Abstract
The human erythrocyte (red blood cell, RBC) demonstrates extraordinary ability to undergo reversible large deformation and fluidity. Such mechanical response cannot be consistently rationalized on the basis of fixed connectivity of the cell cytoskeleton that comprises the spectrin molecular network tethered to phospholipid membrane. Active topological remodeling of spectrin network has been postulated, although detailed models of such dynamic reorganization are presently unavailable. Here we present a coarse-grained cytoskeletal dynamics simulation with breakable protein associations to elucidate the roles of shear stress, specific chemical agents, and thermal fluctuations in cytoskeleton remodeling. We demonstrate a clear solid-to-fluid transition depending on the metabolic energy influx. The solid network's plastic deformation also manifests creep and yield regimes depending on the strain rate. This cytoskeletal dynamics model offers a means to resolve long-standing questions regarding the reference state used in RBC elasticity theory for determining the equilibrium shape and deformation response. In addition, the simulations offer mechanistic insights into the onset of plasticity and void percolation in cytoskeleton. These phenomena may have implication for RBC membrane loss and shape change in the context of hereditary hemolytic disorders such as spherocytosis and elliptocytosis.
Keywords: cytoskeleton remodeling, computer simulation, fluidization, plasticity, spectrin–actin dissociation
During its 120-day life span, a red blood cell (RBC) circulates a million times in human body, often squeezing through narrow capillaries. The erythrocyte's remarkable mechanical properties originate from the unique architecture of its cell wall, which is the main load bearing component as there are no stress fibers inside the cell. The cell wall comprises a spectrin tetramer network tethered to a phospholipid bilayer (1, 2). It is very flexible, and yet resilient enough to recover the biconcave shape whenever the cell is quiescent (see Fig. 1). It has been proposed that the spectrin network connectivity is not fixed and can experience extensive remodeling as the RBC undergoes large deformation. Experiments show that, under physiological conditions, the spectrin tetramers in an unstressed intact cell wall exist in rapid dynamic equilibrium with the dimers, and that shear-induced cell wall deformation displaces the balance in favor of the dimers (3). Here, the cell wall behaves as a weak elastic solid at low strain levels, whereas it can be fluidized beyond a certain level of shear deformation, similar to soft matter such as emulsions and colloidal pastes (4, 5). Upon unloading, the dynamic stability is shifted toward tetramers and the stiffness of the cell wall increases. Experiments also suggest that cytoskeleton remodeling can be regulated by biochemical factors such as Ca2+ and ATP (adenosine 5′-triphosphate) concentrations (6, 7). It has been postulated that phosphorylation affects spectrin–actin binding and produces changes in the shape and deformability of human RBC (8–11); this is equivalent to the injection of metabolic energy specifically to spectrin–actin junctions, thereby inducing local rupture between actin and spectrin. Prior analytical modeling addressed the impact of ATP-induced cytoskeletal defects arising from spectrin–actin dissociation on RBC shape and membrane fluctuation (12). Activation of these biochemical processes can result in lower connectivity and, consequently, a softer or even fluidized network. Therefore, the confluence of mechanical strain energy, nonspecific thermal fluctuation energy, and specific biochemical activation energy fundamentally influences the behavior of the cytoskeleton and the membrane tethered to it.
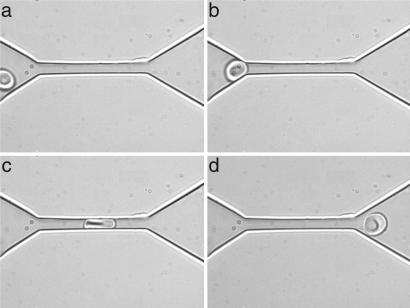
Fig. 1.
An experimental in vitro demonstration of the “fluidization” of a healthy human RBC through a microfluidic channel at room temperature. The series of images show the shape of an RBC as it is squeezed through a 4 μm × 4 μm channel made of polydimethyloxysilane (PDMS), under a pressure differential of 1.5 mm of water. Note the recovery of shape upon egress from the channel. Images a, b, c, and d were taken at relative times of 0, 0.4, 0.8, and 1.4 s, respectively.
Optical tweezers (OT) experiments (13, 14) have shown that the membrane with cytoskeletal attachment can sustain significant shear, with an estimated shear modulus μ = 4–10 μN/m. Indeed, the in-plane shear energy that an RBC can apparently store in these OT experiments is ≈200 pN × 8 μm/2 = 5,000 eV, which far exceeds the total bending energy it stores (≈8πκ = 50 eV), for bending modulus κ ≈ 2 × 10−19 J (15). Classic Canham–Helfrich explanation of the biconcave equilibrium shape of erythrocyte (16, 17) requires minimization of total bending energy only, without considering the much larger in-plane shear energy. Equilibrium shape calculations with inclusion of in-plane energies, at both continuum (14, 18) and discrete spectrin network (14, 19) levels, thus exhibit a seemingly paradoxical behavior. At the experimental range of μ and κ values, the biconcave equilibrium shape is rarely obtained in numerical simulations without careful tuning of the stress-free reference state for the in-plane energy.
