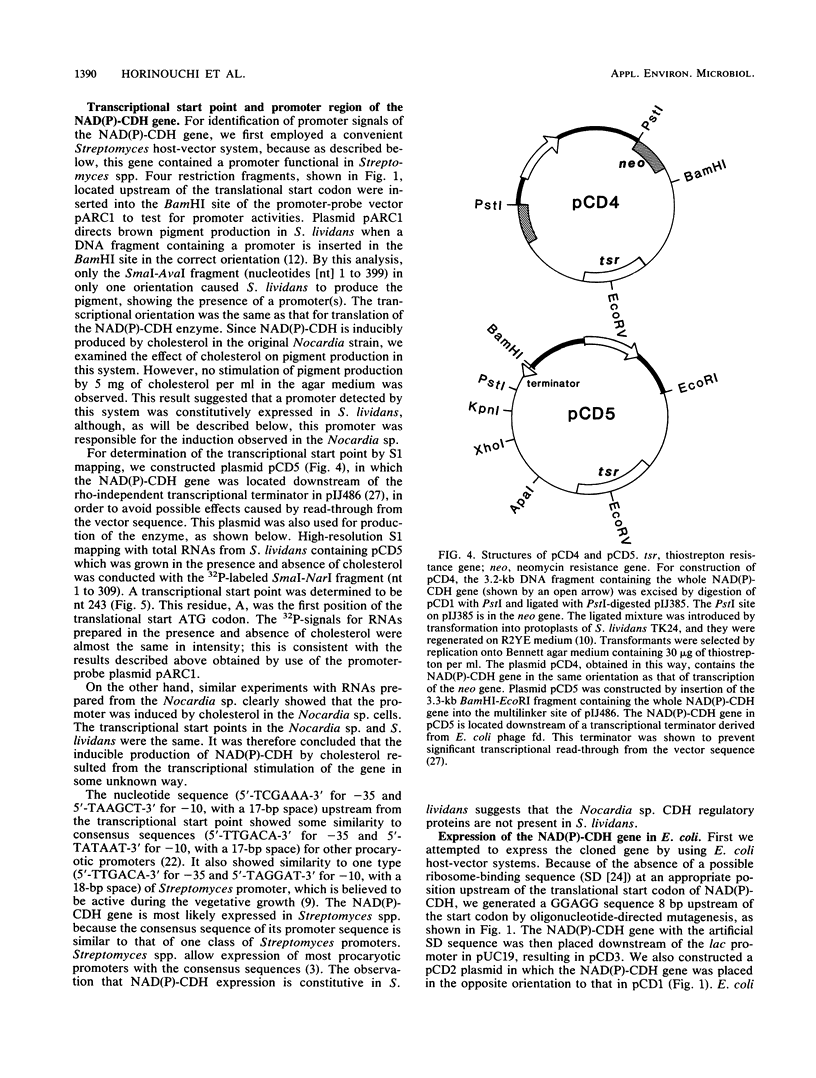
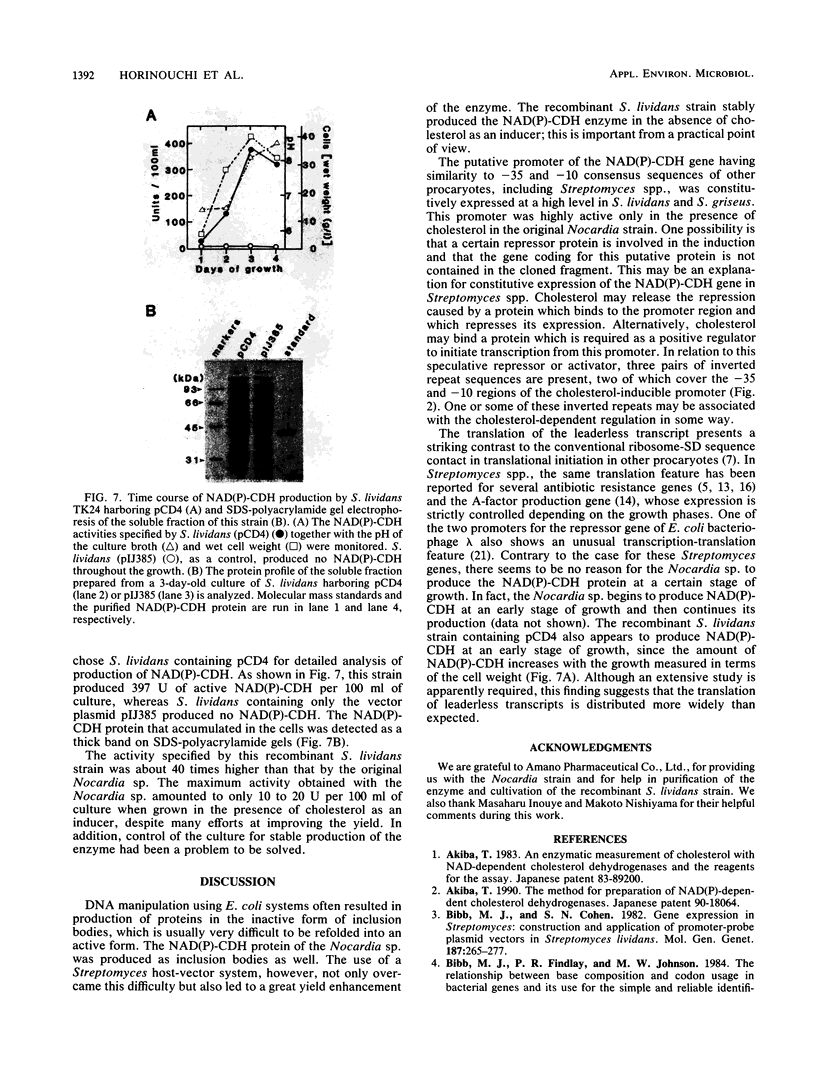
Abstract
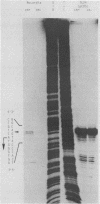
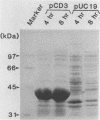
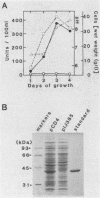
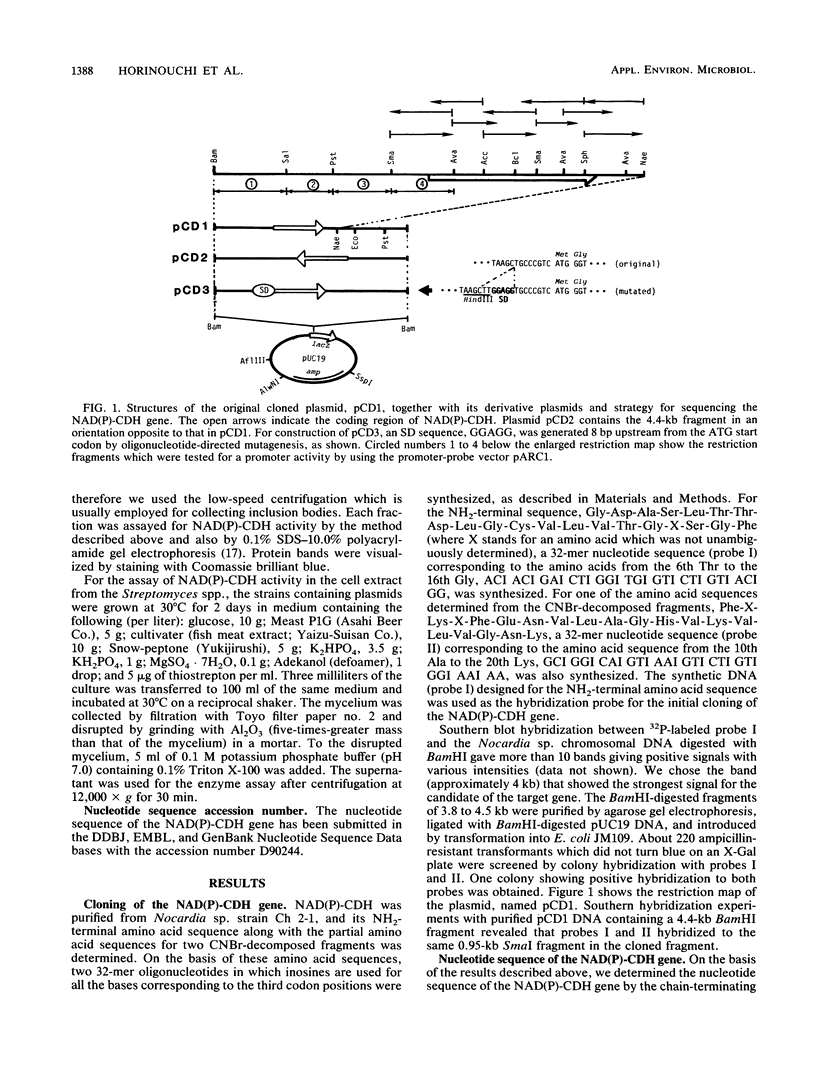
NAD(P)-dependent cholesterol dehydrogenases [NAD(P)-CDH], which allow easier quantification of cholesterol by means of directly measuring the A340 of NAD(P)H, are useful for clinical purposes. The amino acid sequences of the NH2 terminus and the fragments obtained by CNBr decomposition of the NAD(P)-CDH from a Nocardia sp. were determined for preparation of synthetic oligonucleotides as hybridization probes. A 4.4-kbp BamHI fragment hybridizing to these probes was cloned on pUC19 in Escherichia coli. The nucleotide sequence together with the determined amino acid sequences revealed that this enzyme consists of 364 amino acids (Mr, 39,792) and contains an NAD(P)-binding consensus sequence at its NH2-terminal portion. High-resolution S1 nuclease mapping suggested that in NAD(P)-CDH of both Nocardia and Streptomyces spp. transcription initiates at the adenine residue, which is the first position of the translational initiation triplet (AUG) of this protein. The S1 mapping experiments also showed that cholesterol-dependent regulation in the Nocardia sp. occurred at the level of transcription. In Streptomyces lividans containing the cloned fragment, however, this promoter was expressed constitutively. DNA manipulation of the cloned gene in E. coli, including