein was incubated with a 10-fold excess of NBD-Cl or 1,000-fold excess of dimedone. This final sample was named “ascorbate.” In all samples, excess NBD-Cl or dimedone were removed by ultrafiltration. Other modification was performed by incubation of reduced yPrx1 with a 100-fold excess of NEM for 3 h at room temperature. The excess NEM was washed away by ultrafiltration (by using a Microcon YM10; Millipore).
Protein Expression and Purification.
yPrx1 and all other Prx used in this work were purified from bacteria by specific affinity chromatography as described by the respective researcher who kindly provided us with each one of the 1-Cys Prx overexpression vectors (described in Experimental Procedures in SI Text): yPrx1 (S. cerevisiae), yeast recombinant enzyme in fusion with T7-tag for immunoaffinity purification; AhpE (M. tuberculosis), recombinant enzyme in fusion with histidine tag for nickel-affinity purification; ArPer 1 (A. thaliana), recombinant enzyme in fusion wiith biotin; rPf1-Cys-Prx (P. falciparum), recombinant enzyme in fusion with GST-tag, purified by using a GST-Glutathione Affinity System with the AKTA Prime Liquid Chromatography system (Amersham Biosciences); Dpx-2540 and Dpx-6005 (D. melanogaster), recombinant proteins in fusion with histidine tags for nickel-affinity chromatography purification; and Prdx6 (Rattus norvegicus), native protein purified by anion exchange (mono Q; Amersham Biosciences) followed by cation exchange (carboxymethyl Sepharose fast flow column; Amersham Biosciences). All the enzymes were analyzed by SDS/PAGE to determine their purity and integrity.
Enzymatic Kinetic Parameters.
Ascorbate peroxidase activities from yPrx1 (1 nM) and Prdx6 (0.5 nM) were assayed in a standard reaction mixture. The reaction was started with addition of H2O2 (200 μM) when ascorbate concentration varied from 10 to 80 μM. Ascorbate oxidation was monitored spectrophotometrically (ε265 = 14,500 M−1 cm−1) at 37°C.
Peroxide Consumption by Prdx6 in the Presence of Rat Homogenates.
The soluble fraction of rat kidney, pancreas, liver, and brain (R. norvegicus, strain: Sprague–Dawley) were obtained through potter homogenization of the organs in cold PBS. The homogenates were centrifuged for 45 min at 4°C and 16,000 × g. Purified Prdx6 (10 μM) was incubated with H2O2 (300 μM), and the rat organ homogenates (90 μg of protein per ml) were used as reductants. Consumption of H2O2 (≈150 μM) by Prdx6 was measured by FOX assay during 15 min at 37°C. In identical incubation conditions, homogenates were treated with ascorbate oxidase (1 unit) to deplete the reducing power derived from ascorbate, and these treated extracts were used as reductants of Prdx6. The effects of ascorbate oxidase or of extracts alone were analyzed as controls, and, in these conditions, minimal consumption was detected.
Supplementary Material
Acknowledgments
We thank S. Kawazu (Research Institute, International Medical Center of Japan, Tokoyo, Japan), A.B. Fisher (University of Pennsylvania School of Medicine, Philadelphia, PA), R. B. Aalen (University of Oslo, Oslo, Norway), B. G. Guimarães (Brazilian Synchrotron Light Laboratory, São Paolo, Brazil), and W. C. Orr (Southern Methodist University, Dallas, TX) for kindly providing expression clones; Earl R. Stadtman and Barbara Berlett (National Heart, Lung, and Blood Institute, National Institutes of Health) who kindly provided glutamine synthetase purified in their laboratory; Walter S. Sheppard, Marilene Demasi, and Maria C. Arias for helpful suggestions and critical reading of this manuscript; Dr. M. A. de Oliveira, Dr. E. Linares, Dr. A. J. Kowaltowski, and C. C. Caldeira for technical advice and assistance; S. Marana for helpful discussions about enzymatic procedures; and J. Hesson for revision of English grammar. This research was supported by grants from Fundação de Amparo a Pesquisa no Estado de São Paulo and Conselho Nacional de Pesquisas, as part of the Instituto do Milênio Redoxoma.
