Abstract
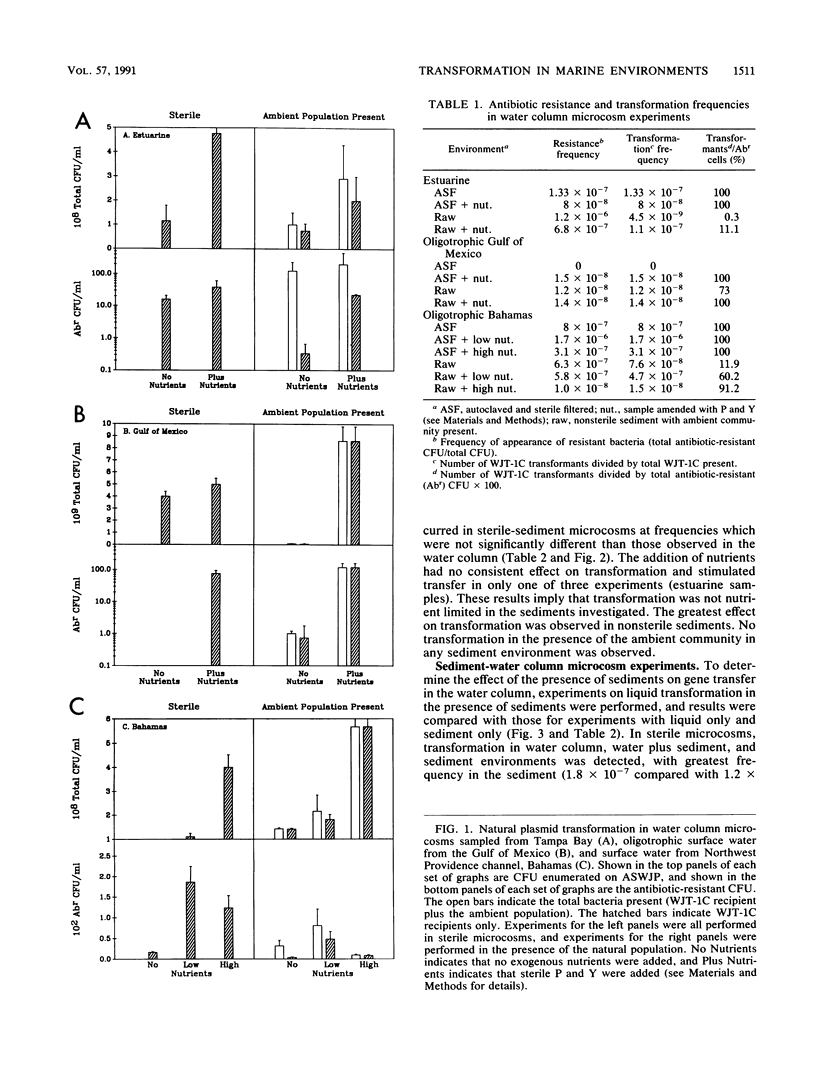
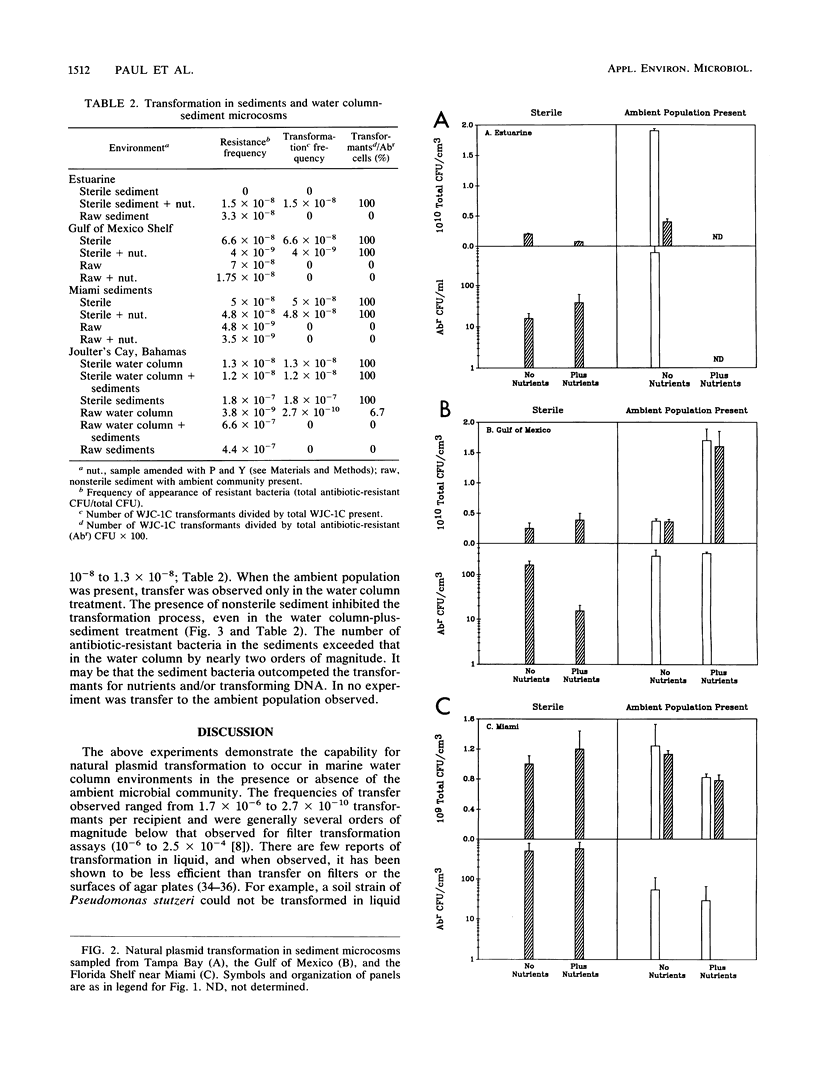
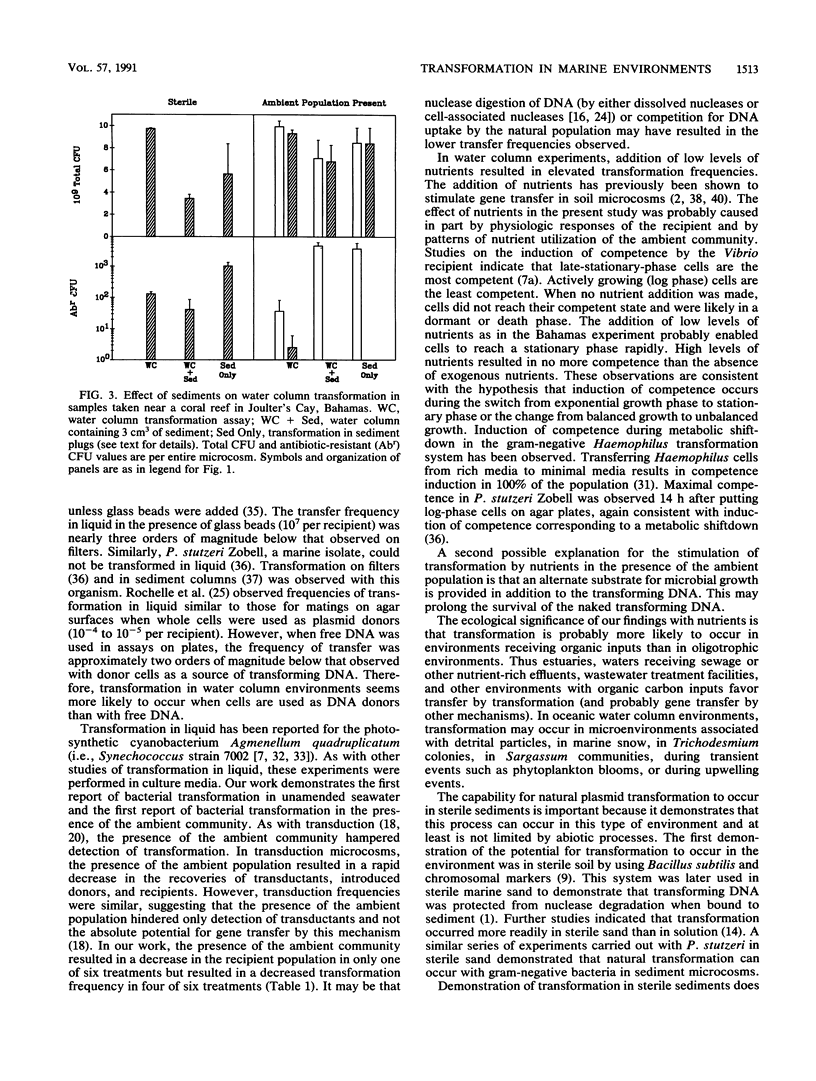
We investigated the possibility for natural transformation in the marine environment by using broad-host-range plasmid multimers and a high-frequency-of-transformation (HFT) Vibrio strain as the recipient. Water and sediment samples were taken from Tampa Bay, the eastern Gulf of Mexico, the Florida Shelf near Miami, and the Bahamas Bank. In water column microcosms, transformation frequencies ranged from 1.7 × 10-6 to 2.7 × 10-10 transformants per recipient, with highest frequencies occurring when low levels of nutrients (peptone and yeast extract) were added. The presence of the ambient community either reduced transformation frequency by an order of magnitude or had no effect. In sterile sediments, nutrient additions had no consistent effect on transformation, with transfer frequencies similar to those observed in the water column. Transformation was not observed in any sediment experiment when the ambient microbial community was present. These findings are the first report of natural plasmid transformation in seawater and in the presence of the ambient microbial community. This process may be a mechanism for the acquisition of small, nonconjugative plasmids, which are commonly found in aquatic bacteria. Our data also suggest that natural transformation may be more likely to occur in the water column than in native marine sediments, contradicting prior conclusions based on studies with sterile sediments.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aardema B. W., Lorenz M. G., Krumbein W. E. Protection of sediment-adsorbed transforming DNA against enzymatic inactivation. Appl Environ Microbiol. 