Abstract
Hypoxia-inducible factor 1 (HIF-1) is a transcription factor that mediates cellular and systemic homeostatic responses to reduced O2 availability in mammals, including angiogenesis, erythropoiesis, and glycolysis. HIF-1 activity is controlled by the O2-regulated expression of the HIF-1α subunit. Under nonhypoxic conditions, HIF-1α protein is subject to ubiquitination and proteasomal degradation. Here we report that missense mutations and/or deletions involving several different regions of HIF-1α result in constitutive expression and transcriptional activity in nonhypoxic cells. We demonstrate that hypoxia results in decreased ubiquitination of HIF-1α and that missense mutations increase HIF-1α expression under nonhypoxic conditions by blocking ubiquitination.
The ability to sense O2 concentration is a fundamental biological property, and reduced O2 availability (hypoxia) has profound effects on developmental and physiological processes in mammals. Hypoxia-inducible factor 1 (HIF-1) is expressed in response to hypoxia in most cell types and activates the transcription of genes encoding proteins that function to increase O2 delivery [erythropoietin, vascular endothelial growth factor (VEGF)], allow metabolic adaptation (glucose transporters, glycolytic enzymes), or promote cell survival (insulin-like growth factor 2) under hypoxic or ischemic conditions (1). HIF-1 expression is required for embryogenesis and physiological responses to hypoxia (1–4). Thus, the molecular mechanisms by which HIF-1 activity is induced represent an essential component of cellular and systemic O2 homeostasis. HIF-1 is a heterodimeric basic helix-loop-helix–PER-ARNT-SIM (bHLH-PAS) domain protein that consists of HIF-1α and HIF-1β subunits (5). HIF-1β can dimerize with several different bHLH-PAS proteins, whereas HIF-1α is the specific and O2-regulated subunit of HIF-1 that determines its biological activity (1, 5–7). The regulation of HIF-1α by cellular O2 concentration is complex and involves increased mRNA expression, protein stabilization, nuclear localization, and transactivation function in response to hypoxia (1). Perhaps most striking is the induction of HIF-1α protein expression in response to hypoxia and its rapid degradation on reoxygenation, both in cultured cells and in vivo (5, 6, 8, 9). Under nonhypoxic conditions HIF-1α protein is subject to ubiquitination and proteasomal degradation (10–12). However, decreased ubiquitination in response to hypoxia has not been demonstrated and thus the regulated step in the hypoxic induction of HIF-1α expression has not been determined. In renal carcinoma cell lines, the von Hippel–Lindau (VHL) tumor-suppressor protein, a putative ubiquitin–protein ligase (13, 14), appears to play a role in this process, as loss of VHL function results in constitutive expression of HIF-1α (15), but VHL-mediated ubiquitination of HIF-1α has not been demonstrated.
Structure–function analyses of HIF-1α have revealed that the N-terminal half of the molecule containing the bHLH-PAS domain is required for dimerization and DNA binding (16). The C-terminal half of HIF-1α contains domains required for hypoxia-induced nuclear localization, protein stabilization, and transactivation (10, 16–18). A recent study suggested that amino acids 401–603 of HIF-1α constituted an oxygen-dependent degradation domain, as presence or absence of this region had significant effects on protein stability under nonhypoxic conditions (10). However, effects on ubiquitination were not established and thus it was unclear whether this region represented a target for ubiquitination, proteasomal degradation, or an upstream signaling event. Here we demonstrate that multiple domains of HIF-1α regulate its expression, that specific point mutations can increase HIF-1α expression, and that they do so by altering the ubiquitination of HIF-1α under nonhypoxic conditions.
Materials and Methods
HIF-1α Mutagenesis.
Pfu DNA polymerase (Stratagene) was used for PCR. C-terminal deletions were made by using pCEP4/HIF-1α (16) as template, a common forward primer (FP), and a unique reverse primer (RP) corresponding to the specific deletion (supplemental Tables 1 A and 2; see www.pnas.org). The RP also incorporated a stop codon and a BamHI restriction site. Internal deletions were made by using a FP corresponding to the specific deletion (supplemental Tables 1 B and 2) with an AflII restriction site and a common RP containing a BamHI restriction site. PCR products were digested with AflII and BamHI and inserted into AflII/BamHI-digested pCEP4/HIF1α. Point mutations between residues 512 and 521 were generated by using pCEP4/HIF1α (1–391/512–826) as template, a common RP (containing a BamHI restriction site), and specific mutant FP (supplemental Tables 1 C and 2). S551G and T552A mutations were made by a two-step PCR method. The template for PCR was pCEP4/HIF1α(1–391/512–826) or one of the internal deletion mutants (supplemental Tables 1 B and 2). A common FP + mutant RP was used in one PCR, and a mutant FP + common RP (containing a BamHI site) was used in a second PCR (supplemental Tables 1 C and 2). After gel purification, the two PCR products were mixed and used as template for PCR with the common FP + common RP. The final product was digested with AflII and BamHI and inserted into AflII/BamHI-digested pCEP4/HIF1α.