Based on extensive parametric studies, the following mechanistic hypothesis was proposed (14) to resolve the paradox. In an ideal limit, for any RBC shape, the cytoskeleton always undergoes remodeling in topological connectivity at a certain rate to relax its in-plane shear elastic energy to zero. Therefore in considering the equilibrium shape of an RBC, one does not need to account for the in-plane shear energy, even though in OT experiments it can be injected temporarily into the cytoskeleton. The remodeling process has been studied heuristically in (14) by simulating simple liquids confined on a surface and interacting via the Lennard–Jones or Stillinger–Weber potentials (20). This produces cytoskeleton that appears similar to atomic force microscopy scans (21–23), akin to reverse Monte Carlo (24) modeling. Despite these considerations, detailed simulations of spectrin network dynamics promoting plasticity and fluidization of RBC are presently unavailable.
In this paper, a cytoskeletal dynamics (CD) simulation framework is developed to study the RBC spectrin network remodeling process and fluidization. The framework comprises the following key ingredients: (i) a minimal representation of the cytoskeleton geometry that can self-organize and dynamically evolve, and (ii) mechanisms for nonthermal energies such as the strain energy or specific biochemical energy to influence and regulate structural evolution. By abstracting out many details, such a cytoskeletal dynamics simulation provides detailed mechanistic insights into the mechanics and physics of RBC deformation. This model could then be used in studying RBCs with cytoskeletal defects which are typical in blood disorders such as hereditary spherocytosis and elliptocytosis (25, 26). The present work thus seeks to provide a significant advance over previous models that were either formulated at the continuum scale (18), thereby ignoring the discrete cytoskeletal structure, or that assumed a fixed cytoskeleton connectivity (14, 19, 27–30), thereby useful only to study elastic cell deformation without any dynamic cytoskeletal remodeling due to mechanical, thermal, or chemical driving forces.
Computational Model
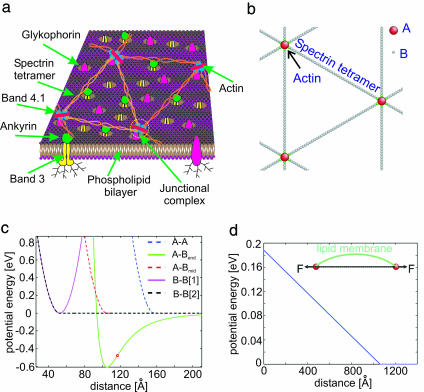
The erythrocyte cell wall consists of a lipid bilayer, cytoskeleton network, and integral membrane proteins. The intramembrane proteins band-3 and glycophorin tether the cytoskeleton network to the bilayer via additional binding proteins (e.g., ankyrin and 4.1). A schematic representation is shown in Fig. 2a. Spectrin is a protein tetramer formed by head-to-head association of two identical heterodimers (31). Each heterodimer has an α-chain, which has 22 triple-helical segments, and a β-chain, which has 17 triple-helical segments. A minimalistic representation of spectrin is therefore chosen to be S = 39 spherical beads (B in Fig. 2b) connected by S − 1 unbreakable springs. The spring potential, VBB(r) = k0(r − r0)2/2, is plotted as the magenta curve in Fig. 2c, with equilibrium distance r0 = Lmax/(S − 1), where Lmax is the contour length of the spectrin (chosen to be 200 nm). Thus, r0 = 5.26 nm. The spring constant k0 is chosen later. Beads that are not topologically connected on the chain, or are on different chains, repel each other sterically. Their interaction potential is taken to be simply VBB(r)H(r0 − r), shown as the black dash curve in Fig. 2c, where H(x) is the Heaviside step function.
Fig. 2.
Coarse-grained cytoskeletal dynamics computer simulation. (a) Schematic of cytoskeleton of the human RBC. (b) A minimal physically realistic model with breakable actin–spectrin interaction. A (red sphere) represents an actin protofilament, and B (green and gray spheres) represents a spectrin segment. Only the two ending spectrin units (green spheres) of a spectrin chain can bind to A. (c) Interaction potentials between A–A (steric repulsion), A–Bend (Lennard–Jones potential), A–Bmid (steric repulsion), B–B[1] (spring potential between nearest neighbor spectrin units), and B–B[2] (steric repulsion between any other spectrin units). Bmid represents a nonending spectrin unit, and Bend represents an ending spectrin unit. Note that A–Bend forms breakable linkage. (d) Lipid membrane introduces additional effective repulsion between A–A (12), modeled by a linear potential with cutoff.
Among the S beads of a spectrin chain, there are two special beads colored in green in Fig. 2b that represent the free ends. They can bind noncovalently to junction complexes (2), composed of a short actin proto-filament and protein band 4.1. Experiments indicate that, without protein 4.1, spectrin only binds weakly to actin, with equilibrium association constant K1 ≈ 5 × 103 M−1. With 4.1, however, strong binding occurs, forming ternary complexes with equilibrium constant K2 ≈ 1 × 1012 M−1 (32). A separate measurement indicated that the equilibrium association constant of protein 4.1 with actin itself is K3 ≈ 1 × 107 M−1 (33). In the schematic of the simulation model, the large red bead (A in Fig. 2b) denotes the junction complex. The terminal green beads of spectrin are attracted to A via the Lennard–Jones potential, VAB(r) = 4ε((σ/r)12 − (σ/r)6), plotted as the green curve in Fig. 2c. The A–B association energy ε is chosen to be 14 kcal/mol or 0.6 eV. The characteristic interaction length scale σ, on the other hand, is chosen so that the equilibrium A–B distance 21/6σ is precisely 2r0. The “capture radius” of A–B attraction is ≈20 nm (Fig. 2c). The capture diameter is therefore ≈40 nm, which matches the actual actin proto-filament length of ≈35 nm. To eliminate a free parameter, k0 is chosen to be identical to the curvature of VAB(r) at its well bottom, k0 = 36 (2)2/3εσ−2. The molecular mass of a spectrin tetramer is almost 1,000 kDa (1). Thus, the B beads are chosen to have a mass of 1,000/S kDa each. The mass of A bead is chosen to be 121 kDa, as the sum of the masses of actin proto-filament and band 4.1 (1).