the generation of a ribosome-binding sequence at an appropriate position by oligonucleotide-directed mutagenesis, led to production of this protein in a very large amount but in the enzymatically inactive form of inclusion bodies. On the other hand, a Streptomyces host-vector system was successfully used for producing 40 times as much enzymatically active NAD(P)-CDH as that produced by the original Nocardia sp.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bibb M. J., Cohen S. N. Gene expression in Streptomyces: construction and application of promoter-probe plasmid vectors in Streptomyces lividans. Mol Gen Genet. 1982;187(2):265–277. doi: 10.1007/BF00331128. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Chen C. W., Thomas C. A., Jr Recovery of DNA segments from agarose gels. Anal Biochem. 1980 Jan 15;101(2):339–341. doi: 10.1016/0003-2697(80)90197-9. [DOI] [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood D. A., Kieser T., Wright H. M., Bibb M. J. Plasmids, recombination and chromosome mapping in Streptomyces lividans 66. J Gen Microbiol. 1983 Jul;129(7):2257–2269. doi: 10.1099/00221287-129-7-2257. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Beppu T. Construction and application of a promoter-probe plasmid that allows chromogenic identification in Streptomyces lividans. J Bacteriol. 1985 Apr;162(1):406–412. doi: 10.1128/jb.162.1.406-412.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Furuya K., Nishiyama M., Suzuki H., Beppu T. Nucleotide sequence of the streptothricin acetyltransferase gene from Streptomyces lavendulae and its expression in heterologous hosts. J Bacteriol. 