GAL4/HIF1α Expression Vectors.
A PCR product encoding residues 429–608 was amplified from pCEP4/HIF1α (supplemental Tables 1 D and 2), digested with BamHI, and inserted into pGAL0 (19). The S551G/T552A, S551G, and T552G constructs were generated by two-step PCR by using pGAL/HIF1α (429–608) as template (supplemental Tables 1 D and 2). The final PCR products were digested with BamHI and inserted into pGAL0.
His6-HIF-1α Expression Vector.
PCR was performed by using template pCEP4/HIF1α (supplemental Tables 1 E and 2). The product was NcoI/BglII-digested, inserted into pBluescriptSK/HIF1α (5), excised by KpnI/NotI digestion, and inserted into pCEP4 (Invitrogen).
Transient Transfection Assays.
293 cells were maintained in DMEM with 10% FBS and transfected by calcium phosphate coprecipitation. Hep3B cells were maintained in MEM with Earle's salts and 10% FBS and transfected by electroporation, and COS cells were maintained in DMEM with 10% FBS and transfected by using DEAE-dextran (5, 16).
Immunoblot Assays.
Nuclear extracts were prepared from transfected cells as described (16). For whole cell lysates, cells were incubated for 30 min in cold immunoprecipitation buffer (0.1% SDS/1% Nonidet P-40/5 mM EDTA/0.5% deoxycholate/150 mM NaCl/50 mM Tris⋅HCl, pH 8) containing 2 mM DTT, 0.4 mM PMSF, 2 μg/ml leupeptin, 2 μg/ml aprotinin, 2 μg/ml pepstatin, and 1 mM NaVO3. Samples were centrifuged at 10,000 × g to pellet cell debris. Aliquots were fractionated by SDS/PAGE and subjected to immunoblot assay (6) by using an affinity-purified rabbit-polyclonal Ab raised against HIF-1α residues 807–826 (C terminus) at 1:500 dilution, or protein G-purified mouse monoclonal Ab H1α67 (Novus Biologicals Inc., Littleton, CO) raised against a GST fusion protein containing HIF-1α residues 432–528 (20) at 1:1,000 dilution. The secondary Ab was horseradish peroxidase-conjugated anti-rabbit or -mouse Ig (Amersham), respectively. Signal was developed by using ECL reagents (Amersham). GAL4/HIF-1α fusion proteins were detected by using H1α67 or a polyclonal anti-GAL4 Ab (Santa Cruz Biotechnology).
His6-HIF-1α Purification.
Transfected cells were lysed in denaturing buffer (8 M urea/20 mM Tris⋅HCl, pH 8/100 mM NaCl). Aliquots of total protein [≈250 μg normalized to β-galactosidase (β-gal) activity] were incubated with 50 μl of 50% Talon metal-affinity resin (CLONTECH) for 30 min and centrifuged at 800 × g for 1 min. The resin was washed five times with denaturing buffer. Samples were boiled in SDS-gel loading buffer and fractionated by 7% SDS/PAGE. Immunoblot assays were performed with the anti-HIF-1α polyclonal Ab.
Detection of His6-Ubiquitinated HIF-1α.
Cells were transfected with pSVβgal (Promega), plasmid encoding wild-type or mutant HIF-1α, and plasmid encoding no protein or His6-ubiquitin (21). After 36 h at 20% O2, cells were exposed to 20% or 1% O2 for 4 h and lysed in immunoprecipitation buffer containing 75 mM imidazole. Aliquots of total protein (750–1,000 μg normalized to β-gal activity) were incubated for 30 min with 100 μl of 50% Probond resin (Invitrogen) and washed with immunoprecipitation buffer containing 75 mM imidazole. Samples were boiled in SDS-gel loading buffer and fractionated by 7% SDS/PAGE. Immunoblots were analyzed with anti-HIF-1α polyclonal Ab.
Results
Although HIF-1α mRNA expression is induced by hypoxia or ischemia in vivo (9), in cultured cells HIF-1α mRNA is constitutively expressed, and regulation occurs exclusively at the level of protein expression via changes in half life (8). Deletion of HIF-1α residues 391–826 generated a polypeptide that was constitutively expressed at high levels in both hypoxic and nonhypoxic cells, indicating that HIF-1α protein stability was under negative regulation in nonhypoxic cells (16). To localize sequences involved in this negative regulation, a series of C-terminal and internal deletion mutants of HIF-1α were generated, starting from residues 826 and 391, respectively.