The novelty of this model over prior spectrin-based simulations (14, 19, 27–30) is that the A–B association is breakable, as well as reformable, with the network topology changing dynamically. Under increasing tensile force, the A–B association breaks when their distance reaches the inflexion point of VAB(r), rinflexion = (26/7)1/6σ, indicated as the red circle in Fig. 2c. This corresponds to 10.9% bond strain and 24.9 pN force. A saturation effect exists for the A–B affinity because most of the actin proto-filaments have, at most, six slots to hook up with spectrin ends (via band 4.1) (30). This is implemented in the simulation quite naturally as follows. The attractive part of the Lennard–Jones potential, −4ε(σ/r)6, is turned on for an ending B, if and only if there are less than six other ending Bs already within the inflexion radius rinflexion. In other words, when six ending Bs are already associated with A (judged by their distances to that A), all other ending Bs only see the repulsive part of the potential 4ε(σ/r)12.
Nonending Bs have only steric repulsion with A, taken to be simply VBB(r−r0)H(2r0−r), shown as the red dash curve in Fig. 2c. Similarly, any two As would repel each other at short range with VBB(r − 2r0)H(3r0 − r), shown as the blue dash curve. This cytoskeletal interaction potential model has the maximum simplicity in terms of choice of parameters. Essentially, once the A–B association energy ε is known, there are no free parameters to choose.
The lipid bilayer exerts additional forces on cytoskeleton. The fluid-like lipid bilayer is slightly crumpled at equilibrium (12) (see Fig. 2d) pinned by the junction complexes (34), and it exerts an effective repulsion between two neighboring junction complexes. It can be shown that the repulsive force due to a bent elastic cylindrical shell is constant with respect to end-to-end distance, until the shell is fully straightened out. To model this effect, we add an additional repulsive potential between pairs of As that are nearest neighbors
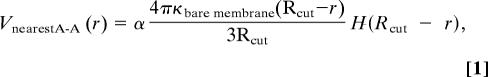 |
where Rcut is the distance at which the membrane becomes flat. We take Rcut = 106.1 nm, which is 20% larger than the thermally averaged A–A distance, chosen to be R0 = 88.4 nm. With the estimated bare membrane bending modulus κbare membrane = 2 × 10−20 J (12), α = 0.36, so that the pressure of the entire system is nearly zero at T = 300 K and R0 = 88.4 nm.
A and B are also confined vertically by the lipid bilayer. Soft harmonic confinement potentials Vconfine(z) = kconfinez2/2 are used in the z direction to mimic this effect without explicitly simulating the fluid bilayer. Because the junction complex is attached to the bilayer, A's confinement potential is in both +z and –z directions (Vconfine(z)), whereas B is confined on the +z side only (Vconfine(z)H(z)). In other words, the spectrin chain can freely swing down, but will be hampered when swinging up. The stiffness of both confinement potentials are quite arbitrarily chosen to be kconfine = 0.1k0.
We perform 3D coarse-grained molecular dynamics simulations governed by the aforementioned potentials. Its fundamental time scale (chain oscillations) is at the nanosecond level, and it is typical for a whole CD simulation run to cover a few microseconds. The Berendsen thermostat (35) is used to control the system's temperature, and the instantaneous stress tensor τij is calculated by using the Virial formula (36). Hydrodynamic interactions (37) are ignored [see supporting information (SI) Text]. We begin with the configuration in Fig. 2b that is a perfect triangular network in a periodic supercell, containing 7 × 8 = 56 junction complexes, 168 chains, and 6,552 spectrin beads, spanning an area ≈1.5 μm × 1.5 μm at T = 0 K. Although discrete spectrin-based whole-cell simulations have been performed before (14), and it is not out of the question even for the present cytoskeletal dynamics model, it is much more appealing to begin with a study of some unit processes in a representative area element.
Results
Chemically Induced Spectrin–Actin Dissociation.
Starting with perfect triangular network at T = 0 K, where A–A nearest-neighbor distance is Lmax + 27/6σ = 221 nm, the network is gradually heated while maintaining the internal pressure near zero. As expected, the system shrinks because of entropic elasticity, to an average nearest-neighbor A–A distance of 88 nm at T = 300 K and total area ≈0.6 μm × 0.6 μm (Fig. 3a). The vertical fluctuations of spectrin beads are ≈40 nm. The topology of network remains nearly perfect at T = 300 K. The mean degree of the network 〈d〉 is taken to be the average number of spectrin ends attached to a junction complex. 〈d〉 = 6 if the network topology is perfect. Upon further heating, broken-link, degree-5 defects are spontaneously created (12).
