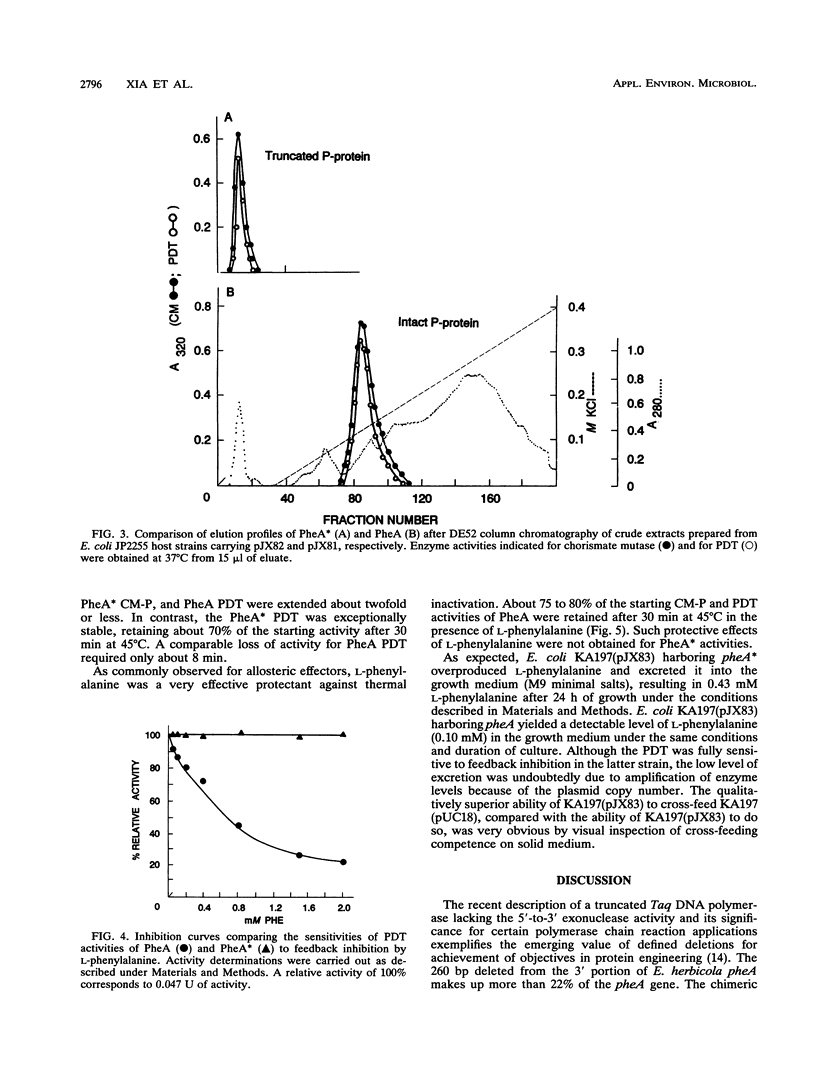
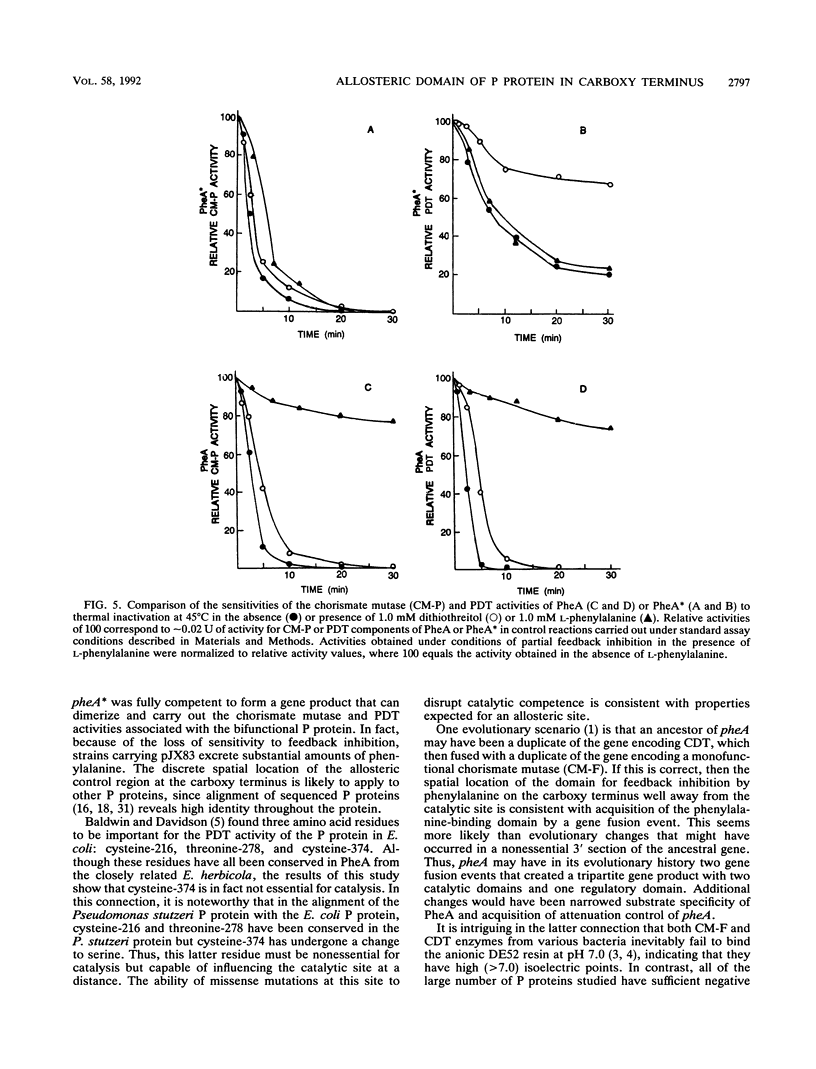
Abstract
A bifunctional protein denoted as the P protein and encoded by pheA is widely present in purple gram-negative bacteria. This P protein carries catalytic domains that specify chorismate mutase (CM-P) and prephenate dehydratase. The instability of a recombinant plasmid carrying a pheA insert cloned from Erwinia herbicola resulted in a loss of 260 bp plus the TAA stop codon from the 3' terminus of pheA. The plasmid carrying the truncated pheA gene (denoted pheA*) was able to complement an Escherichia coli pheA auxotroph. pheA* was shown to be a chimera composed of the residual 5' part of pheA (901 bp) and a 5-bp fragment from the pUC18 vector. The new fusion protein (PheA*) retained both chorismate mutase and prephenate dehydratase activities. PheA* had a calculated subunit molecular weight of 33,574, in comparison to the 43,182-molecular-weight subunit size of PheA. The deletion did not affect the ability of PheA* to assume the native dimeric configuration of PheA. Both the CM-P and prephenate dehydratase components of PheA* were insensitive to L-phenylalanine inhibition, in contrast to the corresponding components of PheA. L-Phenylalanine protected both catalytic activities of PheA from thermal inactivation, and this protective effect of L-phenylalanine upon the PheA* activities was lost. PheA* was more stable than PheA to thermal inactivation; this was more pronounced for prephenate dehydratase than for CM-P. In the presence of dithiothreitol, the differential resistance of PheA* prephenate dehydratase to thermal inactivation was particularly striking.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmad S., Jensen R. A. The prephenate dehydrogenase component of the bifunctional T-protein in enteric bacteria can utilize L-arogenate. FEBS Lett. 1987 May 25;216(1):133–139. doi: 10.1016/0014-5793(87)80771-8. [DOI] [PubMed] [Google Scholar]