1987 May;169(5):1929–1937. doi: 10.1128/jb.169.5.1929-1937.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Suzuki H., Nishiyama M., Beppu T. Nucleotide sequence and transcriptional analysis of the Streptomyces griseus gene (afsA) responsible for A-factor biosynthesis. J Bacteriol. 1989 Feb;171(2):1206–1210. doi: 10.1128/jb.171.2.1206-1210.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen G. R., Ward J. M., Bibb M. J. Unusual transcriptional and translational features of the aminoglycoside phosphotransferase gene (aph) from Streptomyces fradiae. Genes Dev. 1989 Mar;3(3):415–429. doi: 10.1101/gad.3.3.415. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Ptashne M., Backman K., Humayun M. Z., Jeffrey A., Maurer R., Meyer B., Sauer R. T. Autoregulation and function of a repressor in bacteriophage lambda. Science. 1976 Oct 8;194(4261):156–161. doi: 10.1126/science.959843. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Scrutton N. S., Berry A., Perham R. N. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature. 1990 Jan 4;343(6253):38–43. doi: 10.1038/343038a0. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Ward J. M., Janssen G. R., Kieser T., Bibb M. J., Buttner M. J., Bibb M. J. Construction and characterisation of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol Gen Genet. 1986 Jun;203(3):468–478. doi: 10.1007/BF00422072. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]