Abstract
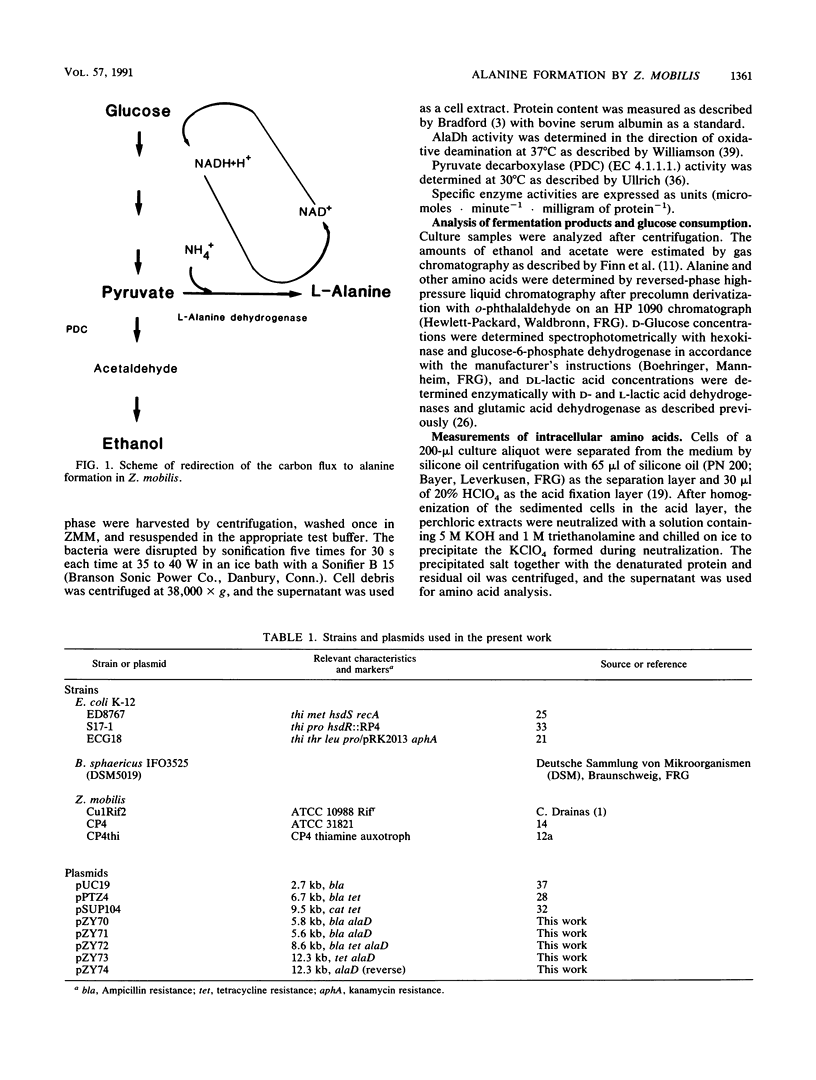
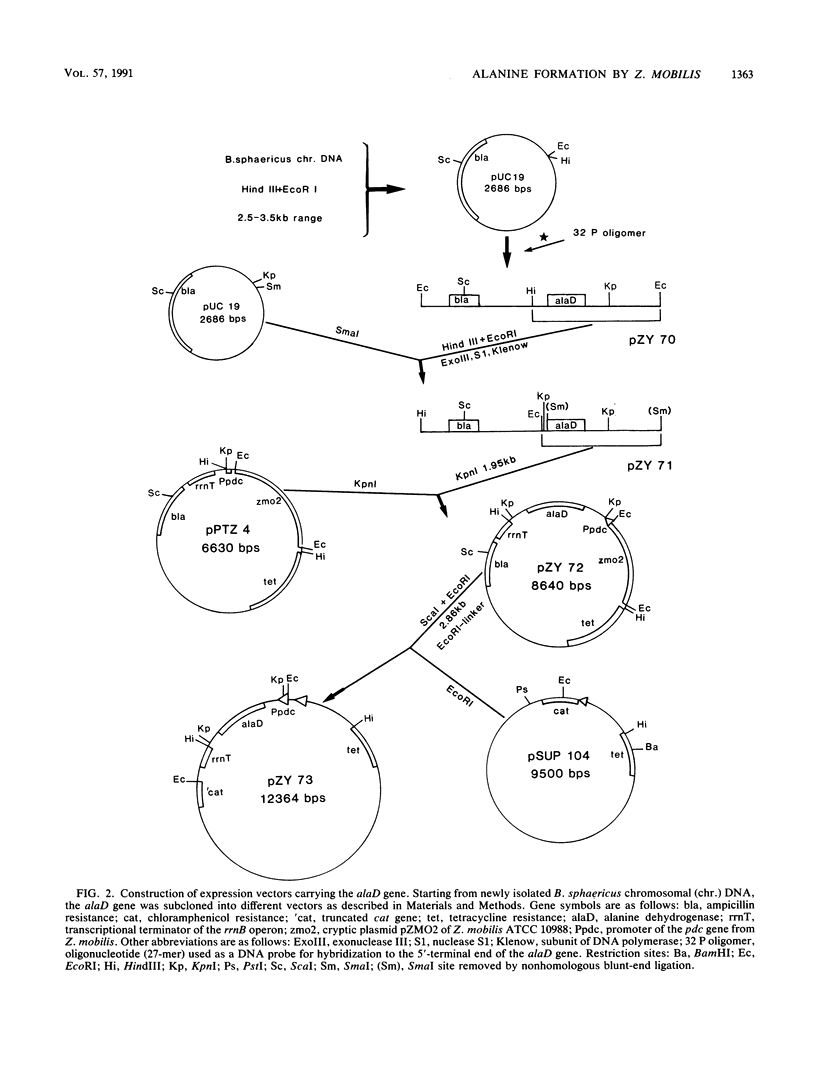
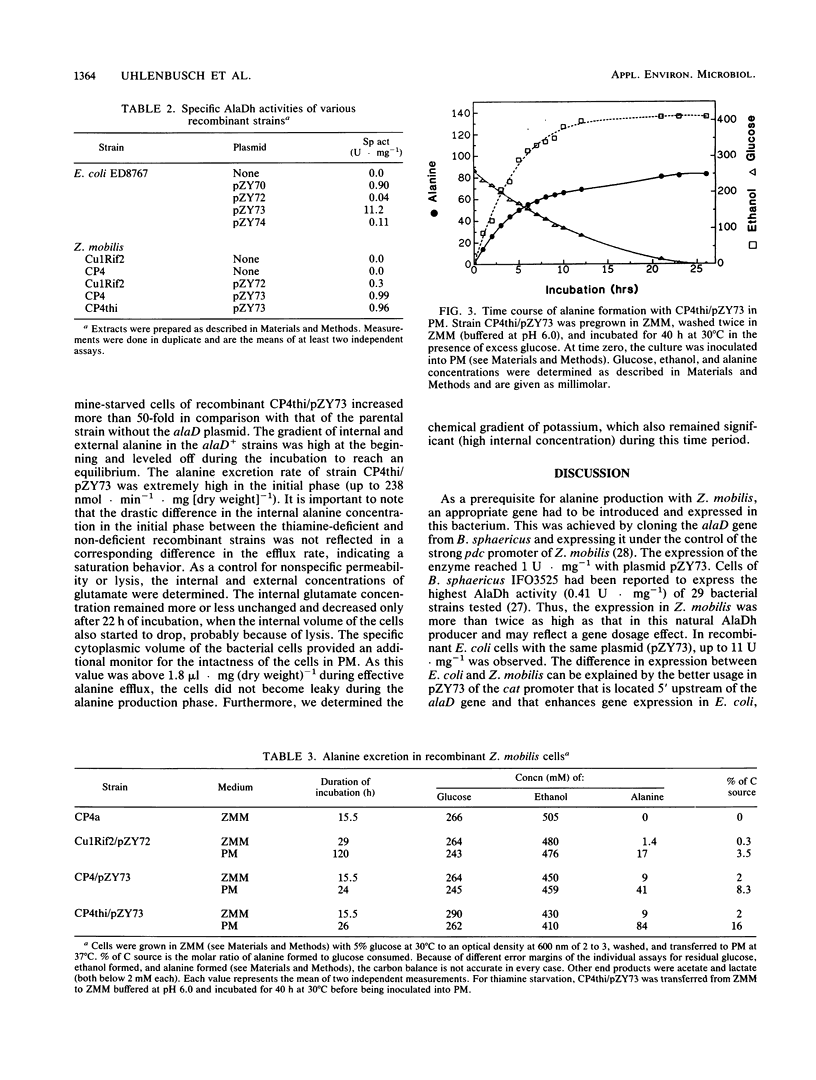
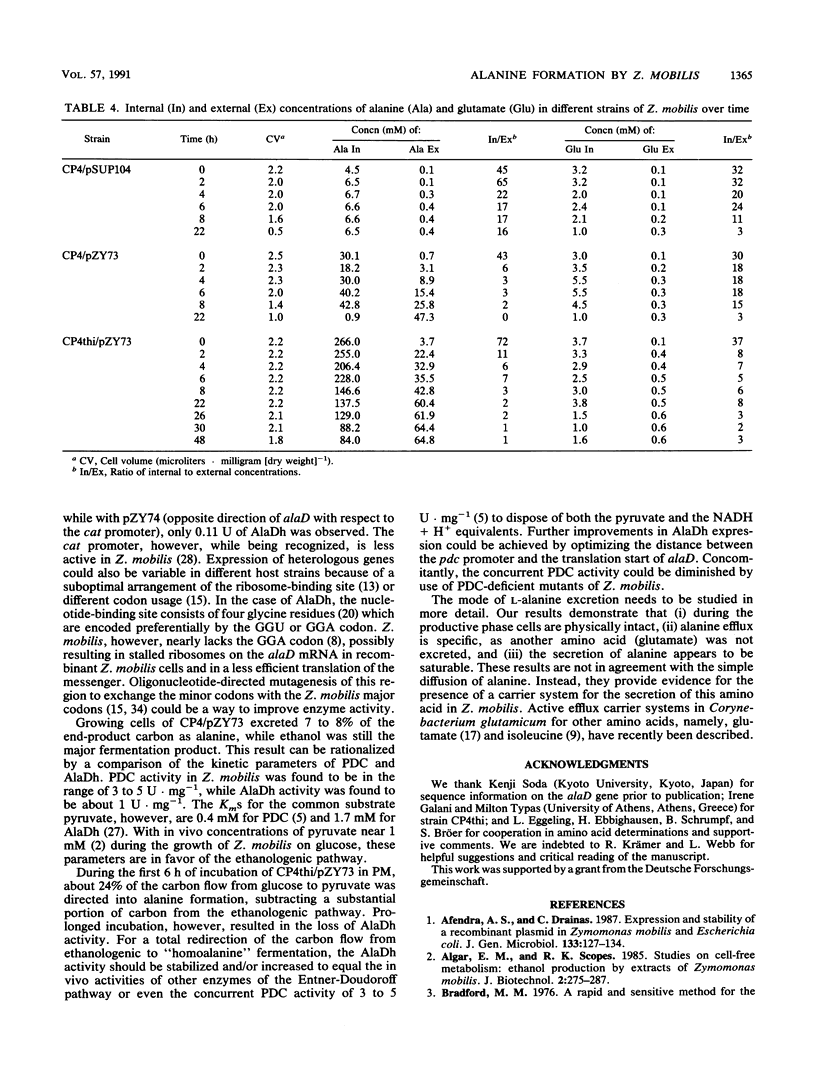
An approach to broaden the product range of the ethanologenic, gram-negative bacterium Zymomonas mobilis by means of genetic engineering is presented. Gene alaD for L-alanine dehydrogenase (EC 1.4.1.1.) from Bacillus sphaericus was cloned and introduced into Z. mobilis. Under the control of the strong promoter of the pyruvate decarboxylase (pdc) gene, the enzyme was expressed up to a specific activity of nearly 1 mu mol . min -1 . mg of protein -1 in recombinant cells. As a results of this high L-alanine dehydrogenase activity, growing cells excreted up to 10 mmol of alanine per 280 mmol of glucose utilized into a mineral salts medium. By the addition of 85 mM NH4+ to the medium, growth of the recombinant cells stopped, and up to 41 mmol alanine was secreted. As alanine dehydrogenase competed with pyruvate decarboxylase (PDC) (EC 4.1.1.1.) for the same substrate (pyruvate), PDC activity was reduced by starvation for the essential PDC cofactor thiamine PPi. A thiamine auxotrophy mutant of Z. mobilis which carried the alaD gene was starved for 40 h in glucose-supplemented mineral salts medium and then shifted to mineral salts medium