Abstract
Mycobacterium tuberculosis synthesizes specific polyketide lipids that interact with the host and are required for virulence. Using a mass spectrometric approach to simultaneously monitor hundreds of lipids, we discovered that the size and abundance of two lipid virulence factors, phthiocerol dimycocerosate (PDIM) and sulfolipid-1 (SL-1), are controlled by the availability of a common precursor, methyl malonyl CoA (MMCoA). Consistent with this view, increased levels of MMCoA led to increased abundance and mass of both PDIM and SL-1. Furthermore, perturbation of MMCoA metabolism attenuated pathogen replication in mice. Importantly, we detected increased PDIM synthesis in bacteria growing within host tissues and in bacteria grown in culture on odd-chain fatty acids. Because M. tuberculosis catabolizes host lipids to grow during infection, we propose that growth of M. tuberculosis on fatty acids in vivo leads to increased flux of MMCoA through lipid biosynthetic pathways, resulting in increased virulence lipid synthesis. Our results suggest that the shift to host lipid catabolism during infection allows for increased virulence lipid anabolism by the bacterium.
Keywords: lipid virulence factor, metabolic flux, pathogenesis, PDIM, sulfolipid-1
Mycobacterium tuberculosis, the causative agent of tuberculosis, synthesizes and secretes a wide array of biologically active polyketide lipids that interact with the host (1). Genes involved in the synthesis and export of surface-exposed lipid virulence factors such as phthiocerol dimycocerosate (PDIM) and sulfolipid-1 (SL-1) are required for bacterial growth and virulence in mice (2–5). Surface-exposed lipids provide protection against host induced damage as well as modulate the immune response to infection (6, 7).
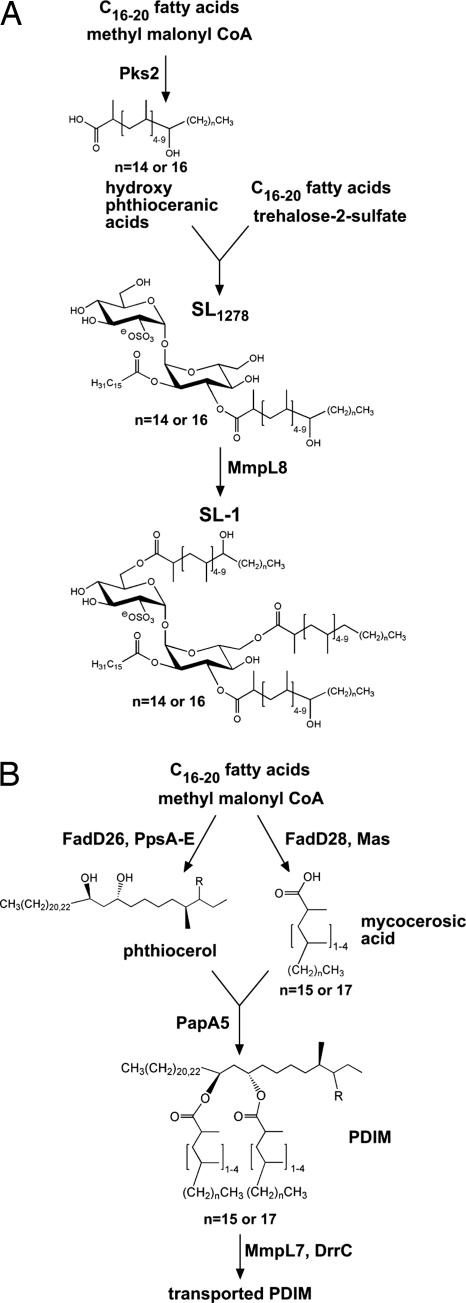
Most surface-exposed lipids are synthesized by specialized polyketide synthases that elongate straight chain fatty acids by the stepwise addition short acyl chains. PDIM is the product of two such systems (Fig. 1). Mas synthesizes methyl branched mycocerosic acids through successive additions of methyl malonyl CoA (MMCoA) (8, 9), whereas PpsA-E synthesize phthiocerol (10). PDIM is exported to the cell surface via the MmpL7 transporter (2, 11). Similarly, Pks2 incorporates methyl branches into the virulence lipid SL-1 (12). MmpL8 is required for the complete synthesis of SL-1 and a biosynthetic precursor, SL1278, accumulates in mmpL8− cells (4).
Fig. 1.