1983 Aug;46(2):417–420. doi: 10.1128/aem.46.2.417-420.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton N. F., Day M. J., Bull A. T. Distribution of bacterial plasmids in clean and polluted sites in a South Wales river. Appl Environ Microbiol. 1982 Nov;44(5):1026–1029. doi: 10.1128/aem.44.5.1026-1029.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughter J. P., Stewart G. J. Genetic exchange in the environment. Antonie Van Leeuwenhoek. 1989;55(1):15–22. doi: 10.1007/BF02309615. [DOI] [PubMed] [Google Scholar]
- Essich E., Stevens S. E., Jr, Porter R. D. Chromosomal transformation in the cyanobacterium Agmenellum quadruplicatum. J Bacteriol. 1990 Apr;172(4):1916–1922. doi: 10.1128/jb.172.4.1916-1922.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischer M. E., Thurmond J. M., Paul J. H. Natural plasmid transformation in a high-frequency-of-transformation marine Vibrio strain. Appl Environ Microbiol. 1990 Nov;56(11):3439–3444. doi: 10.1128/aem.56.11.3439-3444.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J. B., Istock C. A. Genetic exchange in Bacillus subtilis in soil. Mol Gen Genet. 1978 Nov 9;166(3):287–290. doi: 10.1007/BF00267620. [DOI] [PubMed] [Google Scholar]
- Hada H. S., Sizemore R. K. Incidence of Plasmids in Marine Vibrio spp. Isolated from an Oil Field in the Northwestern Gulf of Mexico. Appl Environ Microbiol. 1981 Jan;41(1):199–202. doi: 10.1128/aem.41.1.199-202.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobori H., Sullivan C. W., Shizuya H. Bacterial plasmids in antarctic natural microbial assemblages. Appl Environ Microbiol. 