- Ahmad S., Weisburg W. G., Jensen R. A. Evolution of aromatic amino acid biosynthesis and application to the fine-tuned phylogenetic positioning of enteric bacteria. J Bacteriol. 1990 Feb;172(2):1051–1061. doi: 10.1128/jb.172.2.1051-1061.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin G. S., Davidson B. E. A kinetic and structural comparison of chorismate mutase/prephenate dehydratase from mutant strains of Escherichia coli K 12 defective in the PheA gene. Arch Biochem Biophys. 1981 Oct 1;211(1):66–75. doi: 10.1016/0003-9861(81)90430-6. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Byng G. S., Whitaker R. J., Gherna R. L., Jensen R. A. Variable enzymological patterning in tyrosine biosynthesis as a means of determining natural relatedness among the Pseudomonadaceae. J Bacteriol. 1980 Oct;144(1):247–257. doi: 10.1128/jb.144.1.247-257.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COTTON R. G., GIBSON F. THE BIOSYNTHESIS OF PHENYLALANINE AND TYROSINE; ENZYMES CONVERTING CHORISMIC ACID INTO PREPHENIC ACID AND THEIR RELATIONSHIPS TO PREPHENATE DEHYDRATASE AND PREPHENATE DEHYDROGENASE. Biochim Biophys Acta. 1965 Apr 12;100:76–88. doi: 10.1016/0304-4165(65)90429-0. [DOI] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Thomas M., Cameron J., St John T. P., Scherer S., Padgett R. A. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65(1):404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich H. A., Gelfand D., Sninsky J. J. Recent advances in the polymerase chain reaction. Science. 1991 Jun 21;252(5013):1643–1651. doi: 10.1126/science.2047872. [DOI] [PubMed] [Google Scholar]
- Fischer R. S., Zhao G., Jensen R. A. Cloning, sequencing, and expression of the P-protein gene (pheA) of Pseudomonas stutzeri in Escherichia coli: implications for evolutionary relationships in phenylalanine biosynthesis. J Gen Microbiol. 1991 Jun;137(6):1293–1301. doi: 10.1099/00221287-137-6-1293. [DOI] [PubMed] [Google Scholar]
- Fischer R., Jensen R. Arogenate dehydratase. Methods Enzymol. 1987;142:495–502. doi: 10.1016/s0076-6879(87)42061-2. [DOI] [PubMed] [Google Scholar]
- Hudson G. S., Davidson B. E. Nucleotide sequence and transcription of the phenylalanine and tyrosine operons of Escherichia coli K12. J Mol Biol. 1984 Dec 25;180(4):1023–1051. doi: 10.1016/0022-2836(84)90269-9. [DOI] [PubMed] [Google Scholar]
- Humphreys G. O., Willshaw G. A., Anderson E. S. A simple method for the preparation of large quantities of pure plasmid DNA. Biochim Biophys Acta. 1975 Apr 2;383(4):457–463. doi: 10.1016/0005-2787(75)90318-4. [DOI] [PubMed] [Google Scholar]