with 85 mM NH4+ and 280 mmol of glucose. The recombinants excreted up to 84 mmol of alanine (7.5 g/liter) over 25 h. Alanine excretion proceeded at an initial velocity of 238 nmol . min-1 . mg [dry weight]-1. Despite this high activity, the excretion rate seemed to be a limiting factor, as the intracellular concentration of alanine was as high as 260 mM at the beginning of the excretion phase and decreased to 80 to 90 mM over 24 h.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Afendra A. S., Drainas C. Expression and stability of a recombinant plasmid in Zymomonas mobilis and Escherichia coli. J Gen Microbiol. 1987 Jan;133(1):127–134. doi: 10.1099/00221287-133-1-127. [DOI] [PubMed] [Google Scholar]
- BECKER E. L. Inhibition of complement activity by di-isopropyl fluorophosphate. Nature. 1955 Dec 3;176(4492):1073–1073. doi: 10.1038/1761073a0. [DOI] [PubMed] [Google Scholar]
- Conway T., Byun M. O., Ingram L. O. Expression Vector for Zymomonas mobilis. Appl Environ Microbiol. 1987 Feb;53(2):235–241. doi: 10.1128/aem.53.2.235-241.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Sewell G. W., Ingram L. O. Glyceraldehyde-3-phosphate dehydrogenase gene from Zymomonas mobilis: cloning, sequencing, and identification of promoter region. J Bacteriol. 1987 Dec;169(12):5653–5662. doi: 10.1128/jb.169.12.5653-5662.