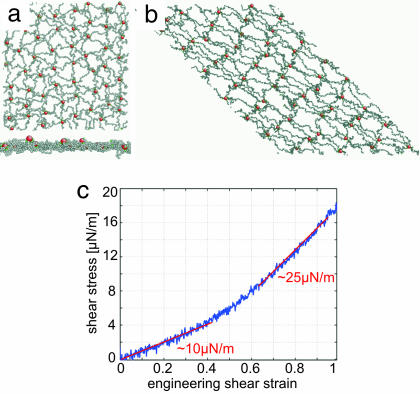
Fig. 3.
CD simulations with only mechanical energy input. (a) Snapshot of the equilibrium network structure at 300 K, top view and side view. (b) Snapshot of the sheared (γ = 1) network structure at 300 K, top view. (c) Shear stress-strain response at strain rate γ̇ = 3 × 105 per s.
Fixing T = 300 K, we then apply simple shear to the supercell with strain rate γ̇ = 3 × 105 per s, where γ is the engineering shear strain. Final sheared network configuration is shown in Fig. 3b, and the shear stress versus shear strain curve is plotted in Fig. 3c. Linear elastic shear modulus μ0 ≈ 10 μN/m agrees well with literature values from OT and micropipette aspiration experiments (13, 14). Using the worm-like chain model for the spectrin molecule (14), this μ0 corresponds to a persistence length of p ≈ 6 nm, which is close to the B bead diameter (5.26 nm). This numerical agreement in μ0 is perhaps fortuitous without any intentional fitting to the shear response.
At large shear strains, γ = 80%, the tangent shear modulus stiffens to μf ≈ 25 μN/m. This value then amounts to a prediction of the simulation. Even at γ = 1, few topological defects appear in the simulation. The stress–strain response is nearly perfectly reversible and elastic. Again, the agreement is good with OT experiments (13, 38). To match the experiments up to 100% RBC elongation with a finite-element model, a third-order hyperelasticity constitutive relation is needed for cell wall material with stiffening behavior (13, 38). The value of μf estimated from finite element simulations is 20 μN/m at γ = 1.
Until now, the cytoskeleton behaves just like an ordinary inert material. Room-temperature thermal fluctuations do not overcome A–B binding to effect topology change. Because of homeostasis (nearly constant body temperature), cytoskeletal changes cannot arise from significant temperature changes. However, the CD could be driven by chemical energy flux, pushing the system out of equilibrium (39). To explore this effect, we mimic biochemically activated actin–spectrin dissociation by an instantaneous kinetic energy transfer of X = 0.7 eV to an ending B bead. The magnitude of X matches roughly the energy release from ATP hydrolysis reaction, which is ≈20–25kBT per molecule (40). The direction of kinetic energy transfer is random in the 4π spherical angle. Because the binding energy between A–B is ε = 0.6 eV, it is highly likely that such a hit would lead to dissociation of the particular AB junction. The schedule of kinetic energy hits conforms to a random Poisson process, with equal probability for all spectrin ends, irrespective of their hit histories. The center-of-mass of the cytoskeleton is held fixed at all times.
Initially, we assign a hit rate of H = 100 per μs per spectrin end without simultaneously shearing the cytoskeleton. The network quickly becomes proliferated with topological defects, as indicated by the 〈d〉 statistics (see SI Fig. 6a). Larger and larger holes appear, which eventually percolate through the supercell (41). The network then loses shear-bearing capability; that is, the solid is completely fluidized at H = 100 per μs without mechanical stress (gel-to-sol transition). The fluidization process reflects a dynamic percolation of broken bonds across the network (41), which could lead to mass and momentum transport even below the static percolation limit. The importance of the percolation limit of broken bonds to shear elasticity was pointed out in ref. 12.
To see the “annealing” or “recovery” behavior (sol-to-gel transition), we then turn off the kinetic energy hits (H = 0), and the cytoskeleton reconstitutes readily, as indicated by the 〈d〉 statistics (see SI Fig. 6b), within the CD simulation time scale. The network that reforms from fluidized state does not have a perfect triangular-net topology, it contains topological “mistakes,” but structural similarity to a triangular network is strong. The shear stress–strain response (not shown) is similar as well. To further confirm this finding, we plot the potential energy U of reformed network (SI Fig. 6b Inset), which approaches that of the perfect network reference value over time. Thus the cytoskeleton “re-gels” from fluidized state after biochemical energy influx is turned off, with a time constant τg ≈ 0.1 μs, taken from the half life of 〈d〉 and U recoveries in SI Fig. 6b.
The characteristic recovery time τg in present simulation is perhaps much faster than that in reality. A–B association is represented by a smooth potential VAB(r) with no activation energy barrier (Fig. 2c). Thus, once A loses one B association and its d is <6, it turns on a smooth attractive force field with capture diameter ≈40 nm for all free spectrin ends. In reality, both association and dissociation reactions have activation barriers (42), catalyzed by protein kinases (7) as well. We now nondimensionalize all time and rate labels by τg. The fluidization at H = 100 per μs is thus attributed to Hτg ≫ 1. In other words, the rate of biochemical activation and induced dissociation far exceeds that of intrinsic recovery, whereby a disconnected spectrin chain reconnects somewhere to reduce the free energy. The recovery rate depends on applied stress and temperature, which change driving force and kinetics of reattachment (forming a stretched chain with two constrained ends versus a coiled chain with one or two free ends).