Abbreviations
- Prx
peroxiredoxin(s)
- RSH
thiol
- ROS
reactive oxygen species
- RNS
reactive nitrogen species
- SOH
sulfenic acid
- RS−
thiolate group
- Cysp
peroxidatic cysteine
- Cysr
resolving cysteine
- GS
glutamine synthetase
- DTT
dithiothreitol
- t-BOOH
tert-butyl hydroperoxide
- NEM
N-ethylmaleimide.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700481104/DC1.
References
- 1.Imlay JA. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 2.Rumsey SC, Levine M. J Nutr Biochem. 1998;9:116–130. [Google Scholar]
- 3.Wood ZA, Schroder E, Harris JR, Poole LB. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 4.Konig J, Baier M, Horling F, Kahmann U, Harris G, Schurmann P, Dietz KJ. Proc Natl Acad Sci USA. 2002;99:5738–5743. doi: 10.1073/pnas.072644999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neumann CA, Krause DS, Carman CV, Das S, Dubey DP, Abraham JL, Bronson RT, Fujiwara Y, Orkin SH, Van Etten RA. Nature. 2003;424:561–565. doi: 10.1038/nature01819. [DOI] [PubMed] [Google Scholar]
- 6.Sherman DR, Mdluli K, Hickey MJ, Arain TM, Morris SL, Barry CE, III, Stover CK. Science. 1996;272:1641–1643. doi: 10.1126/science.272.5268.1641. [DOI] [PubMed] [Google Scholar]
- 7.Park SH, Chung YM, Lee YS, Kim HJ, Kim JS, Chae HZ, Yoo YD. Clin Cancer Res. 2000;6:4915–4920. [PubMed] [Google Scholar]
- 8.Chung YM, Yoo YD, Park JK, Kim YT, Kim HJ. Anticancer Res. 2001;21:1129–1133. [PubMed] [Google Scholar]
- 9.Choi MH, Lee IK, Kim GW, Kim BU, Han YH, Yu DY, Park HS, Kim KY, Lee JS, Choi C, et al. Nature. 2005;435:347–353. doi: 10.1038/nature03587. [DOI] [PubMed] [Google Scholar]
- 10.Vivancos AP, Castillo EA, Biteau B, Nicot C, Ayte J, Toledano MB, Hidalgo E. Proc Natl Acad Sci USA. 2005;102:8875–8880. doi: 10.1073/pnas.0503251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jang HH, Lee KO, Chi YH, Jung BG, Park SK, Park JH, Lee JR, Lee SS, Moon JC, Yun JW, et al. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Biteau B, Labarre J, Toledano MB. Nature. 2003;425:980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- 13.Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 14.Rhee SG, Chae HZ, Kim K. Free Radical Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 15.Demasi AP, Pereira GAG, Netto LES. FEBS J. 2006;273:805–816. doi: 10.1111/j.1742-4658.2006.05116.x. [DOI] [PubMed] [Google Scholar]
- 16.Huang ME, Rio AG, Nicolas A, Kolodner RD. Proc Natl Acad Sci USA. 2003;100:11529–11534. doi: 10.1073/pnas.2035018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rand JD, Grant CM. Mol Biol Cell. 2006;17:387–401. doi: 10.1091/mbc.E05-06-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zee JVD, Broek PJA. Free Radical Biol Med. 1998;25:282–286. doi: 10.1016/s0891-5849(98)00049-5. [DOI] [PubMed] [Google Scholar]
- 19.Pedrajas JR, Miranda-Vizuete A, Javanmardy N, Gustafsson JA, Spyrou G. J Biol Chem. 2000;275:16296–16301. doi: 10.1074/jbc.275.21.16296. [DOI] [PubMed] [Google Scholar]
- 20.Choi HJ, Kang SW, Yang CH, Rhee SG, Ryu SE. Nat Struct Biol. 1998;5:400–406. doi: 10.1038/nsb0598-400. [DOI] [PubMed] [Google Scholar]
- 21.Chae HZ, Chung SJ, Rhee SG. J Biol Chem. 1994;269:27670–27678. [PubMed] [Google Scholar]