Pathways of SL-1 and PDIM biosynthesis. (A) The structure of SL1278 (4, 5) and SL-1 as proposed by Goren (33) is shown. The trehalose-2-sulfate core is esterified with two hydroxy-phthioceranic groups, one phthioceranic group and a palmitate or stearate. (B) The structures of phthiocerol, mycocerosic acids and PDIM are shown. Pps, phthiocerol synthase; Mas, mycocerosic acid synthase. PDIM consists of two mycocerosic acids esterified to phthiocerol. Mas synthesizes the methyl branched mycocerosic acids through successive additions of MMCoA (8, 9), whereas PpsA-E synthesize phthiocerol (10). FadD26 and FadD28 are responsible for activating straight chain fatty acids for transfer to Pps and Mas, respectively (34). MmpL7 and DrrC are required for transport of PDIM to the cell wall (2, 3).
Host lipids also play an important role during infection as they appear to be the primary carbon source for M. tuberculosis in vivo. Bacteria harvested from infected tissues preferentially metabolize fatty acids over carbohydrates (13) and the icl1 and icl2 genes, which are required for growth on lipids as a sole carbon source, are required for growth in vivo (14, 15). Furthermore, icl1 and icl2, along with genes involved in the β-oxidation of fatty acids, are up-regulated during M. tuberculosis infection (16–21).
In this work, we analyzed the global lipid profile of M. tuberculosis using Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR MS) (4, 22) and discovered that the size and abundance of PDIM and SL-1 are controlled by the availability of a common precursor, MMCoA, and that this regulation occurs during infection. Our results suggest that growth of M. tuberculosis on fatty acids during infection leads to increased flux of MMCoA through lipid biosynthetic pathways, resulting in increased virulence lipid synthesis.
Results
For further details, see supporting information (SI) Text, SI Figs. 5–12, and SI Table 1.
Global Analysis of M. tuberculosis Lipids.
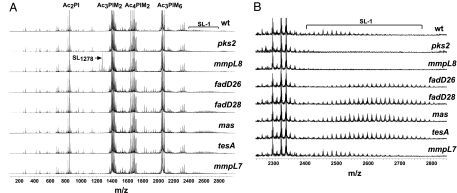
To comprehensively analyze lipid regulation in M. tuberculosis, we used FT-ICR MS to simultaneously measure the abundance of the complex array of lipid species produced by the bacterium. We directly infused total lipid extracts of M. tuberculosis cells into the ionization source of the mass spectrometer and observed reproducible spectra containing hundreds of unique lipid species in both negative mode (Fig. 2A) and positive mode (data not shown). Accurate mass measurements permitted identification of a number of complex lipid species, including phosphatidylinositol mannosides (PIMs), SL-1, SL1278 (Fig. 2A), and PDIM (Fig. 3B and SI Fig. 8).
Fig. 2.
Increased abundance and mass of SL-1 in PDIM synthesis mutants. (A) Crude lipid extracts prepared from M. tuberculosis cultures from the indicated strains were analyzed in negative ion mode by FT-ICR MS. SL-1, sulfolipid-1; PI, phosphatidyl inositol; PIM, phosphatidyl inositol mannoside; Ac, acyl chain. The phosphatidylinositol (PI) linkers esterified with two acyl chains (Ac2PI, m/z 835.5355 for PI esterified to palmitate and oleate, and m/z 851.5638 for PI esterified to palmitate and tuberculostearate) were observed within 2 ppm of their theoretical masses (24, 35). The dimannose species esterified to three acyl chains (Ac3PIM2, m/z 1413.8988) and the hexamannose species esterified to three acyl chains (Ac3PIM6, m/z 2062.1079) were also observed. Multiple lipoforms of SL-1 (m/z 2401.1544 to 2639.3480) and SL1278 (m/z 1277.9394) are also indicated. (B) The SL-1 regions of the spectra obtained in A are shown. SL-1 is observed as a broad set of peaks separated by 14 amu corresponding to differing numbers of CH2 units.
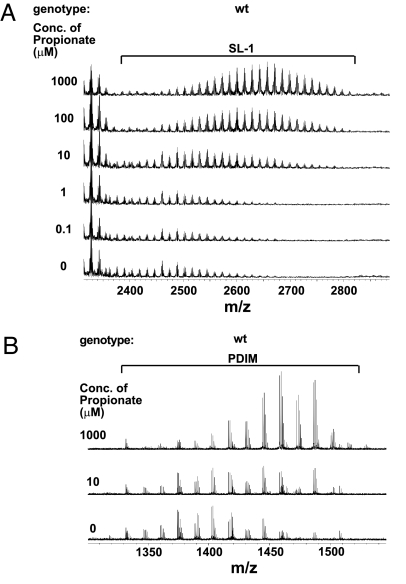
Fig. 3.