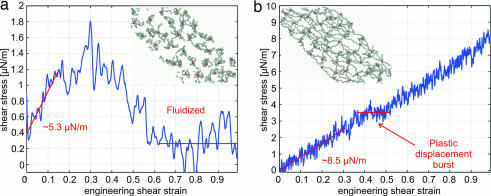
We next choose the borderline case of H = 10 per μs, Hτg = 1. The cytoskeleton shear stress vs. shear strain is plotted in Fig. 4a, at the same strain rate γ̇ = 3 × 105 per s or γ̇τg = 0.03. The linear modulus μ0 is softened from 10 to 5.3 μN/m. More dramatically, the shear stress vanishes above γ = 60%. A look at the structure indicates that the cytoskeleton has indeed fluidized (41), with large holes percolating through the simulation supercell (Fig. 4a Inset). This behavior is in stark contrast to Fig. 3 with no biochemical activation, where the network deforms elastically, and even stiffens to μf ≈ 25 μN/m. Thus, at hit rate Hτg ≈ 1, a softened, but still solid, cytoskeleton is obtained at small strain, which transforms into fluid at critical strain γfluidize ≈ 50%. RBCs commonly encounter such strains in circulation (43).
Fig. 4.
Shear stress-strain response of network at strain rate γ̇ = 3 × 105 per s. (a) Energy hit rate 10 per μs and final network structure at γ = 1 (Inset). (b) Energy hit rate 2.5 per μs and final network structure at γ = 1 (Inset).
The mechanism for this fluidization transition could be summarized as follows. Under shear strain, some spectrin chains are elongated and some shortened, depending on their alignment to the principal axes of stress. The elongated chains exert an effective tensile force FWLC at the A–B junction roughly according to worm-like chain entropic force (14). At small γ, the maximum elongation is small, so even after dissociation the probability of reattachment is large. However, at large γ, the initial elongation is too large (see Fig. 4b Inset). Therefore, once a spectrin link is broken, the chain recoils and becomes exponentially unlikely for the spectrin chain to fluctuate back to the original length to reestablish the connection.
The response seen in the above CD simulation comes from a nonequilibrium system, which cannot be made similar to an inert system by simple scaling (law of corresponding states). This is because one essential element, FWLC, is proportional to kBT, where T is the thermodynamic temperature; but the other element, the rate of A–B association/dissociation, is related to the rate of phosphorylation. This is analogous to the concept of two temperatures (39), one for general degrees of freedom including chain vibrations and bending (thermodynamic temperature) and one just for topological change (effective or “structural evolution” temperature refs. 5, 44, and 45). When the thermodynamic temperature is raised, due to equi-partition theorem, all degrees of freedom tend to be equally excited. In contrast, biochemical activation is highly targeted due to the specific biochemical machinery required, e.g., for phosphorylation; this is reflected in our model, because when H is raised, only the A–B associations (strategic nodes of the network) are strongly excited. The effective temperature concept is generic in soft glassy matter (45), irradiated materials (46), and generally nonequilibrium materials (39) under mechanical, chemical, or radiative energy influxes. However, it is perhaps in the biological context that the concept is put to use in a very sophisticated manner (47). Biochemical activation is specific and local. Only certain cytoskeletal proteins can be activated to elicit certain structural response.
We then examine the other limit, Hτg ≪ 1, with H = 2.5 per μs. The shear stress–strain response is plotted in Fig. 4b, with final configuration at γ = 100% (Fig. 4b Inset). The cytoskeleton is a solid throughout the entire range of strain, because although voids exist, they never percolate through the network. Its linear modulus μ0 (8.5 μN/m) is very close to that with no energy hits. The difference with the zero chemical energy influx situation (Fig. 3) is that there is a plastic displacement burst at γ = 35–50%, similar to incipient plasticity events in metals (48). However, the system is still essentially a solid. Therefore, we conclude that, at Hτg ≪ 1, the cytoskeleton behaves like a plastically deforming solid, with the rate of plastic deformation governed by H and stress.
The strain γF at the first plastic displacement burst depends on the energy hit-rate H, the energy per hit X, and the spectrin–actin association energy ε. SI Table 1 shows that γF and the shear moduli (at the initial elastic response) decrease as the energy hit-rate increases from 0 to 10 per μs per spectrin. The constant strain rate used in all of the deformation simulations, γ̇ = 3 × 105 per s, is very high in terms of absolute value. However, if scaled by τg, the nondimensionalized strain rate γ̇τg is actually quite reasonable with a clear physical meaning. Take the last case of Hτg = 0.1: combined with γ̇τg = 0.03, this would mean that during the course of straining the cytoskeleton to 100%, each spectrin end would receive 3.3 biochemical activation hits on average. We have also investigated the effect of strain rate on γF (SI Table 2). When H = 0, γF is large (>100%) and nearly independent of strain rate. At finite H, we discovered two regimes of behavior. Take the case of Hτg = 0.1. When γ̇τg < 0.014, γF is sensitive to γ̇ in almost linear relationship; but when γ̇τg > 0.014, γF becomes much less sensitive to γ̇. The reason is that, at small γ̇, the first plastic displacement burst is governed more by damage accumulation caused by H than by mechanical stress τ. The time tD to accumulate enough damage to facilitate a displacement burst depends more on H, and less on τ or γ, so γF = γ̇tD can be approximated by γF ≈ γ̇tD(H), and thus the linear relationship. But when γ̇ is large, stress ramping is fast compared with damage by H, and the high stress begins to play a much more substantial role in inducing displacement burst, and the strain threshold for this to happen becomes more constant. These two plastic deformation regimes are analogous to the creep and yield regimes of engineering materials, which are sensitive (much less sensitive) to the strain rate at small (large) strain rates, respectively, with the latter regime possessing a much more clear-cut yield strain threshold behavior.