- 22.Ellman GL. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 23.You K-S, Benitez LV, McConachie WA, Allison WS. Biochim Biophys Acta. 1975;384:317–330. doi: 10.1016/0005-2744(75)90033-9. [DOI] [PubMed] [Google Scholar]
- 24.Li S, Peterson NA, Kim MY, Kim CY, Hung LW, Yu M, Lekin T, Segelke BW, Lott JS, Baker EN. J Mol Biol. 2005;346:1035–1046. doi: 10.1016/j.jmb.2004.12.046. [DOI] [PubMed] [Google Scholar]
- 25.Segel HI. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. New York: Wiley; 1975. pp. 606–621. [Google Scholar]
- 26.Manevich Y, Fisher AB. Free Radical Biol Med. 2005;38:1422–1432. doi: 10.1016/j.freeradbiomed.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 27.Hancock RD, Galpin JR, Viola R. FEMS Microbiol Lett. 2000;186:245–250. doi: 10.1111/j.1574-6968.2000.tb09112.x. [DOI] [PubMed] [Google Scholar]
- 28.Wilkinson SR, Kelly JM. Biol Chem. 2003;384:517–525. doi: 10.1515/BC.2003.060. [DOI] [PubMed] [Google Scholar]
- 29.Smirnoff N. Curr Opin Plant Biol. 2000;3:229–235. [PubMed] [Google Scholar]
- 30.Pignocchi C, Foyer CH. Curr Opin Plant Biol. 2003;6:379–389. doi: 10.1016/s1369-5266(03)00069-4. [DOI] [PubMed] [Google Scholar]
- 31.Guaiquil VH, Farber CM, Golde DW, Vera JC. J Biol Chem. 1997;272:9915–9921. doi: 10.1074/jbc.272.15.9915. [DOI] [PubMed] [Google Scholar]
- 32.Krauth-Siegel RL, Ludemann H. Mol Biochem Parasitol. 1996;80:203–208. doi: 10.1016/0166-6851(96)02689-8. [DOI] [PubMed] [Google Scholar]
- 33.Washburn MP, Wells WW. Biochem Biophys Res Commun. 1999;257:567–571. doi: 10.1006/bbrc.1999.0508. [DOI] [PubMed] [Google Scholar]
- 34.Shimaoka T, Miyake C, Yokota A. Eur J Biochem. 2003;270:921–928. doi: 10.1046/j.1432-1033.2003.03452.x. [DOI] [PubMed] [Google Scholar]
- 35.Del Bello B, Maellaro E, Sugherini L, Santucci A, Comporti M, Casini AF. Biochem J. 1994;304:385–390. doi: 10.1042/bj3040385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishikawa T, Casini AF, Nishikimi M. J Biol Chem. 1998;273:28708–28712. doi: 10.1074/jbc.273.44.28708. [DOI] [PubMed] [Google Scholar]
- 37.May JM, Mendiratta S, Hill KE, Burk RF. J Biol Chem. 1997;272:22607–22610. doi: 10.1074/jbc.272.36.22607. [DOI] [PubMed] [Google Scholar]
- 38.Wells WW, Xu DP, Yang YF, Rocque PA. J Biol Chem. 1990;265:15361–15364. [PubMed] [Google Scholar]
- 39.Whitbread AK, Masoumi A, Tetlow N, Schmuck E, Coggan M, Board PG. Methods Enzymol. 2005;401:78–99. doi: 10.1016/S0076-6879(05)01005-0. [DOI] [PubMed] [Google Scholar]
- 40.Poole LB, Karplus PA, Claiborne A. Annu Rev Pharmacol Toxicol. 2004;44:325–347. doi: 10.1146/annurev.pharmtox.44.101802.121735. [DOI] [PubMed] [Google Scholar]
- 41.Stadtman ER, Smyrniotis PZ, Davis JN, Wittenberger ME. Anal Biochem. 1979;95:275–285. doi: 10.1016/0003-2697(79)90217-3. [DOI] [PubMed] [Google Scholar]
- 42.Kim K, Rhee SG, Stadtman ER. J Biol Chem. 1985;260:15394–15397. [PubMed] [Google Scholar]
- 43.Levine RL. J Biol Chem. 1983;258:11828–11833. [PubMed] [Google Scholar]
- 44.Jiang ZY, Hunt JV, Wolff SP. Anal Biochem. 1992;202:384–389. doi: 10.1016/0003-2697(92)90122-n. [DOI] [PubMed] [Google Scholar]
- 45.Birkett DJ, Price NC, Radda GK, Salmon AG. FEBS Lett. 1970;6:346–348. doi: 10.1016/0014-5793(70)80095-3. [DOI] [PubMed] [Google Scholar]
- 46.Ellis HR, Poole LB. Biochemistry. 1997;36:15013–15018. doi: 10.1021/bi972191x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.