Addition of propionate leads to increased abundance and mass of SL-1 and PDIM. Increasing concentrations of propionate were added to wild-type M. tuberculosis cells over the course of three doublings and SL-1 (A) and PDIM (B) were monitored by using FT-ICR MS.
In pks2− mutant cells, which are unable to synthesize SL-1 (12), SL-1 peaks were completely absent (Fig. 2B). In mmpL8− cells, SL-1 peaks were also absent (Fig. 2B), but there was a dramatic increase in peaks at m/z 1277.9394 corresponding to SL1278 (Fig. 2A) (4). Likewise, in fadD26−, fadD28−, mas−, and tesA− cells, which are defective in the synthesis of PDIM (2), all PDIM peaks were absent (SI Fig. 8, data not shown).
Increased Production of SL-1 in PDIM Synthesis Mutants.
Surprisingly, in cells defective for PDIM synthesis, we observed greatly increased amounts of SL-1 (Fig. 2B). This increase was unexpected because PDIM synthesis mutants have been well characterized by other methods, but these differences had not been previously detected. In addition to being more abundant, SL-1 from PDIM mutants was also of higher average mass as compared with that from wild-type bacteria. The increase in abundance and average mass was specific to SL-1, with the exception of a slight increase in the abundance of several unknown species at m/z 1826.73, 1854.77, 1942.89, and 1970.91 (data not shown). We quantified SL-1 abundance and found that, on average, there was a 2- to 3-fold increase in the amount of SL-1 in PDIM synthesis mutants as compared with wild-type (SI Fig. 5A). We also calculated a weighted average mass of SL-1 and found a dramatic shift in the average mass of SL-1 of >100 amu (SI Fig. 5B). Interestingly, SL-1 production in mmpL7− cells is similar to wild type (Fig. 2B), demonstrating that SL-1 is regulated in response to inhibition of PDIM synthesis and not to cell wall perturbations due to absence of PDIM.
We hypothesized that inhibition of PDIM production may lead to transcriptional activation of SL-1 synthesis genes. To test this, we performed microarray experiments to compare the global gene expression of wild-type, fadD26− and pks2− cells (data not shown). Significance analysis of microarrays (23) on data from three repetitions of the experiment revealed that fadD26 was the only mRNA consistently reduced >3-fold in fadD26− cells as compared with wild type, and similarly, pks2 was the only transcript differentially expressed in pks2− cells. Therefore, the increased SL-1 production in PDIM synthesis mutants is likely due to posttranscriptional regulation.
Production of SL-1 and PDIM Increase with MMCoA Concentration.
Because both PDIM and SL-1 biosynthetic pathways share a common precursor, MMCoA (12, 24), a simple model is that in the absence of PDIM synthesis, greater amounts of MMCoA are available to the SL-1 biosynthetic pathway, leading to increased SL-1 production. To test this, we added increasing concentrations of a precursor of MMCoA, propionate, to wild-type cells (over the course of three doublings) and found that they responded by making 2- to 5-fold more SL-1 that was of higher average mass by >100 amu (Fig. 3A and SI Fig. 6 B and C). Likewise, propionate addition to mmpL8− cells, led to a 2-fold increase in the amount of SL1278 and an increase of ≈24 amu in the average mass, consistent with the SL-1 result (SI Fig. 7).
We also added propionate to the mas mutant to test whether we could further increase the already elevated SL-1 production in these cells. Although there was no significant increase in SL-1 abundance, there was an increase of >50 amu in the average mass of SL-1 at higher concentrations of propionate (SI Fig. 6A). These results indicate that MMCoA is limiting for SL-1 synthesis during growth in culture and its availability regulates production of SL-1.
We also observed an increase in both abundance and average mass of PDIM upon addition of propionate (Fig. 3B and SI Fig. 8). Interestingly, pks2− and mmpL8− cells do not make significantly more PDIM than wild-type (SI Fig. 8). Because SL-1 is much less abundant than PDIM (unpublished observations), it is possible that inhibition of SL-1 synthesis does not alter the concentration of MMCoA enough to affect PDIM synthesis. Taken together, our results indicate that production of PDIM and SL-1 are coupled via the metabolic flux of a common precursor, MMCoA, and that increased flux of MMCoA through both pathways leads to an increase in PDIM and SL-1 production.
Increased Average Mass of PDIM Is Due To Increased Chain-Length of Mycocerosic Acids.