C-Terminal Domain Involved in the Regulation of HIF-1α Expression.
Plasmids encoding full-length or C-terminal-deleted HIF-1α were transfected into 293 cells and exposed to 20% or 1% O2 for 4 h, followed by nuclear extract preparation and immunoblot assay. Transfected full-length HIF-1α (1–826) and HIF-1α (1–754) were expressed at higher levels in hypoxic relative to nonhypoxic cells, similar to endogenous HIF-1α, but HIF-1α (1–681) and HIF-1α (1–608) were constitutively expressed (Fig. 1A). Further analysis revealed that expression of HIF-1α (1–729) was regulated, but HIF-1α (1–703) was constitutively expressed (Fig. 1B). HIF-1α (1–726) was analyzed next because it lacks the only phosphorylatable residue between 703 and 729 (S727), but this protein demonstrated regulated expression. These results indicate that amino acids between residues 703 and 726 are necessary for O2-regulated expression of HIF-1α.
Figure 1.
Effect of C-terminal deletions on HIF-1α protein expression. 293 cells were transfected with expression vectors encoding HIF-1α constructs and 36 h later were exposed to 20% O2 or 1% O2 (hypoxia) for 4 h before nuclear extract preparation. Fifteen-microgram aliquots were fractionated by SDS/PAGE and subjected to immunoblot assay by using an anti-HIF-1α Ab. The constructs analyzed encoded the following residues of HIF-1α: (A) 1–826, 1–754, 1–681, 1–608; and (B) 1–729, 1–726, 1–703. Migration of full-length (FL) and deleted (Δ) forms of HIF-1α are indicated (Right).
Internal Domain Involved in the Regulation of HIF-1α Expression.
Internal deletion mutants were constructed involving deletions extending toward the C terminus from residue 391. HIF-1α constructs consisting of residues 1–391 fused to 404–826, 415–826, 429–826, 469–826, 494–826, 508–826, or 512–826 were expressed in an O2-regulated manner, whereas constructs consisting of residues 1–391 fused to 521–826, 549–826, or 576–826 were constitutively expressed (Fig. 2). Missense mutations were introduced at residues between 512 and 521 in the context of HIF-1α(1–391/512–826). These mutations included elimination of: a potentially redox-sensitive cysteine (C520 M) residue (Fig. 3A); potentially phosphorylatable serine (S515A, S517A) and tyrosine (Y519F) residues (Fig. 3B); proline (P513G, P516G) and glutamate (E512G, E518G) residues (Fig. 3C). None of these missense mutations, nor deletion of residues 512–516 (Fig. 3C, lanes 3 and 4), eliminated O2-regulated expression, whereas deletion of residues 512–520 (Fig. 3C, lanes 1 and 2) resulted in constitutive expression. Thus, amino acids between residues 512 and 521 are necessary for O2-regulated expression of HIF-1α but no single residue is absolutely required for this regulation (Fig. 3D).
Figure 2.
Summary of the effect of internal deletions on the O2-dependent regulation of HIF-1α expression. Expression of deletion constructs that were wild type (WT) or contained S551G/T552A missense mutations is summarized (Right). Regulation refers to the presence of increased protein expression under hypoxic as compared with nonhypoxic conditions. Experimental results are shown in Figs. 3 and 4.
Figure 3.
Effect of missense mutations on expression of HIF-1α(1–391/512–826). (A) Immunoblot assay of nuclear extracts from 293 cells transfected with expression vectors encoding HIF-1α(1–391/512–826) (lanes 1–2), HIF-1α(1–391/512–826/C520 M) (lanes 3–4), or HIF-1α(1–391/521–826) (lanes 5–6). (B) Analysis of nuclear extracts from cells transfected with empty vector (EV; lanes 1–2) or HIF-1α constructs consisting of residues 1–826 (lanes 3–4) or 1–391/512–826 that was either wild type (lanes 5–6) or contained S515A/S517A (lanes 7–8) or Y519F (lanes 9–10) missense mutations. (C) Analysis of nuclear extracts from cells transfected with HIF-1α constructs consisting of residues 1–391/521–826 (lanes 1–2), 1–391/517–826 (lanes 3–4), or 1–391/512–826 that was either wild type (lanes 5–6) or contained P513G/P516G (lanes 7–8) or E512G/E518G (lanes 9–10) missense mutations. (D) Summary of the data. The amino acid (AA) sequence of HIF-1α residues 512–521 and the effect of missense mutations and deletions (−) on O2-regulated expression are shown.