Dimer–Dimer Dissociation Due To Shearing.
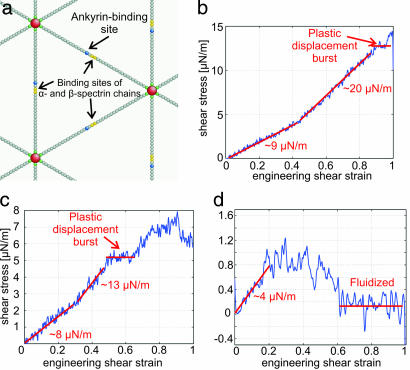
In Fig. 2b, two features of cytoskeleton are ignored: the confinements due to ankyrins and band 3 proteins (2) that also bind the spectrin to the membrane, and the possibility that a spectrin tetramer can dissociate into two heterodimers (3). These two aspects are considered here (Fig. 5a), where interest is shifted from spectrin–actin dissociation (due to locally delivered chemical energy) to the effects of dimer–dimer dissociation during shearing of the network. The strength of dimer–dimer association is known to be weaker than the actin–4.1–spectrin association, with equilibrium constant K4 ≈ 1.5 × 106 M−1 (42). In Fig. 5a, the confinement due to ankyrin and band 3 protein is modeled by applying the same confinement potential Vconfine(z) to the blue beads, which were randomly placed two beads ahead of or behind the center of each chain. Rupturing of the chain (symbolizing dissociation of a spectrin tetramer into dimers) was allowed to occur in the middle between the two yellow beads. The original harmonic potential k0(r − r0)2/2 between the yellow beads B′ was substituted by a Lennard–Jones potential VB′B′ = 4εB′B′ ((σ/(r + r0))12 − (σ/(r + r0))6), which has similar characteristics as VAB. The only difference is that the minimum of VB′B′ is shifted to r0, which is the equilibrium distance between all spectrin beads.
Fig. 5.
Refined CD simulation. (a) A model with breakable actin–spectrin and/or α-spectrin–β-spectrin interaction. Red spheres represent actin protofilaments, and green, gray, blue, and yellow spheres represent a spectrin segment. Only the two ending spectrin units (green spheres) of a spectrin chain can bind to red sphere. (b) Shear stress-strain response of network at γ̇ = 3 × 105 per s and dimer–dimer association energy of 0.6 eV. Corresponding shear stress-strain response of network at 3 × 105 per s and 0.56 eV (c) and 3 × 105 per s and 0.47 eV (d) are shown.
As above, the perfect triangular network was initially heated from T = 0 to 300 K while the internal pressure was kept close to zero. εB′B′ was varied from 0.6 eV to 0.47 eV (that is, from 100% to 78% of the actin–spectrin association energy ε). This choice resulted in an equilibrated network of reduced integrity. In particular, although all of the actin–spectrin connections were unbroken (〈d〉 = 6), the integrity at T = 300 K of the dimer–dimer associations varies from 100% to ≈90%. Next, the same simple shear deformation was applied to model networks of various integrities at a strain rate of γ̇= 3 × 105 per s. No specific biochemical energy hits were injected at the network associations, and only mechanical energy was supplied.
In the first simulation (Fig. 5b), εB′B′ = 0.6 eV, whereas the integrity of dimer–dimer connections at T = 300 K was 100%. The response of the network was initially similar to the response shown in Fig. 3c, where no dimer–dimer dissociation was allowed. At small strains, it exhibited an elastic shear modulus of ≈9 μN/m. Then, at ≈ 45% strain, it became stiffer with a shear modulus of 20 μN/m. At 90% strain, where the shear stress reached the value of ≈12.5 μN/m, a plastic type displacement burst occurred. This burst was caused by dimer–dimer dissociations, because the integrity of actin–spectrin associations was 100% throughout the numerical experiment. At εB′B′ = 0.56 eV (7% lower than ε) the response of the network is shown in Fig. 5c. It initially exhibited a shear modulus of ≈8 μN/m, slightly lower than in the previous case. At 25% strain, the network shows a larger tangent modulus of 13 μN/m. At ≈45% strain, a plastic displacement burst occurred. Then the response was elastic with a tangent shear modulus of 13 μN/m, until 80% shear strain where another plastic displacement burst started.
At εB′B′ = 0.47eV (22% lower than ε), which corresponds to 91% integrity of the dimer–dimer connections, the response of the spectrin network to shear deformation changed dramatically (Fig. 5d). Initially, the spectrin network showed a linear elastic behavior with a reduced shear modulus of ≈4 μN/m. From 20% until 50%, the shear stress was constant. Finally, at 60% strain, the shear stress response almost vanished and the cytoskeleton fluidized.