To determine the differences between low and high mass forms of PDIM, we performed collision-induced dissociation on the different PDIM ions. We found that although the mass of mycocerosic acids increased with higher mass forms of PDIM, the mass of the phthiocerol moiety remained constant (SI Fig. 9). The 1402 and 1486 forms of PDIM differ by a mass of 42 amu in each of their mycocerosic acids, which corresponds exactly to the addition of a single propionate molecule. Thus, it is likely that the mycocerosic acids in the 1486 form have an additional methyl branch as compared with the 1402 form due to increased incorporation of MMCoA during their synthesis (25). These data suggest that, although the Pps enzymes are precise in the number of functional groups they incorporate, Mas is more flexible and is able to polymerize a variable number of methyl branches as a function of intracellular MMCoA concentration.
Perturbations in MMCoA Regulation Lead to Altered PDIM and SL-1 Production and Attentuation in Vivo.
We also tested whether altering endogenous MMCoA production in vivo would lead to changes in PDIM and SL-1 production. We overexpressed MMCoA mutase, which catalyzes the conversion of the citric acid cycle intermediate succinyl CoA to MMCoA (26), by introducing an episomal plasmid carrying the M. tuberculosis mutAB genes into wild-type bacilli (Fig. 4A). This resulted in a marked increase in the average mass of SL-1 and PDIM but, in contrast to the addition of propionate, led to a decrease in their total abundance (reduction of 30% for PDIM and 80% for SL-1, Fig. 4B and SI Fig. 10). Although the reason for this is unclear, it is possible that chronic overexpression of MutAB depletes citric acid cycle intermediates, leading to pleiotropic effects on cell metabolism. In the case of propionate addition, propionate is most likely converted directly from propionyl CoA to MMCoA via a propionyl CoA carboxylase. Several propionyl CoA carboxylase homologs exist in M. tuberculosis and one of these has recently been characterized (27). PDIM and SL-1 production are specifically affected while the rest of the lipid mass-spectrum observed in MutAB overexpressing cells was similar to wild-type (data not shown). MutAB overexpressing cells also doubled with same kinetics in culture as wild-type cells carrying the empty vector (doubling time of 25 h), showing that MutAB overexpression did not affect cell growth and viability.
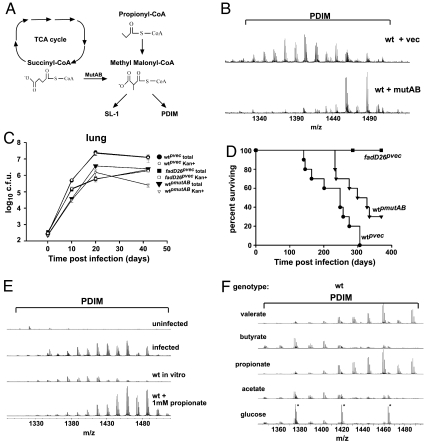
Fig. 4.
Regulation of PDIM and SL-1 synthesis during infection. (A) Pathway depicting the production and incorporation of MMCoA into PDIM and SL-1. MMCoA can be synthesized from propionyl CoA or from succinyl CoA via the action of MutAB. (B) PDIM region of FT-ICR mass spectra of crude extracts from wild-type M. tuberculosis containing either the MutAB expression construct or an empty vector as a control. (C and D) Overexpression of MutAB leads to attenuated growth in a mouse model of infection. BALB/C mice were infected via the aerosol route with bacteria containing either an empty vector (circles) or the MutAB expression construct (inverted triangles), or fadD26− bacteria containing an empty vector (squares). The data shown here is from one representative experiment of two. (C) Growth in the lung was monitored by harvesting at the indicated timepoints and plating dilutions on solid media containing kanamycin (open symbols) or without antibiotic (filled symbols). Growth on media without kanamycin revealed the total number of bacteria, whereas growth on media containing kanamycin revealed the number of bacteria that retained the plasmid. Each data point represents the average colony-forming units (cfu) from four to five infected mice, and error bars indicate the standard deviation of the means. (D) Survival of mice (n = 10 per group). (E) PDIM is produced in higher mass forms in a mouse model of M. tuberculosis infection. BALB/C mice were infected with a high dose (≈1,400 cfu) of wild-type M. tuberculosis, and lungs were harvested 19 days after infection. Bacillary loads in the lung at this timepoint were 8 × 108 colony-forming units (cfu). Lipids were extracted from lungs and analyzed by FT-ICR MS. The data shown here are from one representative experiment of two. FT-ICR mass spectra from wild-type M. tuberculosis cells grown in vitro, with or without propionate, as described in Fig. 3B are also shown here for comparison. Average mass of PDIM in vivo is 1,430 amu as compared with 1,403 amu for cells grown in vitro without propionate and 1,456 amu for cells grown in the presence of 1 mM propionate. (F) PDIM region of FT-ICR mass spectra of crude extracts from wild-type cells grown on 0.1% glucose, or 10 mM acetate, propionate, butyrate or valerate as a sole carbon cource. Three contaminating peaks of polyethylene glycol from residual Tween-80 in the sample are marked with an asterisk in the spectra for cells grown on glucose.