We considered the possibility that the domain encompassing residues 512–521 might serve as a binding site for a kinase/phosphatase that reacts with a more distant residue. Residues S551, T552, and Q553 are conserved in human and mouse HIF-1α and HIF-2α (22). We mutated the two potentially phosphorylatable residues (S551G and T552A) in each of the deletion constructs that had previously demonstrated regulated expression (Fig. 2). Remarkably, the S551G/T552A mutation resulted in constitutive expression of all of these constructs (Figs. 2 and 4A). If S551 or T552 were sites of phosphorylation, then their replacement by negatively charged aspartate residues might mimic a constitutively phosphorylated state. Unlike His6-HIF-1α(1–391/512–826/S551G/T552A), the expression of His6-HIF-1α(1–391/512–826/S551D/T552D) was not increased in nonhypoxic cells (Fig. 4B), which was consistent with the hypothesis that S551 and/or T552 were phosphorylated under these conditions. However, expression of His6-HIF-1α(1–391/512–826/S551D/T552D) was still induced under hypoxic conditions (lane 8), indicating that O2-dependent (de)phosphorylation of these residues was not required for regulated expression. Thus, deletions or missense mutations at three distinct sites (residues 703–726, 512–521, and 551–552) interfere with the regulated expression of HIF-1α under nonhypoxic conditions, and there is no evidence indicating that any of these sites are subject to O2-regulated (de)phosphorylation.
Figure 4.
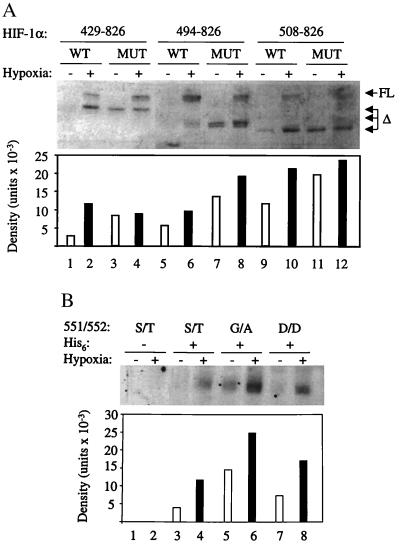
Effect of internal deletions and missense mutations on HIF-1α expression. (A) Expression vectors encoding HIF-1α deletion constructs, consisting of residues 1–391 fused to 429–826, 494–826, or 508–826, either wild type (WT) or containing the S551G/T552A missense mutations (MUT), were transfected into 293 cells and exposed to 20% or 1% O2 for 4 h. Nuclear extracts were prepared, and 15-μg aliquots were subjected to immunoblot assay, which detected both endogenous full-length (FL) and recombinant deleted (Δ) HIF-1α. The expression of the deleted HIF-1α proteins was quantified by densitometry. (B) Cells were cotransfected with pSVβgal and vectors encoding HIF-1α(1–391/512–826) (lanes 1–2), His6-HIF-1α(1–391/512–826) (lanes 3–4), His6-HIF-1α(1–391/512–826/S551G/T552A) (lanes 5–6), or His6-HIF-1α(1–391/512–826/S551D/T552D) (lanes 7–8). Transfected cells were exposed to 20% or 1% O2 for 4 h, and His-tagged proteins were purified from lysates by metal-affinity resin binding. Aliquots of purified protein (normalized to β-gal expression) were subjected to immunoblot assay by using an anti-HIF-1α polyclonal Ab, and protein expression was quantified by densitometry.
To determine whether these mutations result in increased transcriptional activity of HIF-1, Hep3B cells, which have no detectable HIF-1α expression under nonhypoxic conditions (5, 23), were cotransfected with: control plasmid pSVβgal; luciferase reporter plasmid p2.1, which contains a 68-bp hypoxia-response element from the human ENO1 gene (7); and either empty expression vector or vector encoding HIF-1α, HIF-1α(S551G/T552A), or HIF-1α(1–391/521–826). The transfected cells were exposed to 20% O2 for 24 h and lysates were analyzed for β-gal and luciferase activity. Compared with cells transfected with empty vector, p2.1 expression was increased 8.2-, 22-, and 78-fold in the presence of HIF-1α, HIF-1α (S551G/T552A), and HIF-1α(1–391/521–826), respectively (Fig. 5). Thus, increased expression of the mutant HIF-1α proteins (Figs. 3 and 4) is associated with increased transactivation of target genes (Fig. 5) under nonhypoxic conditions. The increased transcriptional activity mediated by HIF-1α(1–391/521–826) as compared with HIF-1α(S551G/T552A) is consistent with the maximal and submaximal expression of these proteins under nonhypoxic conditions. In addition, transactivation function, which is also under negative regulation in nonhypoxic cells (17), may be increased specifically by the deletion.