1984 Sep;48(3):515–518. doi: 10.1128/aem.48.3.515-518.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. G., Aardema B. W., Wackernagel W. Highly efficient genetic transformation of Bacillus subtilis attached to sand grains. J Gen Microbiol. 1988 Jan;134(1):107–112. doi: 10.1099/00221287-134-1-107. [DOI] [PubMed] [Google Scholar]
- Lorenz M. G., Wackernagel W. Natural genetic transformation of Pseudomonas stutzeri by sand-adsorbed DNA. Arch Microbiol. 1990;154(4):380–385. doi: 10.1007/BF00276535. [DOI] [PubMed] [Google Scholar]
- Meyer R., Laux R., Boch G., Hinds M., Bayly R., Shapiro J. A. Broad-host-range IncP-4 plasmid R1162: effects of deletions and insertions on plasmid maintenance and host range. J Bacteriol. 1982 Oct;152(1):140–150. doi: 10.1128/jb.152.1.140-150.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Jeffrey W. H., DeFlaun M. F. Dynamics of extracellular DNA in the marine environment. Appl Environ Microbiol. 1987 Jan;53(1):170–179. doi: 10.1128/aem.53.1.170-179.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H. Use of hoechst dyes 33258 and 33342 for enumeration of attached and planktonic bacteria. Appl Environ Microbiol. 1982 Apr;43(4):939–944. doi: 10.1128/aem.43.4.939-944.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul John H., Deflaun Mary F., Jeffrey Wade H., David Andrew W. Seasonal and Diel Variability in Dissolved DNA and in Microbial Biomass and Activity in a Subtropical Estuary. Appl Environ Microbiol. 1988 Mar;54(3):718–727. doi: 10.1128/aem.54.3.718-727.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochelle P. A., Day M. J., Fry J. C. Occurrence, transfer and mobilization in epilithic strains of Acinetobacter of mercury-resistance plasmids capable of transformation. J Gen Microbiol. 1988 Nov;134(11):2933–2941. doi: 10.1099/00221287-134-11-2933. [DOI] [PubMed] [Google Scholar]