We assessed virulence of the MutAB overexpressing strain by infecting BALB/C mice via the aerosol route. We found that, during infection, bacterial growth of the MutAB overexpressing strain is significantly attenuated (≈10-fold) compared with wild-type cells by 3 weeks (P < 0.0001), similar to what we observed for fadD26− cells (Fig. 4C and SI Fig. 11). We also found that, although there was no plasmid loss in the wild-type or fadD26− cells carrying the empty vector, there was 90% plasmid loss in the MutAB overexpressing strain in mouse lungs by 6 weeks, demonstrating a strong selective advantage for cells that had lost the plasmid (Fig. 4C). Furthermore, mice infected with the MutAB overexpressing strain survived significantly longer than those infected with wild-type (Fig. 4D, P < 0.02). These data clearly show that the strain overexpressing MutAB is attenuated relative to wild-type cells and that proper regulation of MMCoA in the bacterial cell is critical for virulence.
PDIM Is Produced in Higher Mass Forms During Infection.
Because M. tuberculosis catabolizes fatty acids during infection (28), a consequent increase in the concentration of citric acid cycle intermediates or an increase in odd-chain fatty acid metabolism could lead to increased production of MMCoA in vivo leading to increased virulence factor production during infection. To test this possibility, we analyzed lipids extracted from M. tuberculosis infected lungs during infection. Although SL-1 levels were below the limit of detection, PDIM was readily observed by FT-ICR MS (Fig. 4E). Importantly, we found that M. tuberculosis produced high mass forms of PDIM in vivo, with an average mass of 1,430 amu compared with 1,403 amu for PDIM extracted from in vitro grown cells. The increase in mass was similar to that observed from wild-type cultures grown in the presence of propionate (Fig. 4E), suggesting that this regulation is due to increased concentration of MMCoA in vivo.
Increased Synthesis of PDIM and SL-1 During Growth on Odd-Chain Fatty Acids.
The increased mass of PDIM during infection suggests a link between anabolism of virulence polyketides and catabolism of host lipids. Therefore, we grew M. tuberculosis in culture on different short-chain fatty acids as a sole carbon source and monitored lipid production. There was no increase in PDIM or SL-1 production upon growth on acetate and the even-chain fatty acid butyrate as compared with glucose, but we observed a significant increase during growth on propionate and the odd-chain fatty acid valerate (Fig. 4F and SI Fig. 12). These data suggest that the increased mass of PDIM observed in vivo during infection was due to released propionyl CoA from β-oxidation of odd-chain fatty acids.
Discussion
FT-ICR MS Methodology.
In these studies, we used FT-ICR MS to monitor the lipid profile of M. tuberculosis quantitatively. This method provided a broad view of the lipid profile and allowed us to monitor hundreds of species simultaneously, aiding in the identification of cellular lipid changes in mutants. FT-ICR MS also offered a highly sensitive way of analyzing lipid virulence factors under different conditions and allowed us to discriminate between species with varying lengths of lipid chains. Ion trapping and subsequent collision-induced dissociation also allowed us to elucidate more detailed structural information about the molecules in question. In this way, we were able to determine that increased synthesis of PDIM under high MMCoA conditions was due exclusively to increased length of the mycocerosic acids, whereas the phthiocerol portion of the lipid was insensitive to precursor concentration. These results suggest that the Pps enzymes are precise over the range of MMCoA concentrations inside the cell but the processivity of Mas is influenced by substrate availability. This flexibility was also apparent for Pks2 during SL-1 synthesis.
Metabolic Coupling of Virulence Lipid Synthesis.
We have gained insight into the compensatory cellular lipid changes that occur upon genetic perturbation. Our experiments show that interconnections between seemingly disparate pathways can complicate standard genetic analysis of lipid biosynthetic mutants. Using FT-ICR MS, we made the unexpected discovery that the production of two lipid virulence factors of M. tuberculosis, PDIM and SL-1, are coupled via the metabolic flux of a common precursor, MMCoA. We suggest that, in the absence of PDIM synthesis, there is increased concentration of the MMCoA precursor, which is used in the SL-1 biosynthetic pathway, leading to the observed increase in abundance and mass of SL-1. It is important to note that mmpL7− cells have a comparable virulence phenotype to PDIM biosynthetic mutants (2), indicating that SL-1 up-regulation cannot compensate for the loss of PDIM during infection. Exogenous addition of a direct precursor of MMCoA, propionate, to wild-type cells results in increased abundance and mass of both PDIM and SL-1. This finding further supports the notion that increased flux of MMCoA through both pathways leads to an increase in PDIM and SL-1 production.