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FREESE E., PARK S. W., CASHEL M. THE DEVELOPMENTAL SIGNIFICANCE OF ALANINE DEHYDROGENASE IN BACILLUS SUBTILIS. Proc Natl Acad Sci U S A. 1964 Jun;51:1164–1172. doi: 10.1073/pnas.51.6.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Kuroda S., Tanizawa K., Sakamoto Y., Tanaka H., Soda K. Alanine dehydrogenases from two Bacillus species with distinct thermostabilities: molecular cloning, DNA and protein sequence determination, and structural comparison with other NAD(P)(+)-dependent dehydrogenases. Biochemistry. 1990 Jan 30;29(4):1009–1015. doi: 10.1021/bi00456a025. [DOI] [PubMed] [Google Scholar]
- Ludwig R. A. Gene tandem-mediated selection of coliphage lambda-receptive Agrobacterium, Pseudomonas, and Rhizobium strains. Proc Natl Acad Sci U S A. 1987 May;84(10):3334–3338. doi: 10.1073/pnas.84.10.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie K. F., Conway T., Aldrich H. C., Ingram L. O. Expression of Zymomonas mobilis adhB (encoding alcohol dehydrogenase II) and adhB-lacZ operon fusions in recombinant Z. mobilis. J Bacteriol. 1989 Sep;171(9):4577–4582. doi: 10.1128/jb.171.9.4577-4582.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCowen S. M., Phibbs P. V., Jr Regulation of alanine dehydrogenase in Bacillus (licheniformis). J Bacteriol. 