Figure 5.
Effect of deletion or missense mutations on HIF-1α-mediated reporter gene transcription. Hep3B cells were transfected with 5 μg of pSVβgal, 10 μg of p2.1 (containing a hypoxia response element upstream of an SV40 promoter-luciferase reporter gene), and 125 ng of expression vectors encoding either no protein (EV), full-length HIF-1α (FL), full-length HIF-1α with S551G/T552A missense mutations (MIS), or HIF-1α(1–391/521–826) (DEL). Transfected cells were incubated under 20% O2 for 24 h. The luciferase:β-gal activity ratio was determined and normalized to the value obtained from cells transfected with empty vector (EV) to obtain the relative luciferase activity. Results shown represent the mean and standard error for three plates of transfected cells.
To determine whether the internal domain was sufficient for O2-regulated expression of a heterologous protein, a fusion protein consisting of the DNA-binding domain of the yeast GAL4 transcription factor fused to HIF-1α residues 429–608 was constructed. The expression of this protein in COS cells was hypoxia inducible (Fig. 6, lanes 3 and 4), whereas introduction of the S551G/T552A mutations resulted in constitutive expression (lanes 5 and 6). The single mutation S551G or T552G did not completely eliminate regulated expression (data not shown).
Figure 6.
Expression of GAL4/HIF-1α constructs. COS cells were transfected with pSVβgal and expression vectors encoding the GAL4 DNA-binding domain (DBD) and either no additional residues (lanes 1–2) or HIF-1α amino acid sequences 429–608 that were either wild type (lanes 3–4) or contained the S551G/T552A mutations (lanes 5–6). Transfected cells were exposed to 20% or 1% O2 for 4 h. Lysates were prepared and 60-μg aliquots were fractionated by SDS/PAGE and subjected to immunoblot assays by using an Ab against endogenous HIF-1α (Top) or the GAL4 DBD (Middle). The isolated GAL4 DBD (Middle, lanes 1–2) migrated faster than the GAL4/HIF-1α fusion proteins and thus is not within the portion of the immunoblot displayed. Expression of GAL4/HIF-1α fusion proteins was quantified by densitometry (Bottom).
O2-Regulated Ubiquitination of HIF-1α.
Changes in the ubiquitination of HIF-1α in response to hypoxia have not been demonstrated. To investigate this possibility, 293 cells were transfected with a HIF-1α expression vector and either empty vector or vector encoding His6-tagged ubiquitin (His6-ubi) and exposed to 20% or 1% O2. Lysates were prepared and aliquots were subject to immunoblot assay before and after affinity purification to compare the levels of total and ubiquitinated HIF-1α, respectively (Fig. 7A). As in other systems (21), ubiquitinated forms are not detected by analysis of total HIF-1α, presumably because of rapid degradation after ubiquitination. However, the ability to specifically capture His6-ubi-tagged molecules allows the detection of a ladder of isoforms extending up the gel from monoubiquitinated to polyubiquitinated forms of HIF-1α. Because of the marked increased in HIF-1α protein levels in hypoxic cells, it was necessary to compare the relative rather than absolute levels of ubiquitination. Densitometric quantitation revealed that the proportion of HIF-1α that was ubiquitinated decreased by >7-fold under hypoxic conditions (Fig. 7B). Thus, exposure of cells to 1% O2, which represents a submaximal hypoxic stimulus (6), reduced the ubiquitination of HIF-1α. To determine whether mutations that result in constitutive expression of HIF-1α also affect its ubiquitination, 293 cells were transfected with either empty vector or vector encoding His6-ubi and vector encoding HIF-1α(1–391/512–826) or HIF-1α(1–391/512–826/S551G/T552A). Compared with wild type, the proportion of mutant protein that was ubiquitinated in nonhypoxic cells was 18-fold lower (Fig. 7 C and D). These results demonstrate that the S551G/T552A mutations increase the expression of HIF-1α under nonhypoxic conditions by decreasing its ubiquitination.
Figure 7.