- Maruya A., O'Connor M. J., Backman K. Genetic separability of the chorismate mutase and prephenate dehydrogenase components of the Escherichia coli tyrA gene product. J Bacteriol. 1987 Oct;169(10):4852–4853. doi: 10.1128/jb.169.10.4852-4853.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prober J. M., Trainor G. L., Dam R. J., Hobbs F. W., Robertson C. W., Zagursky R. J., Cocuzza A. J., Jensen M. A., Baumeister K. A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science. 1987 Oct 16;238(4825):336–341. doi: 10.1126/science.2443975. [DOI] [PubMed] [Google Scholar]
- Ray J. M., Yanofsky C., Bauerle R. Mutational analysis of the catalytic and feedback sites of the tryptophan-sensitive 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase of Escherichia coli. J Bacteriol. 1988 Dec;170(12):5500–5506. doi: 10.1128/jb.170.12.5500-5506.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rood J. I., Perrot B., Heyde E., Morrison J. F. Characterization of monofunctional chorismate mutase/prephenate dehydrogenase enzymes obtained via mutagenesis of recombinant plasmids in vitro. Eur J Biochem. 1982 Jun;124(3):513–519. doi: 10.1111/j.1432-1033.1982.tb06623.x. [DOI] [PubMed] [Google Scholar]
- Xia T. H., Ahmad S., Zhao G. S., Jensen R. A. A single cyclohexadienyl dehydratase specifies the prephenate dehydratase and arogenate dehydratase components of one of two independent pathways to L-phenylalanine in Erwinia herbicola. Arch Biochem Biophys. 1991 May 1;286(2):461–465. doi: 10.1016/0003-9861(91)90066-r. [DOI] [PubMed] [Google Scholar]
- Xia T., Jensen R. A. Monofunctional chorismate mutase from Serratia rubidaea: a paradigm system for the three-isozyme gene family of enteric bacteria. Arch Biochem Biophys. 1992 Apr;294(1):147–153. doi: 10.1016/0003-9861(92)90149-q. [DOI] [PubMed] [Google Scholar]
- Xia T., Zhao G., Fischer R. S., Jensen R. A. A monofunctional prephenate dehydrogenase created by cleavage of the 5' 109 bp of the tyrA gene from Erwinia herbicola. J Gen Microbiol. 1992 Jul;138(7):1309–1316. doi: 10.1099/00221287-138-7-1309. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]