In the case of dimer–dimer dissociation due to shearing, the yield strain depends on the association energy between the dimers. The spectrin network was first heated from 0 to 300 K and then equilibrated. SI Table 3 shows that, during the above procedure, the resulting spectrin network integrity at the equilibrium reduced from almost 100% at εB′B′ = 0.6 eV to 91% at εB′B′ = 0.47 eV. The yield strain was also reduced from 90% to 20%, respectively. The “thermal equilibrium” dimer content depends on kBT/εB′B′. The plastic strain threshold is very sensitive to the above dimer content, because when kBT/εB′B′ is raised, not only do we have more dimers, but even the existing tetramers are severely weakened by entropic elasticity. The trend of our results matches very well with the experimental data presented in ref. 3. There, it was shown that the cell wall stability decreases when the dimer content of the RBC increases. The most striking outcome of the above work (3) is the observation that shear deformation induces dissociation of tetramers into dimers. This finding matches our numerical results, which show a progressive decrease of the dimer–dimer association integrity during shearing (SI Fig. 7).
Discussion and Concluding Remarks
We have developed a cytoskeletal dynamics simulation framework that allows active remodeling of the 3D cytoskeleton via breakable as well as reformable associations of the junction complex and spectrin tetramer. Possible roles of external shear stress (related to deformability of healthy RBC at large strains), specific chemical energy (i.e., ATP, as a possible means to actively adjust RBC deformability in circulation), and protein association strength (possibly related to RBC disease states with weakened/broken cytoskeletal connectivity) versus temperature and strain rate can be studied parametrically by using the proposed model. These results offer mechanistic rationale for the erythrocyte cytoskeleton mechanical function under loading. The model demonstrates that cytoskeletal remodeling through specific biochemical activation is a possible means for RBC deformability to be actively “tuned” during its circulation in microvasculature. On one hand, structural integrity of the cell demands certain minimal properties of the cytoskeleton. For instance, the largest flaw in the RBC cytoskeleton cannot exceed a critical size, otherwise lipid membrane loss will occur via the vesiculation instability (49). On the other hand, RBC also encounters different environments (artery, capillary, splenic sinus, etc.) and mechanical conditions in circulation, and its mechanical properties may need to change “on the fly” (50). The relative influence of mechanical stress during constrained shear flow, energy exchange and biochemical modification of dimer–dimer or actin–spectrin binding energy is largely unknown at this time, and could depend on the natural variations in the cytoskeletal structures in a given population of cells and the local biochemical environment. Our parametric CD model is a first step toward unraveling possible mechanistic underpinnings of these complex, intertwined pathways.
Supplementary Material
Acknowledgments
We thank David J. Quinn for preparing the images in Fig. 1 from microfluidics experiments conducted in the laboratory of S.S. This work was supported by National Institutes of Health/National Institute of General Medical Sciences Grant 1-R01-GM076689-01.
Abbreviations
- OT
optical tweezer
- CD
cytoskeletal dynamics.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700257104/DC1.
References
- 1.Bennett V. Annu Rev Biochem. 1985;54:273–304. doi: 10.1146/annurev.bi.54.070185.001421. [DOI] [PubMed] [Google Scholar]
- 2.Mohandas N, Evans E. Annu Rev Biophys Biomol Struct. 1994;23:787–818. doi: 10.1146/annurev.bb.23.060194.004035. [DOI] [PubMed] [Google Scholar]
- 3.An XL, Lecomte MC, Chasis JA, Mohandas N, Gratzer W. J Biol Chem. 2002;277:31796–31800. doi: 10.1074/jbc.M204567200. [DOI] [PubMed] [Google Scholar]
- 4.Cloitre M, Borrega R, Leibler L. Phys Rev Lett. 2000;85:4819–4822. doi: 10.1103/PhysRevLett.85.4819. [DOI] [PubMed] [Google Scholar]
- 5.Bursac P, Lenormand G, Fabry B, Oliver M, Weitz DA, Viasnoff V, Butler JP, Fredberg JJ. Nat Mater. 2005;4:557–561. doi: 10.1038/nmat1404. [DOI] [PubMed] [Google Scholar]
- 6.Pinder JC, Bray D, Gratzer WB. Nature. 1977;270:752–754. doi: 10.1038/270752a0. [DOI] [PubMed] [Google Scholar]
- 7.Manno S, Takakuwa Y, Mohandas N. J Biol Chem. 2005;280:7581–7587. doi: 10.1074/jbc.M410650200. [DOI] [PubMed] [Google Scholar]
- 8.Weed RI, Lacelle PL, Merrill EW. J Clin Invest. 1969;48:795–809. doi: 10.1172/JCI106038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birchmeier W, Singer SJ. J Cell Biol. 1977;73:647–659. doi: 10.1083/jcb.73.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lutz HU, Liu SC, Palek J. Experientia. 1977;33:796–796. [Google Scholar]