Analysis of HIF-1α ubiquitination. (A) 293 cells were cotransfected with expression vectors encoding HIF-1α and either no protein (lanes 1–2) or His6-ubiquitin (lanes 3–4). Cells were exposed to 20% or 1% O2 for 4 h, and lysates were prepared. Two hundred-microgram aliquots of total protein were incubated with nickel resin and washed with buffer containing 75 mM imidazole. Aliquots of unpurified (Top) and nickel resin-purified (Bottom) lysates were subjected to SDS/PAGE and immunoblot assay by using an anti-HIF-1α polyclonal Ab. (B) Levels of total and ubiquitinated HIF-1α in lanes 3–4 were quantified by densitometry. The ratio of ubiquitinated/total HIF-1α was determined and normalized to the result obtained for the lysates analyzed in lane 4 to yield the relative HIF-1α ubiquitination. (C) Cells were cotransfected with: pSVβgal; expression vector encoding HIF-1α(1–391/512–826) that was either wild type (WT; lanes 1–3) or contained the S551G/T552A missense mutations (M; lane 4); and vector encoding either no protein (lanes 1–2) or His6-ubiquitin (lanes 3–4). Cells were exposed to 20% or 1% O2 for 4 h, and lysates were prepared. Aliquots of total protein (normalized to β-gal) were incubated with nickel resin and washed with RIPA buffer (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) containing 75 mM imidazole. Aliquots of unpurified (Top) or resin-purified (Bottom) lysates were subjected to immunoblot assay by using an anti-HIF-1α polyclonal Ab. (D) Levels of total and ubiquitinated wild-type and mutant HIF-1α (1–391/512–826) in lanes 3–4 were quantified by densitometry. The ratio of ubiquitinated/total HIF-1α was determined and normalized to the result obtained for the lysate analyzed in lane 4 to yield the relative HIF-1α ubiquitination.
Discussion
Regulation of HIF-1α protein levels represents a molecular basis for hypoxia-inducible expression of multiple genes in mammalian cells (1). To rapidly induce HIF-1 activity in response to hypoxia, HIF-1α is subject to negative regulation in nonhypoxic cells. Deletion of HIF-1α sequences involved in this regulation results in constitutive expression. While our study was under way, analysis of GAL4/HIF-1α fusion proteins revealed that, in agreement with our data, HIF-1α residues 401–603 were necessary and sufficient for O2-regulated expression (10). Our analysis of HIF-1α deletion mutants in the context of the native HIF-1 heterodimer, rather than as a fusion protein, demonstrated that a C-terminal domain encompassing residues 703–726 is also required for negative regulation of HIF-1α expression in nonhypoxic cells. Further studies are required to determine whether this deletion affects ubiquitination or a downstream step such as proteasomal degradation. Nuclear localization of a green fluorescent protein/HIF-1α fusion protein was shown to be induced by exposure of cells to the iron chelator dipyridyl, whereas deletion (residues 653–826) or mutation (K719T) of C-terminal residues blocked this response (18). Because we analyzed HIF-1α expression in nuclear extracts, it is possible that loss of hypoxic induction of C-terminal deletion constructs may be caused by decreased nuclear localization under hypoxic conditions as well as increased protein expression under nonhypoxic conditions.
Although HIF-1α residues 401–603 were previously implicated in the regulation of HIF-1α stability, neither specific residues nor a specific molecular mechanism was determined (10). Our data indicate that the protein stabilization domain minimally encompasses residues 517–552. Missense mutation of residues 561–568 was recently shown to eliminate O2-regulated expression of a GAL4/HIF-1α (529–826) fusion protein (24), indicating that the minimal domain includes residues 517–568. Our results suggest that residues S551 and T552 play a particularly important role in the regulation of HIF-1α expression, and their mutation results in markedly decreased ubiquitination and increased protein expression under nonhypoxic conditions. Residues S551 and T552 are conserved in human and mouse HIF-1α as well as human and mouse HIF-2α (22), the expression of which is also O2-regulated (25, 26). Our studies demonstrate that HIF-1α is subject to increased ubiquitination under nonhypoxic as compared with hypoxic conditions and that the S551G/T552A mutations interfere with this process.
The mechanism by which HIF-1α is targeted for ubiquitination is unknown. Multiple examples of phosphorylation-dependent ubiquitination have been described (21, 27). If HIF-1α phosphorylation under nonhypoxic conditions represented a signal for ubiquitination, then mutation or deletion of the phosphorylatable residue(s) should result in constitutive expression. Exposure of Hep3B cells to 10 mM sodium fluoride, a nonspecific inhibitor of serine-threonine protein phosphatases, blocked the induction of HIF-1α expression in response to hypoxia (23). However, the activity of the more specific phosphatase inhibitors calyculin A and okadaic acid in blocking hypoxia-induced HIF-1α expression paralleled their toxicity, thus precluding a definitive conclusion regarding their effects (data not shown). We have performed multiple metabolic labeling experiments in an attempt to determine the phosphorylation status of wild-type and mutant HIF-1α in nonhypoxic and hypoxic cells. Although we have demonstrated that HIF-1α is extensively phosphorylated, primarily if not exclusively, on serine residues, we have not been able to demonstrate a reproducible effect of mutation or oxygenation on the phosphorylation of specific tryptic peptides (data not shown). The results presented above also provide no evidence of O2-regulated (de)phosphorylation of HIF-1α despite the importance of S551 and T552 in the regulation of its ubiquitination. Alternatively, regulation of HIF-1α ubiquitination may be determined by O2-regulated changes in the redox status of HIF-1α or interacting proteins (12, 15, 24, 28), similar to the iron-dependent oxidation, ubiquitination, and degradation of iron regulatory protein 2 (29, 30). A redox-sensitive cysteine residue (C800) has been shown to affect the ability of HIF-1α to interact with the coactivator cAMP-response element-binding protein and activate transcription (25). HIF-1 DNA-binding activity in vitro is also sensitive to redox status (28, 31). The identification of specific residues required for O2-regulated HIF-1α ubiquitination described above and in other recent studies (10, 24) will facilitate the delineation of molecular mechanisms underlying this process.