1974 May;118(2):590–597. doi: 10.1128/jb.118.2.590-597.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. E., Brammar W. J., Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977 Jan 7;150(1):53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- Ohashima T., Soda K. Purification and properties of alanine dehydrogenase from Bacillus sphaericus. Eur J Biochem. 1979 Oct;100(1):29–30. doi: 10.1111/j.1432-1033.1979.tb02030.x. [DOI] [PubMed] [Google Scholar]
- Reynen M., Reipen I., Sahm H., Sprenger G. A. Construction of expression vectors for the gram-negative bacterium Zymomonas mobilis. Mol Gen Genet. 1990 Sep;223(2):335–341. doi: 10.1007/BF00265073. [DOI] [PubMed] [Google Scholar]
- Rottenberg H. The measurement of membrane potential and deltapH in cells, organelles, and vesicles. Methods Enzymol. 1979;55:547–569. doi: 10.1016/0076-6879(79)55066-6. [DOI] [PubMed] [Google Scholar]
- Swings J., De Ley J. The biology of Zymomonas. Bacteriol Rev. 1977 Mar;41(1):1–46. doi: 10.1128/br.41.1.1-46.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen M. A., Kurland C. G., Pedersen S. Codon usage determines translation rate in Escherichia coli. J Mol Biol. 1989 May 20;207(2):365–377. doi: 10.1016/0022-2836(89)90260-x. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]