Although both mmpL8− and pks2− cells do not make SL-1, mmpL8− cells have attenuated growth in vivo, whereas pks2− mutants do not (4). It is possible that compensatory regulation exists in pks2− mutants that is not present in mmpL8− cells. Although there are no significant changes in PDIM in pks2− cells, we observed an increase in abundance of unknown lipid species at m/z 1826.73, 1854.77, 1942.89, and 1970.91 under conditions of high propionate as well as in pks2− cells and PDIM synthesis mutants but not in mmpL8− cells. These are likely to be methyl-branched lipids that, perhaps, play a role in virulence.
Regulation of Virulence Lipid Synthesis During Infection.
FT-ICR MS has allowed us to directly analyze M. tuberculosis lipids from infected tissue. The increased mass of PDIM recovered from infected organs suggests that high levels of MMCoA exist during infection. Although we cannot rule out the possibility that Mas has increased processivity in vivo due to other reasons, the simplest model is that high levels of MMCoA lead to increased flux of this metabolite through polyketide biosynthetic pathways, resulting in greater production of pathogenic lipids by M. tuberculosis. Increased levels of MMCoA could be due to either increased availability of propionate in animal tissues, up-regulation of MMCoA production by the bacteria during growth on fatty acids, or increased propionyl CoA due to β-oxidation of odd-chain fatty acids by the bacteria. Although all three may happen during infection, our data suggest that the last likely occurs as PDIM and SL-1 production was stimulated only during growth on valerate, but not on butyrate (Fig. 4F). Thus, we propose that virulence polyketide anabolism is directly regulated by the metabolic shift to growth on fatty acids, including odd-chain fatty acids, during infection.
Propionyl CoA released from the catabolism of odd chain fatty acids can also be assimilated into the citric acid cycle via the methyl citrate cycle. A recent study showed that this pathway is dispensable for M. tuberculosis virulence in mice and proposed that methyl-branched polyketide production may act as a sink to buffer the increased concentration of propionyl CoA in the absence of this pathway in vivo (29). Our results are consistent with this idea. This finding raises the interesting possibility that additional control mechanisms exist to regulate the flux of propionyl CoA through catabolic or anabolic pathways during M. tuberculosis infection.
Materials and Methods
Strains and Plasmids.
M. tuberculosis cells (Erdman strain) were cultured in 7H9 medium supplemented with 10% OADC, 0.5% glycerol, and 0.05% Tween-80, or on 7H10 solid agar medium with the same supplements plus 50 mg liter−1 cycloheximide and without Tween-80. Kanamycin (20 μg ml−1) was used where necessary. All strains and plasmids used in this study are described in SI Table 1.
Biochemical Extraction of Lipids.
Cells were synchronized to an OD600 of 0.2 and grown to an OD600 of 0.8. Ten milliliters of cells were harvested by centrifugation, and total lipids were extracted by the Bligh–Dyer method (30). For samples analyzed in positive mode, Tween-80 was removed by resuspending extracts in a 1:1 mixture of hexanes and water, mixing thoroughly, and centrifuging. The organic layer was extracted with water five times.
Addition of Propionate to Cultures.
Cultures of M. tuberculosis were synchronized to an OD600 of 0.1 and propionic acid (P1386; Sigma, St. Louis, MO) was added at the indicated concentration daily over the course of three doublings until the cells reached an OD600 of 0.8. Ten milliliters of culture was used to extract lipids as described above. Cells doubled with the same kinetics independent of the propionate concentration used.
Growth of M. tuberculosis on Fatty Acids.
Cultures of M. tuberculosis were synchronized to an OD600 of 0.1 and resuspended in media containing either 0.1% glucose or 10 mM acetate, propionate, butyrate or valeric acid as the sole carbon source, along with 0.47% 7H9, 0.5% albumin, 0.085% NaCl, and 0.05% Tween-80, as described (29). Cells doubled with the same kinetics independent of the media used and 20 ml of culture was harvested at an OD600 of 0.4 for lipid extraction.
Extraction of Bacterial Lipids from Mouse Lung Tissue.
Lungs from infected mice were homogenized in PBS and pelleted by centrifugation. Host lipids were extracted by washing the homogenates three times with 5 ml methanol. The remaining material was extracted with 4 ml 1:1 chloroform/methanol solution and agitated for 8 h. The organic extracts were clarified by centrifugation and analyzed by FT-ICR MS.
FT-ICR MS Methods.