HIF-1α is absolutely required for hypoxia-induced expression of the Vegf gene in mouse embryonic stem cells (2–4). Expression of HIF-1α protein has been shown to parallel that of VEGF mRNA in the heart (32) and retina (33) in response to hypoxia/ischemia in those tissues. Clinical trials of therapeutic angiogenesis involving administration of specific VEGF isoforms or other angiogenic growth factors to patients with ischemic cardiovascular disease are underway (34). Expression of HIF-1 would induce not only all isoforms of VEGF but potentially other angiogenic factors, as well as survival factors such as IGF-2 (35). However, most patients with coronary artery disease do not have reduced myocardial blood flow or ischemia at rest (36), and HIF-1α may be subject to rapid degradation. Thus, a constitutively expressed form of HIF-1α may provide increased efficacy in gene therapy approaches to ischemic cardiovascular disease.
Supplementary Material
Acknowledgments
We thank Chi Dang (Johns Hopkins University, Baltimore) and Dirk Bohman (European Molecular Biology Laboratory, Heidelberg) for providing plasmids, David Feldser for technical assistance, and Atul Bedi for manuscript review. We are grateful to Kailesh Gopalbhai and Steve Levine in the laboratories of Sylvain Meloche and Gary Perdew for their assistance in performing the phosphopeptide analyses that are cited in Discussion. This work was supported by grants from the National Institutes of Health (R01-DK39869 and R01-HL55338) and the Genzyme Corporation to G.L.S. C.H.S. was supported by the Johns Hopkins University Training Program in Hematology (T32-HL07525). Under a licensing agreement between Johns Hopkins University and Genzyme Corporation, G.L.S. is entitled to a share of royalties received by the University from sales of licensed technology. G.L.S. is also a consultant to Genzyme Corporation. The terms of these arrangements are being managed by the University in accordance with its conflict of interest policies.
Abbreviations
- FP
forward primer
- β-gal
β-galactosidase
- HIF-1
hypoxia-inducible factor 1
- RP
reverse primer
- VEGF
vascular endothelial growth factor
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080072497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080072497
References
- 1.Semenza G L. Annu Rev Cell Dev Biol. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- 2.Carmeliet P, Dor Y, Herbert J-M, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, et al. Nature (London) 1998;394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 3.Iyer N V, Kotch L E, Agani F, Leung S W, Laughner E, Wenger R H, Gassmann M, Gearhart J D, Lawler A M, Yu A Y, et al. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryan H E, Lo J, Johnson R S. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang G L, Jiang B-H, Rue E A, Semenza G L. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang B-H, Semenza G L, Bauer C, Marti H H. Am J Physiol. 1996;271:C1172–C1180. doi: 10.1152/ajpcell.1996.271.4.C1172. [DOI] [PubMed] [Google Scholar]
- 7.Semenza G L, Jiang B-H, Leung S W, Passantino R, Concordet J-P, Maire P, Giallongo A. J Biol Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 8.Huang L E, Arany Z, Livingston D M, Bunn H F. J Biol Chem. 1996;271:32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- 9.Yu A Y, Frid M G, Shimoda L A, Wiener C M, Stenmark K, Semenza G L. Am J Physiol. 1998;275:L818–L826. doi: 10.1152/ajplung.1998.275.4.L818. [DOI] [PubMed] [Google Scholar]
- 10.Huang L E, Gu J, Schau M, Bunn H F. Proc Natl Acad Sci USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kallio P J, Wilson W J, O'Brien S, Makino Y, Poellinger L. J Biol Chem. 1999;274:6519–6525. doi: 10.1074/jbc.274.10.6519. [DOI] [PubMed] [Google Scholar]