- 11.Sackmann E. FEBS Lett. 1994;346:3–16. doi: 10.1016/0014-5793(94)00484-6. [DOI] [PubMed] [Google Scholar]
- 12.Gov NS, Safran SA. Biophys J. 2005;88:1859–1874. doi: 10.1529/biophysj.104.045328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mills JP, Qie L, Dao M, Lim CT, Suresh S. Mech Chem Biosyst. 2004;1:169–180. [PubMed] [Google Scholar]
- 14.Li J, Dao M, Lim CT, Suresh S. Biophys J. 2005;88:3707–3719. doi: 10.1529/biophysj.104.047332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Humpert C, Baumann M. Mol Membr Biol. 2003;20:155–162. doi: 10.1080/09687680307080. [DOI] [PubMed] [Google Scholar]
- 16.Canham PB. J Theor Biol. 1970;26:61–81. doi: 10.1016/s0022-5193(70)80032-7. [DOI] [PubMed] [Google Scholar]
- 17.Helfrich W. Z Naturforsch. 1973;C28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 18.Lim HWG, Wortis M, Mukhopadhyay R. Proc Natl Acad Sci USA. 2002;99:16766–16769. doi: 10.1073/pnas.202617299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcelli G, Parker KH, Winlove CP. Biophys J. 2005;89:2473–2480. doi: 10.1529/biophysj.104.056168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stillinger FH, Weber TA. Phys Rev B. 1985;31:5262–5271. doi: 10.1103/physrevb.31.5262. [DOI] [PubMed] [Google Scholar]
- 21.Takeuchi M, Miyamoto H, Sako Y, Komizu H, Kusumi A. Biophys J. 1998;74:2171–2183. doi: 10.1016/S0006-3495(98)77926-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swihart AH, Mikrut JM, Ketterson JB, MacDonald RC. J Microsc (Oxford) 2001;204:212–225. doi: 10.1046/j.1365-2818.2001.00960.x. [DOI] [PubMed] [Google Scholar]
- 23.Liu F, Mizukami H, Sarnaik S, Ostafin A. J Struct Biol. 2005;150:200–210. doi: 10.1016/j.jsb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 24.McGreevy RL. J Phys Cond Mat. 2001;13:R877–R913. [Google Scholar]
- 25.Eber S, Lux SE. Semin Hematol. 2004;41:118–141. doi: 10.1053/j.seminhematol.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Gallagher PG. Semin Hematol. 2004;41:142–164. doi: 10.1053/j.seminhematol.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Hansen JC, Skalak R, Chien S, Hoger A. Biophys J. 1996;70:146–166. doi: 10.1016/S0006-3495(96)79556-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamenovic D, Fredberg JJ, Wang N, Butler JP, Ingber DE. J Theor Biol. 1996;181:125–136. doi: 10.1006/jtbi.1996.0120. [DOI] [PubMed] [Google Scholar]
- 29.Discher DE, Boal DH, Boey SK. Biophys J. 1998;75:1584–1597. doi: 10.1016/S0006-3495(98)74076-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vera C, Skelton R, Bossens F, Sung LPA. Ann Biomed Eng. 2005;33:1387–1404. doi: 10.1007/s10439-005-4698-y. [DOI] [PubMed] [Google Scholar]
- 31.Bennett V, Baines AJ. Physiol Rev. 2001;81:1353–1392. doi: 10.1152/physrev.2001.81.3.1353. [DOI] [PubMed] [Google Scholar]
- 32.Ohanian V, Wolfe LC, John KM, Pinder JC, Lux SE, Gratzer WB. Biochemistry. 1984;23:4416–4420. doi: 10.1021/bi00314a027. [DOI] [PubMed] [Google Scholar]
- 33.Tyler JM, Reinhardt BN, Branton D. J Biol Chem. 1980;255:7034–7039. [PubMed] [Google Scholar]
- 34.Gov N, Safran SA. Phys Rev E. 2004;69 011101. [Google Scholar]
- 35.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 36.Lutsko JF. J Appl Phys. 1988;64:1152–1154. [Google Scholar]
- 37.Ermak DL, McCammon JA. J Chem Phys. 1978;69:1352–1360. [Google Scholar]
- 38.Dao M, Li J, Suresh S. Mater Sci Eng C. 2006;26:1232–1244. [Google Scholar]
- 39.Kurchan J. Nature. 2005;433:222–225. doi: 10.1038/nature03278. [DOI] [PubMed] [Google Scholar]
- 40.Howard J. Mechanics of Motor Proteins and the Cytoskeleton. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- 41.Druger SD, Nitzan A, Ratner MA. J Chem Phys. 1983;79:3133–3142. [Google Scholar]
- 42.Ungewickell E, Gratzer W. Eur J Biochem. 1978;88:379–385. doi: 10.1111/j.1432-1033.1978.tb12459.x. [DOI] [PubMed] [Google Scholar]
- 43.Fung YC. Biomechanics: Mechanical Properties of Living Tissues. New York: Springer; 1993. [Google Scholar]
- 44.Fabry B, Maksym GN, Butler JP, Glogauer M, Navajas D, Taback NA, Millet EJ, Fredberg JJ. Phys Rev E. 2003;68 041914. [Google Scholar]
- 45.Sollich P. Phys Rev E. 1998;58:738–759. [Google Scholar]
- 46.Martin G. Phys Rev B. 1984;30:1424–1436. [Google Scholar]
- 47.Fabry B, Fredberg JJ. Respir Physiol Neurol. 2003;137:109–124. doi: 10.1016/s1569-9048(03)00141-1. [DOI] [PubMed] [Google Scholar]
- 48.Li J, Van Vliet KJ, Zhu T, Yip S, Suresh S. Nature. 2002;418:307–310. doi: 10.1038/nature00865. [DOI] [PubMed] [Google Scholar]
- 49.Knowles DW, Tilley L, Mohandas N, Chasis JA. Proc Natl Acad Sci USA. 1997;94:12969–12974. doi: 10.1073/pnas.94.24.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yap B, Kamm RD. J Appl Physiol. 2005;98:1930–1939. doi: 10.1152/japplphysiol.01226.2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.