Total lipids extracted from cells as described above, were resuspended in 2:1 chloroform/methanol solution and introduced into the mass spectrometer by means of an Apollo (Bruker-Daltonics, Billerica, MA) pneumatically assisted electrospray source operating in positive or negative ion mode. Mass spectra were acquired on a Bruker-Daltonics Apex II FT-ICR mass spectrometer (31) and experiments for structural analysis of lipid components were performed as described previously (22). For details see SI Text. Total abundance of lipid species was calculated by summing the peak intensities as measured by FT-ICR and reported by Xmass. Average mass was calculated as a weighted average using peak intensities as weights.
Microarray Analysis.
RNA was harvested from log phase grown cells as described (32) and reverse transcribed to generate cDNA. Dye-conjugated cDNA samples were competitively hybridized on microarrays containing oligonucleotide spots representing every gene in M. tuberculosis (Qiagen, Valencia, CA). See SI Text for details. Significance Analysis of Microarray was used to detect statistically significant differences in gene transcription between strains (23).
Mouse Infections.
Bacteria were grown to log phase, cup-sonicated by using a Branson Sonifier 250 at 90% for 15 s, spun for 5 min at 40 × g to remove clumps, and diluted to the desired inoculum in PBS. Bacteria were administered to BALB/C mice via nebulization for 15 min using a custom built aerosolization chamber (Mechanical Engineering Shops, University of Wisconsin, Madison). For normal dose infections (Fig. 4 C and D and SI Fig. 11), an OD600 of 0.1 was used, which resulted in an initial seeding of ≈250 bacteria per mouse. For high-dose infections with the wild-type Erdman strain (Fig. 4E), an OD600 of 0.8 was used, resulting in an initial seeding of ≈1,400 bacteria per mouse and an average of 8 × 108 cfu in the lungs at 19 days. BALB/C mice thus infected had severely reduced longevity to ≈3 weeks. Organs from infected mice were homogenized and plated for cfu as described (2). Four to five mice were used per timepoint. Survival times (Fig. 4D) were assessed by euthanizing mice upon 15% weight loss. All mice were housed and treated humanely using procedures described in an animal care protocol approved by University of California, San Francisco, Institutional Animal Care and Use Committee. Statistical analysis was performed on cfu data using the nonparametric Mann–Whitney test and on survival data using a log-rank test.
Supplementary Material
Acknowledgments
We thank H. Madhani and all members of the J.S.C. laboratory for critical reading of the manuscript, all members of the A. Sil and J.S.C. laboratories for helpful discussions, S. Raghavan and S. Stanley for assistance with mouse experiments, and J. McKinney for advice on aerosol infections. M.J. is supported by a Howard Hughes Medical Institute Predoctoral Fellowship. J.S.C. gratefully acknowledges the support of the Pew Scholars Program in the Biomedical Sciences, the Sandler Family Supporting Foundation, and the W.M. Keck Foundation. This work was supported by National Institutes of Health Grant AI68540.
Abbreviations
- PDIM
phthiocerol dimycocerosate
- SL-1
sulfolipid-1
- FT-ICR MS
Fourier transform ion cyclotron resonance mass spectrometry
- PIM
phosphatidylinositol mannoside
- cfu
colony-forming units
- MMCoA
methyl malonyl CoA.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610634104/DC1.
References
- 1.Brennan PJ, Nikaido H. Annu Rev Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 2.Cox JS, Chen B, McNeil M, Jacobs WR., Jr Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 3.Camacho LR, Ensergueix D, Perez E, Gicquel B, Guilhot C. Mol Microbiol. 1999;34:257–267. doi: 10.1046/j.1365-2958.1999.01593.x. [DOI] [PubMed] [Google Scholar]
- 4.Converse SE, Mougous JD, Leavell MD, Leary JA, Bertozzi CR, Cox JS. Proc Natl Acad Sci USA. 2003;100:6121–6126. doi: 10.1073/pnas.1030024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Domenech P, Reed MB, Dowd CS, Manca C, Kaplan G, Barry CE., III J Biol Chem. 2004;279:21257–21265. doi: 10.1074/jbc.M400324200. [DOI] [PubMed] [Google Scholar]