- Sizemore R. K., Colwell R. R. Plasmids carried by antibiotic-resistant marine bacteria. Antimicrob Agents Chemother. 1977 Sep;12(3):373–382. doi: 10.1128/aac.12.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Danner D. B., Deich R. A. Genetic transformation. Annu Rev Biochem. 1981;50:41–68. doi: 10.1146/annurev.bi.50.070181.000353. [DOI] [PubMed] [Google Scholar]
- Stevens S. E., Jr, Porter R. D. Heterospecific transformation among cyanobacteria. J Bacteriol. 1986 Sep;167(3):1074–1076. doi: 10.1128/jb.167.3.1074-1076.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens S. E., Porter R. D. Transformation in Agmenellum quadruplicatum. Proc Natl Acad Sci U S A. 1980 Oct;77(10):6052–6056. doi: 10.1073/pnas.77.10.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G. J., Carlson C. A., Ingraham J. L. Evidence for an active role of donor cells in natural transformation of Pseudomonas stutzeri. J Bacteriol. 1983 Oct;156(1):30–35. doi: 10.1128/jb.156.1.30-35.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G. J., Carlson C. A. The biology of natural transformation. Annu Rev Microbiol. 1986;40:211–235. doi: 10.1146/annurev.mi.40.100186.001235. [DOI] [PubMed] [Google Scholar]
- Stewart G. J., Sinigalliano C. D. Detection of horizontal gene transfer by natural transformation in native and introduced species of bacteria in marine and synthetic sediments. Appl Environ Microbiol. 1990 Jun;56(6):1818–1824. doi: 10.1128/aem.56.6.1818-1824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]