- 12.Salceda S, Caro J. J Biol Chem. 1997;272:22642–22647. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- 13.Lisztwan J, Imbert G, Wirbelauer C, Gstaiger M, Krek W. Genes Dev. 1999;13:1822–1833. doi: 10.1101/gad.13.14.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwai K, Yamanaka K, Kamura T, Minato N, Conaway R C, Conaway J W, Klausner R D, Pause A. Proc Natl Acad Sci USA. 1999;96:12436–12441. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maxwell P H, Wiesener M S, Chang G-W, Clifford S C, Vaux E C, Cockman M E, Wykoff C C, Pugh C W, Maher E R, Ratcliffe P J. Nature (London) 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 16.Jiang B-H, Rue E, Wang G L, Roe R, Semenza G L. J Biol Chem. 1996;271:17771–17778. doi: 10.1074/jbc.271.30.17771. [DOI] [PubMed] [Google Scholar]
- 17.Jiang B-H, Zheng J Z, Leung S W, Roe R, Semenza G L. J Biol Chem. 1997;272:19253–19260. doi: 10.1074/jbc.272.31.19253. [DOI] [PubMed] [Google Scholar]
- 18.Kallio P J, Okamoto K, O'Brien S, Carrero P, Makino Y, Tanaka H, Poellinger L. EMBO J. 1998;17:6573–6586. doi: 10.1093/emboj/17.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dang C V, Barrett J, Villa-Garcia M, Resar L M, Kato G J, Fearon E R. Mol Cell Biol. 1991;11:954–962. doi: 10.1128/mcb.11.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong H, De Marzo A M, Laughner E, Lim M, Hilton D A, Zagzag D, Buechler P, Isaacs W B, Semenza G L, Simons J W. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]
- 21.Treier M, Staszewski L M, Bohman D. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 22.Tian H, McKnight S L, Russell D W. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 23.Wang G L, Jiang B-H, Semenza G L. Biochem Biophys Res Commun. 1995;216:669–675. doi: 10.1006/bbrc.1995.2674. [DOI] [PubMed] [Google Scholar]
- 24.Srinivas V, Zhang L-P, Zhu X-H, Caro J. Biochem Biophys Res Commun. 1999;260:557–561. doi: 10.1006/bbrc.1999.0878. [DOI] [PubMed] [Google Scholar]
- 25.Ema M, Hirota K, Mimura J, Abe H, Yodoi J, Sogawa K, Poellinger L, Fujii-Kuriyama Y. EMBO J. 1999;18:1905–1914. doi: 10.1093/emboj/18.7.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Rourke J F, Tian Y-M, Ratcliffe P J, Pugh C W. J Biol Chem. 1999;274:2060–2071. doi: 10.1074/jbc.274.4.2060. [DOI] [PubMed] [Google Scholar]
- 27.Laney J D, Hochstrasser M. Cell. 1999;97:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 28.Huang L E, Arany Z, Livingston D M, Bunn H F. J Biol Chem. 1996;271:32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- 29.Guo B, Phillips J D, Yu Y, Leibold E A. J Biol Chem. 1995;270:21645–21651. doi: 10.1074/jbc.270.37.21645. [DOI] [PubMed] [Google Scholar]
- 30.Iwai K, Drake S K, Wehr N B, Weissman A M, LaVaute T, Minato N, Klausner R D, Levine R L, Rouault T A. Proc Natl Acad Sci USA. 1998;95:4924–4928. doi: 10.1073/pnas.95.9.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang G L, Jiang B-H, Semenza G L. Biochem Biophys Res Commun. 1995;212:550–556. doi: 10.1006/bbrc.1995.2005. [DOI] [PubMed] [Google Scholar]
- 32.Martin C, Yu A Y, Jiang B-H, Davis L, Kimberly D, Hohimer A R, Semenza G L. Am J Obstet Gynecol. 1998;178:527–534. doi: 10.1016/s0002-9378(98)70433-8. [DOI] [PubMed] [Google Scholar]
- 33.Ozaki H, Yu A Y, Della N, Ozaki K, Luna J D, Yamada H, Hackett S F, Okamoto N, Zack D J, Semenza G L, et al. Invest Ophthalmol Vis Sci. 1999;40:182–189. [PubMed] [Google Scholar]
- 34.Losordo D W, Vale P R, Isner J W. Am Heart J. 1999;138:132–141. doi: 10.1016/s0002-8703(99)70333-9. [DOI] [PubMed] [Google Scholar]
- 35.Feldser D, Agani F, Iyer N V, Pak B, Ferreira G, Semenza G L. Cancer Res. 1999;59:3915–3918. [PubMed] [Google Scholar]
- 36.Uren N G, Melin J A, De Bruyne B, Wijns W, Baudhuin T, Camici P G. N Engl J Med. 1994;330:1782–1788. doi: 10.1056/NEJM199406233302503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.