- 6.Rousseau C, Winter N, Pivert E, Bordat Y, Neyrolles O, Ave P, Huerre M, Gicquel B, Jackson M. Cell Microbiol. 2004;6:277–287. doi: 10.1046/j.1462-5822.2004.00368.x. [DOI] [PubMed] [Google Scholar]
- 7.Reed MB, Domenech P, Manca C, Su H, Barczak AK, Kreiswirth BN, Kaplan G, Barry CE., III Nature. 2004;431:84–87. doi: 10.1038/nature02837. [DOI] [PubMed] [Google Scholar]
- 8.Azad AK, Sirakova TD, Rogers LM, Kolattukudy PE. Proc Natl Acad Sci USA. 1996;93:4787–4792. doi: 10.1073/pnas.93.10.4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trivedi OA, Arora P, Vats A, Ansari MZ, Tickoo R, Sridharan V, Mohanty D, Gokhale RS. Mol Cell. 2005;17:631–643. doi: 10.1016/j.molcel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 10.Azad AK, Sirakova TD, Fernandes ND, Kolattukudy PE. J Biol Chem. 1997;272:16741–16745. doi: 10.1074/jbc.272.27.16741. [DOI] [PubMed] [Google Scholar]
- 11.Jain M, Cox JS. PLoS Pathog. 2005;1:e2. doi: 10.1371/journal.ppat.0010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sirakova TD, Thirumala AK, Dubey VS, Sprecher H, Kolattukudy PE. J Biol Chem. 2001;276:16833–9. doi: 10.1074/jbc.M011468200. [DOI] [PubMed] [Google Scholar]
- 13.Segal W, Bloch H. J Bacteriol. 1956;72:132–141. doi: 10.1128/jb.72.2.132-141.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKinney JD, Honer zu Bentrup K, Munoz-Elias EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs WR, Jr, Russell DG. Nature. 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 15.Munoz-Elias EJ, McKinney JD. Nat Med. 2005;11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Timm J, Post FA, Bekker LG, Walther GB, Wainwright HC, Manganelli R, Chan WT, Tsenova L, Gold B, Smith I, et al. Proc Natl Acad Sci USA. 2003;100:14321–14326. doi: 10.1073/pnas.2436197100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honer Zu Bentrup K, Miczak A, Swenson DL, Russell DG. J Bacteriol. 1999;181:7161–7167. doi: 10.1128/jb.181.23.7161-7167.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sturgill-Koszycki S, Haddix PL, Russell DG. Electrophoresis. 1997;18:2558–2565. doi: 10.1002/elps.1150181411. [DOI] [PubMed] [Google Scholar]
- 20.Graham JE, Clark-Curtiss JE. Proc Natl Acad Sci USA. 1999;96:11554–11559. doi: 10.1073/pnas.96.20.11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubnau E, Chan J, Mohan VP, Smith I. Infect Immun. 2005;73:3754–3757. doi: 10.1128/IAI.73.6.3754-3757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mougous JD, Leavell MD, Senaratne RH, Leigh CD, Williams SJ, Riley LW, Leary JA, Bertozzi CR. Proc Natl Acad Sci USA. 2002;99:17037–17042. doi: 10.1073/pnas.252514899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tusher VG, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolattukudy PE, Fernandes ND, Azad AK, Fitzmaurice AM, Sirakova TD. Mol Microbiol. 1997;24:263–270. doi: 10.1046/j.1365-2958.1997.3361705.x. [DOI] [PubMed] [Google Scholar]
- 25.Rainwater DL, Kolattukudy PE. J Biol Chem. 1985;260:616–623. [PubMed] [Google Scholar]
- 26.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin TW, Melgar MM, Kurth D, Swamidass SJ, Purdon J, Tseng T, Gago G, Baldi P, Gramajo H, Tsai SC. Proc Natl Acad Sci USA. 2006;103:3072–3077. doi: 10.1073/pnas.0510580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munoz-Elias EJ, McKinney JD. Cell Microbiol. 2006;8:10–22. doi: 10.1111/j.1462-5822.2005.00648.x. [DOI] [PubMed] [Google Scholar]
- 29.Munoz-Elias EJ, Upton AM, Cherian J, McKinney JD. Mol Microbiol. 2006;60:1109–1122. doi: 10.1111/j.1365-2958.2006.05155.x. [DOI] [PubMed] [Google Scholar]
- 30.Bligh EG, Dyer WJ. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 31.Cancilla MT, Gaucher SP, Desaire H, Leary JA. Anal Chem. 2000;72:2901–2907. doi: 10.1021/ac991223e. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. Infect Immun. 2002;70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goren MB. Biochim Biophys Acta. 1970;210:127–138. doi: 10.1016/0005-2760(70)90068-8. [DOI] [PubMed] [Google Scholar]
- 34.Trivedi OA, Arora P, Sridharan V, Tickoo R, Mohanty D, Gokhale RS. Nature. 2004;428:441–445. doi: 10.1038/nature02384. [DOI] [PubMed] [Google Scholar]
- 35.Nigou J, Gilleron M, Puzo G. Biochimie. 2003;85:153–166. doi: 10.1016/s0300-9084(03)00048-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.